Abstract
The virion envelope of human cytomegalovirus (HCMV) is complex and consists of an incompletely defined number of glycoproteins. The gM/gN protein complex is the most abundant protein component of the envelope. Studies have indicated that deletion of the viral gene encoding either gM or gN is a lethal mutation. Analysis of the amino acid sequence of gM disclosed a C-terminal acidic cluster of amino acids and a tyrosine-containing trafficking motif, both of which are well-described trafficking/sorting signals in the cellular secretory pathway. To investigate the roles of these signals in the trafficking of the gM/gN complex during virus assembly, we made a series of gM (UL100 open reading frame) mutants in the AD169 strain of HCMV. Mutant viruses that lacked the entire C-terminal cytoplasmic tail of gM were not viable, suggesting that the cytoplasmic tail of gM is essential for virus replication. In addition, the gM mutant protein lacking the cytoplasmic domain exhibited decreased protein stability. Mutant viruses with a deletion of the acidic cluster or alanine substitutions in tyrosine-based motifs were viable but exhibited a replication-impaired phenotype suggestive of a defect in virion assembly. Analysis of these mutant gMs using static immunofluorescence and fluorescence recovery after photobleaching demonstrated delayed kinetics of intracellular localization of the gM/gN protein to the virus assembly compartment compared to the wild-type protein. These data suggest an important role of the glycoprotein gM during virus assembly, particularly in the dynamics of gM trafficking during viral-particle assembly.
Human cytomegalovirus (HCMV) represents the prototype of the betaherpesvirus subfamily of the family Herpesviridae and is the largest and structurally most complex member of the herpesvirus family (53). HCMV is an important human pathogen that contributes to significant morbidity and mortality in immunocompromised individuals (transplant recipients and human immunodeficiency virus-infected individuals) and is a major cause of congenital viral infections that can result in brain damage and hearing loss in newborn infants (6, 10, 43, 59, 67).
The HCMV genome contains ∼230 kbp of linear double-stranded DNA and includes over 200 open reading frames (ORFs) (14, 18, 54). HCMV shares overall structural features with other herpesviruses, including a protein capsid surrounding the genomic DNA, an amorphous structure containing a large number of virus-encoded proteins that has been designated the tegument, and a lipid-containing envelope membrane enriched with viral glycoproteins (53). The HCMV envelope has been shown to contain a set of highly conserved glycoproteins common to all herpesviruses, including gB, gH/gL (gO), and gM/gN, as well as a larger number of incompletely characterized glycoproteins that are thought to be essential for in vivo pathogenesis, including specific cellular tropism (11, 27, 72, 73, 74).
HCMV, like other herpesviruses, replicates its genome in the nucleus. Following encapsidation of the genomic double-stranded DNA in the nucleus, capsids leave the nucleus by an incompletely defined pathway that likely involves envelopment and de-envelopment at the nuclear membrane (51, 52). During these early stages of infection, most of the tegument proteins and glycoproteins are expressed in the cytoplasm and accumulate within a specific cytoplasmic compartment that has been designated the virus assembly compartment (AC) (61). Cytoplasmic capsids traffic to the AC, where they undergo a final step of tegumentation and envelopment, leading to the formation of an infectious, mature particle. The mature virion is thought to be released from the cell through exocytosis or, alternatively, by lysis of the infected cells.
Sequence analysis of the HCMV strain AD169 genome predicted that approximately 50 ORFs could potentially encode glycoproteins (14). Three major families of glycoproteins have been described, and they exist as disulfide linkage complexes within the mature virion: gCI, represented by homodimers of gB; gCII as a protein complex of gM/gN; and finally, gCIII, which contains the gH/gL/gO protein complex (26). Studies suggest that gB, gH/gL/gO, and possibly gM/gN play roles in HCMV cell attachment, entry, and virus assembly (15, 21, 31, 44, 56, 65, 66). Mass spectroscopic studies of the virion have indicated that gM/gN and gB are the most abundant glycoproteins in the virus particle (the gM/gN complex is approximately five times more abundant than gB homodimers) (72). The homologs of these glycoproteins are present in the envelopes of all members of the herpesvirus family, indicating the importance of these glycoproteins in the virus replicative cycle.
The HCMV glycoprotein gM is a product of the UL100 ORF and contains 372 amino acids (aa) with an approximate molecular mass of 42 kDa (45). The predicted structure of gM indicates that it is a highly hydrophobic type III glycoprotein with seven membrane-spanning domains and a single N-linked carbohydrate modification (42). Even though there is limited amino acid sequence conservation between gMs from different herpesviruses, key structural features appear to be conserved among gMs from other members of the herpesvirus family (42, 45, 46). In fact, multimers consisting of HCMV gN and heterologous gMs from herpes simplex virus (HSV) and other herpesviruses can be detected in cells following transient expression of these proteins (data not shown). The intracellular trafficking of gM is dependent on complex formation with the product of the UL73 ORF, glycoprotein N (gN) (45, 46). The gM/gN complex is formed in the endoplasmic reticulum (ER) and is required for efficient transport of these molecules through the secretory pathway. Recent data have indicated that gM/gN complex formation includes both disulfide bond formation (gM44Cys-gN90Cys) between highly conserved Cys residues and yet-undefined noncovalent interactions (46). Consistent with the results from a transient-expression study in which gM and gN coexpression was necessary for transport of gM/gN as a protein complex from the ER, infectious virus cannot be recovered from either UL100 (gM) or UL73 (gN) HCMV deletion mutants (29, 47). This is in contrast to findings in alphaherpesviruses, HSV and pseudorabies virus (PRV), in which gM and gN are not essential for the assembly of the infectious virus (3, 24, 35, 48, 49). However, deletion of the gene encoding gM in Marek's disease virus, an avian alphaherpesvirus, results in loss of infectivity, suggesting that the requirements for this glycoprotein in virus assembly could vary considerably among herpesviruses (69). The requirement for gM and gN coexpression is not unique to HCMV, and complex formation between homologs of gM and gN in Epstein-Barr virus, PRV, equine herpesvirus type 1 (EHV-1), and varicella-zoster virus has been reported to be required for wild-type levels of virus production (36, 41, 58, 60). These results point to a key function of the gM/gN complex in the assembly of infectious herpesvirus particles.
There is only limited information available on the intracellular trafficking and accumulation of gM/gN during HCMV assembly. Interestingly, the C-terminal cytoplasmic domain of gM contains two trafficking motifs, a tyrosine-based YXXØ (X, any amino acid; Ø, an amino acid with a bulky hydrophobic side chain) and a 12-amino-acid-long acidic cluster, similar to those found in the cytoplasmic tails of other viral glycoproteins; however, it is unknown if these signals function in HCMV gM/gN trafficking during virus infection (1, 17, 20, 71). We began our studies of the role of gM/gN in virus assembly by investigation of the intracellular trafficking of the gM/gN complex in transfected cells to determine if the gM cytoplasmic-tail trafficking motifs are functional. In transiently transfected cells, gM/gN complexes colocalized with AP-1, Rab 7, and CD63 (trans-Golgi network [TGN] and endosomal markers), as well as with glycoprotein gB, suggesting that gM/gN and gB share a common intracellular-trafficking pathway and destination. In addition, this result also suggested the possibility that the gM/gN complex accumulated, not only in the TGN, but also in an endosomal/multivesicular-body (MVB) compartment. To more definitively investigate the role of amino acid motifs in the cytoplasmic domain of gM in virus replication, we made a series of mutations in the UL100 ORF within the HCMV genome. Interestingly, recombinant viruses containing a deletion of the entire C-terminal cytoplasmic domain of gM exhibited a null phenotype, suggesting that these trafficking motifs were critical for the assembly of infectious virions. In addition, viral mutants, one with deletion of an acidic cluster and a second with an alanine substitution in the YXXØ motif, were viable, but both viruses exhibited a significantly replication-impaired phenotype. Static immunofluorescence of infected human foreskin fibroblasts (HFs), together with fluorescence recovery after photobleaching (FRAP) analysis, revealed that both mutant viruses exhibited delayed localization of the gM/gN complex to the AC. Our data provided evidence that gM C-terminal trafficking motifs were functional in virus-infected cells and were required for efficient delivery of the gM/gN complex to the AC. Furthermore, our findings also suggested that accumulation of the gM/gN complex in the AC was essential for wild-type levels of virus replication.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
HCMV strain AD169 and recombinant viruses were propagated in primary HFs grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% newborn calf serum and penicillin/streptomycin (4, 12). Infectious virus stocks were prepared from the supernatant of the infected HFs that exhibited 100% cytopathic effect and were stored at −80°C until they were used. To assay the numbers of infectious particles in the samples, HF monolayers grown in 96-well microtiter plates were infected with serial dilutions of the virus samples. After adsorption of the virus at room temperature for 1 h, the cells were washed and fresh medium was added. Virus titers were determined after 24 h of infection by an indirect immunofluorescence assay using a monoclonal antibody (MAb), p63-27, against immediate-early protein 1 (IE-1) of HCMV, as described previously (2). Human embryonic kidney cells expressing the simian virus 40 large T antigen, 293T cells, were cultured in DMEM supplemented with 10% fetal calf serum (FCS), 0.1% nonessential amino acids, and 50 mg/liter G418. Cos7 cells were passaged in DMEM supplemented with 10% FCS and penicillin/streptomycin. For transient expression, gM (UL100 ORF) and gM mutants (gMΔCT [truncation of the 61 aa of the predicted cytoplasmic tail], gMΔAC [truncation of the C-terminal 12-aa acidic cluster], and gMΔYQAL [329YQAL333→AAAL substitution]) were generated as PCR fragments containing BamHI/EcoRI restriction sites, which permitted cloning into the pEF-1myc/his vector (Invitrogen, San Diego, CA). In addition, all gM constructs were cloned into pEGFP-N1 (Clontech, Mountain View, CA) to introduce an enhanced green fluorescent protein (EGFP) tag at the C terminus of the protein. The construction of pcDNAflag-gN (UL73) and pcDNA-gB (UL55) was described previously (45). pEGFP-Rab7 was kindly provided by B. Van Deurs (University of Copenhagen, Denmark), and pcDNA-pp71, used during recovery of virus following electroporation of recombinant bacterial artificial chromosomes (BAC), was a gift from Bodo Plachter (University of Mainz, Germany).
Pulse-chase and immunoprecipitation.
Human embryonic kidney 293T cells were grown on 10-cm dishes and transfected at 80% confluence with 3 μg of plasmid DNA (pEF-1myc/his-gM, -gMΔCT, -gMΔAC, -gMΔYQAL, and -gMΔYQALΔAC) by DNA calcium phosphate precipitation. All gM constructs were cotransfected with full-length gN to ensure authentic complex formation and trafficking. Forty-eight hours after transfection, the cells were starved for 2 h in medium lacking methionine-cysteine and then pulse-labeled for 20 min with 100 μCi/ml of [35S]methionine. The cultures were washed and chased in growth medium for the indicated times. Cells from the labeling reactions were collected 0, 1, and 3 h postlabeling and immediately processed for immunoprecipitation using magnetic beads (Miltenyi Biotec Inc., Auburn, CA) (46). The immune precipitates were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by fluorography.
BAC mutagenesis and reconstitution of recombinant viruses.
Mutagenesis of HCMV BAC HB5 (strain AD169) was performed using linear DNA fragments and a homologous RED locus recombination system, as described earlier (8). In brief, we generated a gM knockout (KO) BAC by replacing ORF100 (nucleotides 145226 to 146344 of the AD169 genome [nomenclature according to GenBank accession number NC_001347]) with PCR fragments containing an Ampr/LacZ cassette flanked by 50-bp-long 3′ and 5′ ORF100 homology sequences. The recombinant gM KO BAC was selected on plates containing ampicillin and exhibited a blue phenotype in the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranosidase) and IPTG (isopropyl-β-d-thiogalactopyranosidase). This region of the gM KO BAC was then tested for sequence integrity by restriction digestion, PCR, and sequencing. The gM KO BAC was then used to generate mutations in the gM UL100 ORF. In brief, the Ampr/LacZ cassette was replaced with gM UL100 generated by PCR and containing the following mutations: a stop codon at position 146161 (gMΔCT), a stop codon at position 146305 (gMΔAC), YQAL→AAAL at positions 146215 to 146227 (gMΔYQAL), and both of the last two mutations (gMΔYQALΔAC). To confirm the integrity of the recombined BAC, digestion of DNA with the appropriate restriction enzymes was carried out and analyzed using agarose gel electrophoresis with comparison to the parental HB5 BAC DNA. To confirm the presence or absence of mutations in the UL100 ORF following recombination, we amplified the UL100 ORF from recombinant BAC by PCR and analyzed the product by DNA sequencing. Infectious virus was recovered from HFs that were electroporated with 5 μg of purified BAC DNA, together with 1 μg of pcDNA-pp71 plasmid. Twenty-four hours after electroporation, the culture medium was replaced by fresh medium, and the cells were grown until a cytopathic effect was observed. Infectious viruses recovered from these cultures were propagated in fibroblasts.
Imaging and FRAP.
Cos-7 cells were grown on 13-mm glass coverslips in 24-well plates and transfected at 60% confluence with 0.5 μg of each plasmid DNA, using LT1 reagent (Mirus, Madison, WI) according to the manufacturer's directions. Similarly, HFs were seeded on 13-mm glass coverslips in 24-well plates and then infected with the viruses. To assay protein expression by static immunofluorescence, the coverslips were washed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde in PBS. The cells were permeabilized with 0.1% Triton X-100 and blocked with 10% goat serum (Invitrogen/Gibco, San Diego, CA). The primary antibodies, including MAb 14-16A (gM/gN specific), MAb 7-17 (gB specific), MAb anti-AP-1 (Sigma, St. Louis, MO), and MAb anti-CD63 (Iowa Developmental Biology Core, University of Iowa), have been described previously (9, 45). Following incubation with the primary antibody and washing with PBS buffer containing 0.1% Tween 20, antibody binding was detected with the appropriate secondary antibody conjugated with either fluorescein isothiocyanate (FITC), Texas Red, or tetramethyl rhodamine isothiocyanate (Southern Biotech, Birmingham, AL). Images were collected using a Leitz Delux 20 fluorescence microscope fitted with a CoolSnap HQ or with an Olympus Fluoview confocal microscope. To measure the fluorescence intensities in the infected cells, all images were captured with the same parameters of time and fluorescence gain, and pixel intensities were measured for identical areas in the centers of the ACs using a software program (Image-Pro AMS5.1; MediaCybernetics, Silver Springs, MD).
For FRAP analysis, HFs were electroporated with 3 μg of pEGFP plasmids expressing gM, gMΔYQAL, or gMΔAC, and the cells were seeded on a 13-mm coverslip glass in a 24-well plate. The electroporated HFs were infected with HCMV AD169 or the mutant viruses 48 h later at a multiplicity of infection (MOI) of 1.0. At day 5 postinfection (p.i.), the infected live HFs were washed with 25 mM HEPES-buffered phenol red-free DMEM, and the coverslip glasses were placed on rubber gaskets (Molecular Probes/Invitrogen, San Diego, CA) containing phenol-free medium supplemented with 50% FCS. Photobleaching was performed on a confocal laser scanning microscope (Leica SP2). Cells expressing EGFP fusion proteins in moderately high amounts were monitored using the 488 laser line of the argon laser at 25% power and bleached at 100% laser power. The images were taken as follows: 5 frames at 1.4-second intervals for prebleaching, 1 frame at a 1.4-second interval for photobleaching, and 30 frames at 10-second intervals for postbleaching. The area of cell bleaching was the same in all experimental cells. The recovered fluorescence intensity in the region of interest was measured and normalized. The intensity in the prebleach image was set to 100%, and the first postbleach image was set as time point zero. The average fluorescence intensity was plotted over time. The recovery curves shown are mean values and standard errors of the recovery curves of at least 10 cells (n ≥ 10) and are representative of two independent experiments.
RESULTS
The gM/gN complex colocalizes with TGN and endosomal markers.
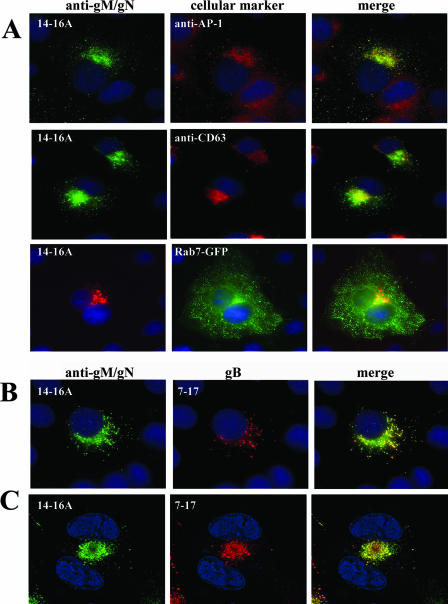
Previous observations suggested that trafficking of the HCMV glycoproteins gM (UL100) and gN (UL73) from the ER is a cooperative process (45). When coexpressed, gM and gN form a complex that is exported from the ER, whereas when expressed independently, both proteins remain in a distinct perinuclear compartment that colocalizes with calreticulin, an ER marker (45). Initially, the intracellular trafficking pathway of the gM/gN complex was studied using static immunofluorescence of transfected cells. In this experiment, we utilized MAb 14-16A, which recognized gN only when the protein was complexed with gM (45). In Cos-7 cells cotransfected with gM and gN, the gM/gN complex could be localized to cytoplasmic vesicles that did not have a typical distribution of the ER markers but colocalized with markers of the most distal compartments of the secretory pathway, including the TGN (AP-1) and the endosomal/MVB compartment (CD63 and Rab 7) (Fig. 1A). In addition, when coexpressed, the gM/gN complex colocalized with glycoprotein gB (UL55) in distinct intracellular vesicles, a finding that suggested that gM/gN and gB share a common intracellular-trafficking pathway (Fig. 1B). This colocalization was even more obvious in virus-infected cells, in which the signal from the gM/gN complex strongly overlapped with anti-gB in the juxtanuclear AC (Fig. 1C) (61). The AC colocalized with a marker of the late endosomal/MVB compartment, Rab7, as well as CD63, and also with the adaptor protein AP-1, a cellular protein that has been associated with protein partitioning between the TGN and endosomes (data not shown). The pattern of colocalization of the gM/gN complex with TGN/endosomal markers was confirmed by additional staining of the gM with anti-myc monoclonal antibodies or of the gN with anti-FLAG antibodies (data not shown).
FIG. 1.
Localization of the gM/gN complex in transfected Cos7 cells and during HCMV infection of HFs. (A) gM/gN was transiently coexpressed in Cos-7 cells as described in Materials and Methods, and the distribution of the gM/gN complex was determined by staining with anti-gM/gN MAb 14-16A and FITC-conjugated anti-mouse immunoglobulin M (IgM). Protein markers of the cellular secretory system were identified with anti-AP-1 and anti-CD63 antibodies, followed by Texas Red-conjugated secondary antibody. To localize Rab 7 expression, EGFP-tagged Rab 7 was cotransfected with gM and gN, and the distribution of gM/gN was determined with the anti-gM/gN MAb 14-16A and a Texas Red-labeled anti-mouse IgM secondary antibody. Nuclei were identified by Hoechst staining (blue). (B) gM/gN and gB colocalize in vesicles in transfected cells. gM/gN were coexpressed together with full-length gB as described in Materials and Methods, and transfected cells were reacted with MAbs 14-16A (gM/gN) and 7-17 (gB), followed by FITC-conjugated anti-IgM (gM/gN) and Texas red-conjugated anti-IgG3 (gB). (C) ACs in HCMV-infected HFs. HFs were infected with HCMV AD169, and 5 days p.i., the cells were fixed and stained with anti-gM/gN MAb 14-16A and anti-gB MAb 7-17, followed by FITC and tetramethyl rhodamine isothiocyanate secondary anti-mouse IgM and IgG3 antibodies.
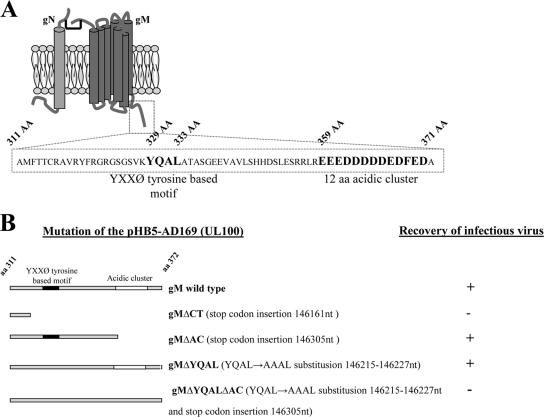
Previous studies have reported that glycoprotein gB accumulated in the TGN/endosomal compartments of transfected cells and that the cellular localization of gB depended on amino acid motifs in the cytoplasmic tail of the gB molecule (16, 33). The cytoplasmic domain of gB contains tyrosine- and dileucine-based motifs, as well as a C-terminal acidic cluster of amino acids. We analyzed the amino acid sequences of the gM and gN glycoproteins to identify potential trafficking motifs that could direct intracellular trafficking and the distribution of the gM/gN complex. There are no identified trafficking motifs in the gN sequence, whereas the C-terminal cytoplasmic tail sequence of the gM contains a tyrosine-based motif, YXXØ (329YQAL333), and a long C-terminal acidic cluster (359 to 371 aa); interestingly, both of these motifs were unique to HCMV gM (Fig. 2A).
FIG. 2.
Proposed model of the gM/gN complex of HCMV and mutations introduced into the cytoplasmic tails of gMs of recombinant viruses. (A) The HCMV glycoprotein gM (UL100) cytoplasmic tail contains the tyrosine-based motif YXXØ and a cluster of acidic amino acids. A cartoon depicting the predicted structure of gM complexed with gN (46) is shown. gM is a type III glycoprotein with a predicted seven transmembrane domains. Sequence analysis of the predicted cytoplasmic domain of gM (aa 311 to 372) revealed two conserved trafficking motifs, (i) the tyrosine-based motif YXXØ and (ii) a cluster of 12 acidic amino acids at the carboxyl terminus of the protein. The amino acid sequences of both predicted motifs are depicted in boldface, and the amino acid positions are listed above the sequence. (B) Cartoon of the gM (UL100) cytoplasmic-tail mutations that were introduced into recombinant viruses. Note that infectious virus was not recovered from the BAC in which the predicted carboxyl-terminal cytoplasmic domain of gM was deleted by insertion of a translational stop codon or from the BAC in which both trafficking motifs were mutated simultaneously. The gM KO BAC was repaired using a wild-type UL100 fragment, and the growth kinetics of the recovered virus was identical to those of the AD169 wild type.
The cytoplasmic tail of gM is essential for HCMV replication.
The predicted cytoplasmic domain of gM includes aa 311 to 372 of the C terminus and, as noted above, contains both a tyrosine-based (YXXØ) and an acidic-cluster trafficking motif. To investigate the roles of these signals in virus assembly and replication, we generated several recombinant viruses with mutations in the cytoplasmic domain of gM. These mutants were constructed using the HB-5 BAC and linear recombination, as previously reported (8). Initially, we replaced the UL100 ORF (nucleotides 145226 to 146344) of the AD169 genome with an Ampr/LacZ cassette. The UL100 gM KO BAC was then used to create series of gM mutants that included (i) deletion of the entire cytoplasmatic tail of UL100 (gMΔCT; insertion of a stop codon at nucleotide position 146161), (ii) deletion of the acidic cluster (gMΔAC; insertion of a stop codon at nucleotide position 146305), (iii) disruption of the YXXØ motif (gMΔYQAL; by substitution of the sequence encoding AAAL at positions 146215 to 146227), and (iv) simultaneous disruption of the YXXØ motif and deletion of the acidic cluster (gMΔYQALΔAC). Mutations of UL100 in the BAC HB5 were confirmed by sequencing, followed by electroporation of the purified mutant BAC DNA into the HFs for recovery of recombinant viruses. Infectious virus was not recovered from HFs electroporated with the gM KO BAC, a finding consistent with previous reports that showed that gM was essential for HCMV replication (19, 29, 75). Furthermore, infectious virus was not recovered from a recombinant BAC in which the entire C-terminal cytoplasmic domain of gM was deleted (gMΔCT) or a recombinant BAC containing mutations in both described trafficking signals within the cytoplasmic tail of gM (gMΔYQALΔAC) (Fig. 2B). To eliminate the possibility that these results could be explained by additional mutations introduced during recombination, we repaired the gM KO BAC by replacing the Ampr/LacZ cassette with the intact UL100 ORF. We recovered infectious virus from the repaired gM KO BAC that exhibited a replication phenotype similar to that of the wild-type AD169 virus (data not shown). These results confirmed previous findings and also indicated that the predicted cytoplasmic domain of glycoprotein gM (aa 311 to 372) was essential for virus replication and formation of infectious particles.
Mutations in the cytoplasmic tail of gM affect HCMV assembly.
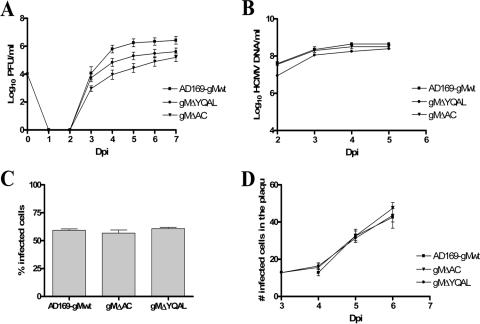
Electroporation of the BAC DNA containing mutations in the UL100 of gMΔAC (acidic-cluster deletion) and gMΔYQAL (YQAL→AAAL) resulted in the recovery of the infectious viruses from the HFs, although significant differences in virus replication were noted. Both recombinant viruses exhibited an impaired-growth phenotype compared with wild-type parental AD169 virus (Fig. 3A). The amount of virus produced by gMΔAC recombinant-virus-infected cells was approximately 2 log units less than that of cells infected with the HCMV AD169 wild type during days 4 to 6 of infection and was approximately 1.5 log units less than that of cells infected with wild-type virus when the experiment was terminated on day 7 p.i. (Fig. 3A). Cells infected with the gMΔYQAL recombinant virus produced about 1 log unit less virus than the parental wild-type AD169 virus (Fig. 3A). These differences were consistent in multiple experiments, and differences in virus yield were observed at both high and low MOIs (0.1 and 1.0). Cells infected with both mutant viruses at a high MOI (5) produced virus in titers nearly equivalent to those of cells infected with wild-type virus (data not shown). The replication defect exhibited by recombinant viruses could be explained by several mechanisms, including defects in DNA replication, virus entry, or virus assembly.
FIG. 3.
Kinetics of the growth, DNA replication, cell entry, and cell-to-cell spread of recombinant viruses containing mutations in the cytoplasmic domain of gM. (A) Growth kinetics in HFs of the wild-type HCMV compared with those of gMΔAC and gMΔYQAL recombinant viruses. HFs were infected with viruses at an MOI of 0.1, and total cultures (supernatant and cells) in 1.5 ml medium were harvested at the indicated time points p.i. (Dpi, days p.i.). The cell supernatants and disrupted cells were combined and assayed for the amounts of infectious particles, using a fluorescence-based infectivity assay (2). The results are expressed as log10 infectious units/ml of sample. The error bars indicate standard errors. (B) DNA replication assay. HFs were infected with the wild-type and recombinant gMΔAC and gMΔYQAL viruses at an MOI of 1.0. Infected cells were harvested at the indicated time points p.i., and total DNA was extracted. The HCMV genomic DNA copies were quantified using real-time PCR with primers specific for the HCMV glycoprotein gB (63). The results are expressed as log10 HCMV genome copies/ml of sample. (C) Cell entry assay. HFs plated on 13-mm glass coverslips in a 24-well plate were infected with wild-type and recombinant gMΔAC and gMΔYQAL viruses at an MOI of 1.0 and at 4°C for 1 h. The cells were then warmed to 37°C with prewarmed medium, and the coverslips were harvested at time zero. No IE-1 expression was detected at time zero. Three hours p.i., a second set of coverslips were fixed in 4% paraformaldehyde and stained with anti-IE-1 antibody to detect the number of cells expressing the IE-1 protein of HCMV. The total number of cells was estimated by Hoechst staining of host cell nuclei. The results are expressed as the percent IE-1-positive cells per 250 cells counted 3 h after being warmed to 37°C. (D) Mutations in the cytoplasmic domain of gM do not inhibit the cell-to-cell spread of HCMV. HFs were plated on 13-mm glass coverslips and then infected with wild-type HCMV AD169 or gMΔAC or gMΔYQAL recombinant virus at the same MOI of 0.1. Three hours after infection, the cells were overlaid with medium containing 0.3% low-melting-point agarose and allowed to solidify at room temperature. At 3 to 7 days p.i., as indicated, the cells were fixed in 4% paraformaldehyde and stained with antibody specific for the IE-1 antigen, as well as with Hoechst nuclear dye. The number of IE-1-positive cells per infectious focus was determined, and the mean and standard error of the number of IE-1-positive cells per focus were plotted versus the day p.i.
Initially, the amount of viral DNA replicated during infection was determined by quantitative PCR at various times following infection. HFs were infected at an MOI of 1.0 with the wild type or a recombinant virus, and DNA was extracted from the cultures at 24-h intervals (Fig. 3B). The levels of viral-DNA replication were similar for all viruses, indicating that the decreased virus yield of the gM mutant viruses was not secondary to a defect in DNA replication. The rate of virus entry was then determined by assaying the expression of the IE-1 antigen in cells infected with either wild-type or mutant virus as a function of the time p.i. Following adsorption at 4°C for 60 min, the infected cells were shifted to 37°C, and IE-1 antigen expression was assayed at 0 and 3 h p.i. The entry of mutant viruses was similar to that of wild-type virus, indicating that the replication defect in these viruses was not due to a defect in entry (Fig. 3C). We also examined the possibly that mutations in the gM cytoplasmic tail could limit the spread of infectious particles to adjacent cells after the production of progeny virions from a focus of infected cells. After infection of a monolayer of HFs, the cultures were overlaid with agarose-containing medium to restrict the spread of supernatant virus. The expansion of foci of virus-infected cells was calculated by determining the number of IE-1-expressing cells in the foci over 3 to 5 days p.i. The rates of expansion of the foci for recombinant viruses and wild-type AD169 virus were similar, indicating that mutations in the tail of gM did not restrict the capacities of these viruses to spread to adjacent cells (Fig. 3D). It is important to note that this immunofluorescence-based assay detected only the capacity of virus to spread from cell to cell and did not quantify the amount of virus released over time. Thus, only qualitative differences in cell-to-cell spread would be identified. Together, these data indicated that the defects in the production of progeny virions from HFs infected with recombinant viruses containing mutations in the cytoplasmic domain of gM were not the result of defects in viral-DNA replication, virus entry, or cell-to-cell spread but were likely secondary to a defect(s) in virus assembly.
Deletion of the gM cytoplasmic tail influences gM protein stability.
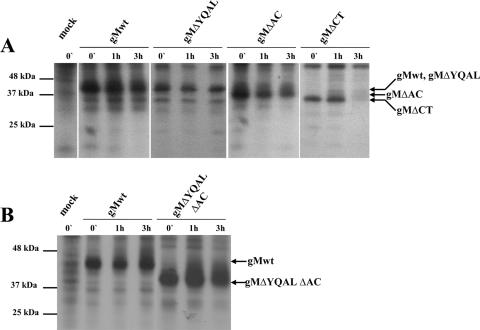
It was possible that mutations introduced into the gM cytoplasmic tail, particularly truncation of the entire cytoplasmic domain, could alter gM protein stability. We determined the stability of gM and the gM mutant proteins in a pulse-chase experiment in transfected HK 293T cells. The cells were transfected with plasmids encoding myc-tagged wild-type gM, gMΔCT, gMΔAC, or gMΔYQAL, together with FLAG-tagged gN to permit gM/gN complex formation and transport of both proteins from the ER. Following transient expression, the cells were pulse-labeled with [35S]methionine/cysteine and then chased in medium containing nonradiolabeled methionine and cysteine for 1 and 3 h. The radiolabeled proteins were immunoprecipitated with magnetic beads conjugated with anti-myc antibody, and the precipitated proteins were analyzed by SDS-PAGE. The mutant gMΔCT protein migrated significantly faster than either the wild type or the mutant gMΔYQAL or mutant gMΔAC protein, a finding consistent with the deletion of approximately 60 aa in this mutant (Fig. 4). Although we did not determine if the gMΔCT mutant protein was glycosylated, it did form a complex with gN and was transported into distal compartments of the secretory pathway, as determined by immunofluorescence (data not shown), indicating that this mutation did not result in ER retention of the protein. However, the gMΔCT mutant protein was significantly less stable than wild-type gM protein and following a 3-h chase interval was nearly undetectable (Fig. 4A). In contrast, the gMΔYQAL and gMΔAC mutant proteins appeared to be nearly as stable as the wild-type gM protein (Fig. 4A). The mutant gM protein gMΔYQALΔAC, with a double mutation, YQAL, that disrupted the tyrosine trafficking signal and deletion of the acidic cluster, exhibited stability similar to that of the wild-type gM protein (Fig. 4B). These results indicated that truncation of the cytoplasmic domain of gM resulted in a significant loss of stability in this glycoprotein. The decreased stability of the gMΔCT protein could have accounted for the lack of recovery of infectious virus from the recombinant HCMV BAC encoding this gM mutation.
FIG. 4.
The stability of the gM mutant protein gMΔCT is decreased. Pulse-chase analysis of the wild-type gM (gMwt) (UL100) and gMΔYQAL, gMΔAC, and gMΔCT recombinant proteins (A) and gMΔYQALΔAC (B) was carried out as described in Materials and Methods. Briefly, HEK 293T cells were transfected with plasmids encoding carboxyl-terminally myc-tagged wild-type gM, gMΔYQAL, gMΔAC, gMΔCT, or gMΔYQALΔAC, together with a plasmid encoding a carboxyl-terminally FLAG-tagged gN. A nontransfected control cell culture was included in the analysis. Following radiolabeling with [35S]methionine/cysteine as described in Materials and Methods, the cells were lysed at different time points and the myc-tagged gM proteins were immunoprecipitated using anti-Myc antibodies conjugated to magnetic beads as previously described (46). Equivalent amounts of the immunoprecipitated proteins were analyzed by SDS-PAGE and fluorography. Molecular mass markers are shown on the left, and the arrows on the right point to migrations of gM-specific proteins.
Trafficking motifs in the gM cytoplasmic tail contribute to the localization kinetics of the gM/gN complex to the AC.
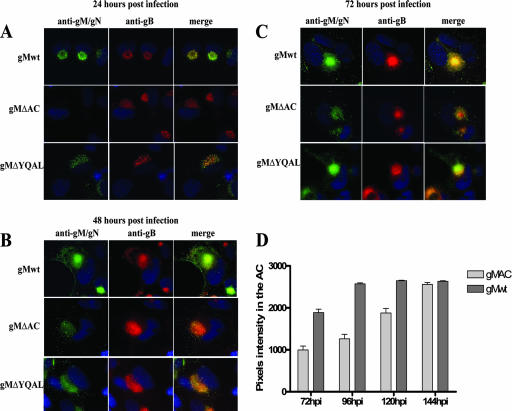
Infection of permissive HFs with HCMV results in the accumulation of the viral glycoproteins and tegument proteins in the juxtanuclear AC of the infected cell (61). Accumulation of the viral proteins in the AC coincides with displacement of Golgi and TGN markers, as well as inclusion of some endosomal markers (63). The maturation of the AC during the progression of the infection is associated with progressive compaction, accumulation of virus-expressed proteins, and, finally, the production of infectious particles. To investigate if differences in the replication of the gM recombinant viruses were reflected in differences in the dynamics of their localization to the AC, we determined the kinetics of localization of viral proteins to the AC at different times p.i. using an immunofluorescence assay. HFs infected with wild-type HCMV AD169 and gMΔAC and gMΔYQAL mutant viruses were fixed at 24-h intervals during infection and stained with MAb 14-16A, specific for the gM/gN complex, and MAb 7-17, which is reactive with gB (Fig. 5A, B, and C.). We observed that the kinetics of localization of the gM/gN complex to the AC and the morphology of the AC differed between cells infected with the recombinant viruses containing mutations in the cytoplasmic domain of gM and the wild-type virus. The accumulation of the gMΔAC/gN complex in the AC of cells infected with gMΔAC virus was delayed, and only a very weak signal could be detected after 48 h of infection compared to cells infected with wild-type virus (Fig. 5B). Interestingly, even 72 h p.i., the fluorescence signal from the gMΔAc/gN complex in the AC was significantly less. At both 48 and 72 h, the AC appeared less compacted than that in cells infected with wild-type virus (Fig. 5B and C). When we compared the amounts of gB localized to the AC in the same experiment, we noted that gB produced in both cells infected with wild-type virus and those infected with gMΔAC mutant virus localized to the AC with nearly identical kinetics, although the intensity of the staining in gMΔAC-infected cells was decreased, presumably secondary to delayed compaction of the AC, possibly associated with the delay in the localization of the gMΔAC/gN complex to the AC (Fig. 5A, B, and C). To quantify the relative amounts of gMΔAc/gN and gM/gN complexes in the ACs of infected cells at various times p.i., we estimated the fluorescence intensity of MAb 14-16A reactivity for the gM/gN complex within the AC by pixel intensity in a series of infected cells (n ≥ 30 cells). Images were captured under identical conditions of exposure time and gain for cells infected with both wild-type and recombinant gMΔAC/gN viruses, and the pixel intensity was determined for the fluorescence signal in the center of the AC. Images were captured beginning at 72 h p.i., which represented the initial time point at which gMΔAC/gN could be consistently detected in the gMΔAC virus-infected cells. The accumulation of gMΔAC/gN in the AC was delayed compared to that of the wild-type gM/gN (Fig. 5D). This delayed accumulation of gMΔAC/gN in the AC correlated with the decreased virus yield in cells infected with this mutant virus (Fig. 3A).
FIG. 5.
Analysis of gM/gN localization in the AC in cells infected with recombinant viruses with mutations in the cytoplasmic domain of gM. (A to C) Kinetics of localization of gM/gN, gMΔAC/gN, and gMΔYQAL/gN in cells infected with the respective recombinant virus. HFs were plated on 13-mm glass coverslips and then infected with wild-type (gMwt) or recombinant gMΔAC or gMΔYQAL virus at an MOI of 1.0. At the indicated time points (24, 48, and 72 h) p.i., cells were fixed and stained with MAb 14-16A, specific for the gM/gN complex, and the gB-specific MAb 7-17 as described in Material and Methods. Note that the AC in the cells infected with the wild-type virus was round and beginning to establish a compacted structure at 24 h p.i., and by 48 to 72 h p.i., the compartment was even more distinct. Also note differences in the morphology and relative intensity of immunofluorescence signals from the wild-type gM/gN complex compared to signals from cells infected with gMΔAC or gMΔYQAL virus. (D) Quantification of the amount of gM/gN in the ACs of infected cells by determination of pixel density. HFs were infected with HCMV AD169 or gMΔAC recombinant virus at an MOI of 1.0, and the coverslips were harvested every 24 h and fixed in 4% paraformaldehyde. The coverslips were reacted with the anti-gM/gN MAb and FITC-conjugated anti-mouse immunoglobulin M. The intensity of the fluorescence in the AC generated by the signal from the anti-gM/gN specific antibody was measured in ≥30 cells using Image Pro software (Media Cybernetics, Silver Spring, MD), and the number of pixels within the AC was determined. The graph shows the intensity of fluorescence as a function of time in the AC (the error bars indicate standard errors). Note that the intensity of fluorescence was measured after 72 h of infection because that represented the initial time point at which the AC could be reproducibly measured in gMΔAC-infected cells.
The kinetics of gMΔYQAL/gN complex localization in the AC in gMΔYQAL virus-infected cells was similar to that observed in cells infected with wild-type virus (Fig. 5A, B, and C). However, the AC in gMΔYQAL virus-infected cells was more diffuse and less compacted at 24 and 48 h p.i. than in cells infected with wild-type virus. In gMΔYQAL virus-infected cells, the AC required about 72 h to achieve a morphological appearance of the AC similar to that observed at 48 h p.i. in cells infected with wild-type virus. These results suggested that mutations of the YXXØ motif in the cytoplasmic tail of gM altered the maturation of the AC and, furthermore, raised the possibility that morphogenesis of the AC required the presence of gM/gN in this intracellular compartment.
Together, these findings argued that trafficking signals in the gM cytoplasmic tail influenced the kinetics of gM/gN complex localization to the AC. Because localization of gM/gN to the AC could be a rate-limiting step in the cytoplasmic envelopment of HCMV, this delay in localization to the AC could have contributed to the lower virus yield of cells infected with either the gMΔAC or gMΔYQAL recombinant virus.
Mutations in the C-terminal trafficking motifs of gM determine the rate of gM transport to the AC.
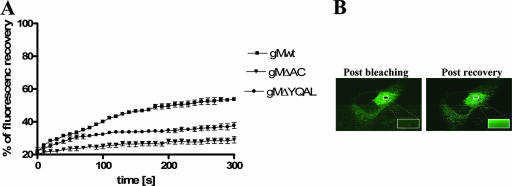
In order to more accurately assay the intracellular-transport kinetics of gM with the mutations in the C-terminal trafficking motifs to the ACs of infected cells, we used FRAP in live cells to measure trafficking of the gM/gN complexes to the AC. HFs were initially transfected with plasmids encoding green fluorescent protein (GFP)-tagged wild-type gM, gMΔAC, or gMΔYQAL and then infected with wild-type virus or with viruses containing the same mutations in the sequence of gM as the corresponding GFP-tagged constructs. The trafficking of the GFP-tagged gMs appeared identical to that of untagged gM, suggesting that the GFP modification did not alter the intracellular localization of gM/gN complexes containing GFP-tagged gMs (data not shown). After 5 days of infection, the cells were subjected to FRAP. For these experiments, we selected virus-infected cells with bright fluorescent signals and compacted ACs formed by accumulation of GFP-tagged gM in the ACs. After photobleaching of the area in the center of the AC, series of images were collected using identical parameters of time, area, and laser intensity in all analyzed cells (Fig. 6A and B). The rate of the refilling of the AC with the GFP-labeled gM was measured for 300 seconds at 10-second intervals. We noted that wild-type gM refilled the AC rapidly, with a half-time of recovery of approximately 100 seconds, and continued to increase in fluorescence recovery throughout the 300-second experiment (Fig. 6A.). In contrast to these findings with wild-type gM, neither the gMΔAC or gMΔYQAL protein reached 50% of the prebleach fluorescence level during the 300 seconds of the experiment. Moreover, both reached a plateau in fluorescence recovery, indicating a significant delay in the trafficking of the mutant gMΔAC/gN and gMΔYQAL/gN complexes to the AC compared to wild-type gM/gN (Fig. 6A.). Interestingly, the delay in refilling of the AC by gMΔAC-containing complexes was much more significant than that observed with gMΔYQAL, a result consistent with differences in the replication phenotypes of these viruses. Together, these findings were consistent with differences in the kinetics of formation and localization of the gM/gN in the AC in the cells infected with recombinant viruses with gM mutations in the cytoplasmic domain compared to wild-type HCMV.
FIG. 6.
Fluorescence recovery after photobleaching (FRAP) analysis of trafficking of the gM/gN, gMΔAC/gN, and gMΔYQAL/gN protein complexes in the ACs of virus-infected cells. Plasmids encoding GFP-tagged wild-type gM (gMwt), gMΔAC, and gMΔYQAL were electroporated into HFs and plated on 13-mm coverslips, and 24 h later, these cells were infected with the respective viruses. Once a well-formed AC was established, the live cells were subjected to FRAP as described in Materials and Methods. (A) Fluorescence recovery following photobleaching of the AC to 20% of initial fluorescence intensity as a function of time (300 s). Note the continued fluorescence recovery of wild-type gM/gN at 300 s compared to the plateau in recovery reached by both gMΔAC/gN and gMΔYQAL/gN at that time point. The error bars indicate standard errors. (B) Examples of the images collected during FRAP analysis of the AC and following fluorescence recovery.
DISCUSSION
Glycoprotein M has been reported to be the most abundant glycoprotein in the envelope of HCMV and has also been shown to be essential for the replication of infectious virus (72). Our results confirmed previous studies that indicated gM was essential for the production of infectious virus and extended these findings by demonstrating that the cytoplasmic tail of gM was required for the function(s) of gM in the assembly of infectious virions. Whether the lethal phenotype of the gMΔCT recombinant virus was secondary to a loss of an essential function of this protein or a result of decreased expression of the gMΔCT protein secondary to decreased stability associated with truncation of the protein cannot be resolved from the data gathered in the current study. Studies of PRV have demonstrated interactions between gM and the tegument protein UL49 during virus assembly, suggesting that the cytoplasmic domain of HCMV gM could also play an essential role in virion assembly through interactions with other viral proteins (23, 39). Alternatively, the phenotype of impaired virus production by recombinant viruses with mutations in specific intracellular-trafficking signals in the cytoplasmic domain of gM raised the possibility that deletion of the cytoplasmic domain of gM resulted in a null phenotype secondary to mislocalization of this essential glycoprotein in the infected cell. The decrease in virus yield from cells infected with recombinant viruses containing mutations in the trafficking signals in the cytoplasmic tail of gM could be explained by several different mechanisms, including decreased virus release and altered function of the virion envelope during early events of infection. A defect in virus release was an unlikely explanation for the observed phenotype because a decrease in virus yield from cells infected with these recombinant viruses was observed regardless of whether virus infectivity was quantified from the supernatant or from disrupted cells (data not shown). Secondly, virions produced by the cells infected with these viral mutants penetrated cells and spread cell to cell as efficiently as wild-type virus. Thus, our findings indicated that recombinant viruses with mutations in the cytoplasmic domain of gM exhibited a replication-impaired phenotype as a result of defective virion assembly, presumably during secondary envelopment in the cytoplasm of the infected cell.
Although a precise role of gM in HCMV assembly cannot be assigned from the findings of the current study, the abundance of this glycoprotein in the virion envelope and the requirement for it in the assembly of infectious virus strongly suggested that gM has a specific role in the morphogenesis of the virion envelope. Our findings were in contrast to proposed functions of the gM homologs of alphaherpesviruses, EHV, HSV, and PRV. The gMs of these viruses have been demonstrated to be nonessential for in vitro replication (3, 13, 38, 49). HSV and PRV gM deletion virus mutants replicated to levels that were similar to those of wild-type virus in the cases of HSV and PRV, whereas replication of EHV was reduced by 1 to 2 log units compared to wild-type virus (64). In addition, and in contrast to the essential function of gN in HCMV virion assembly, the gN homolog of these alphaherpesviruses is not essential for replication (24, 60). However, triple-deletion PRV mutant viruses lacking gM, gE, and gI are noninfectious and cannot assemble a virion envelope in the cytoplasm of the infected cells (7). Characterization of the in vitro phenotypes of these mutant viruses provided direct evidence for cytoplasmic envelopment of these alphaherpesviruses and also strongly suggested that in the cases of EHV and PRV, gM has a direct role in the morphogenesis of the virion envelope (7). Interestingly, a similar phenotype was described for a PRV mutant in which the viral genes encoding both gM and the tegument protein UL11 had been deleted (39). Surprisingly, deletion of PRV gM or UL11 as a single mutation did not result in significant impairment of virus replication, suggesting that both of these viral proteins contributed to secondary envelopment in the cytoplasm of the infected cell (39). In contrast to findings in PRV, an HSV deletion mutant lacking both gM and gE replicated to levels near those of wild-type virus and produced numbers of enveloped cytoplasmic particles similar to those from cells infected with wild-type virus (13). Although we have not characterized particles generated in cells electroporated with the gM KO BAC because of the lack of a complementation system that enabled us to generate a stock of infectious progeny virions, a mutant recombinant virus that was severely replication impaired secondary to a point mutation in gN was shown to exhibit a defect in cytoplasmic envelopment (47). Electron microscopic analysis of cells infected with this gN mutant virus revealed a decrease in the number of enveloped particles (47). Because previous studies indicated that gM and gN must form a complex prior to transport to the AC and that they are found as a protein complex in mature virions, the phenotype of the gN mutant virus was most consistent with a key role of the gM/gN complex in virion envelope assembly and suggested that at least one function of HCMV gM was required for virion envelope morphogenesis. Other potential contributions of gM to virion envelope function during early events of infection, such as attachment/penetration and fusion, cannot be completely excluded. Previous studies have reported that the gM/gN complex binds heparin sulfate, and recent findings have demonstrated that the gM/gN complex and gN are targets of virus-neutralizing antibodies, both findings consistent with a role of the gM/gN complex in early events of infection (37, 65).
We noted that the gM/gN complex localized rapidly to the AC in virus-infected cells with kinetics that appeared more rapid than those of another abundant virion envelope glycoprotein, gB. In addition, we could also demonstrate that the gM/gN complex colocalized with markers of the endosomal/MVB compartment, CD63 and Rab 7, in virus-infected cell, suggesting that the AC was derived from an endosomal compartment and not the TGN as had been reported in earlier studies (30). This result was also consistent with previous findings that demonstrated that the AC was localized to a juxtanuclear site and that markers of the secretory pathway, including mannosidase II (Golgi) and galactosyltransferase (TGN), were radially distributed outside the AC (61, 63). These findings were also consistent with the results of a study using immunoelectron microscopy that demonstrated that during infection, HCMV glycoproteins and newly formed HCMV particles accumulated in MVBs derived from the endosomal vesicles (22). Together, these data suggested that the endosomal/MVB compartment was a potential site of HCMV assembly, but additional localization studies of secretory-pathway markers with HCMV virion proteins will be required to determine the exact relationship between the TGN and MVBs in HCMV envelopment.
In cells cotransfected with plasmids encoding gM, gN, and gB, we could detect cytoplasmic vesicles containing both gM/gN and gB, suggesting that these viral glycoproteins utilized a common intracellular-transport pathway, presumably secondary to similar trafficking signals present in their cytoplasmic domains. As noted previously, both gM and gB contain clusters of acidic amino acids at the C termini of the cytoplasmic tails. Previously, we have shown that mutations in serine 900 (Ser→Ala), which is immediately adjacent to the acidic cluster of gB, had only modest effects on the cellular localization of the glycoprotein and that recombinant viruses with this specific mutation replicated with kinetics identical to those of wild-type virus (33). Interestingly, replacement of serine 900 with an aspartic acid (Ser→Asp) to mimic constitutive phosphorylation led to accumulation of the mutant gB in a post-TGN compartment and increased virus yield (33). This finding was interpreted as evidence that the aspartic acid mutant gB interacted more efficiently with a PACS-1-mediated transport pathway that concentrated the glycoprotein in the TGN and subsequently increased delivery to the AC (33). However, more recent studies have demonstrated that phosphorylation of amino acids proximal to acidic clusters is not required for PACS-1 interactions (5). Thus, the increased accumulation of the aspartic acid-containing gB mutant in the TGN and the increased virus yield in cells infected with this virus could have been secondary to merely increasing the length of the existing acidic cluster (33). This possibility was intriguing when viewed in the context of gM, a glycoprotein with an unusually long (12-aa) acidic cluster and a glycoprotein that appeared to be more efficiently localized to the AC than gB. Interestingly, a gM mutant lacking only 6 of the 12 acidic amino acids has an AC localization phenotype in transient-expression assays that is intermediate between wild-type gM and the mutant that lacked the entire C-terminal acidic cluster (data not shown). It is possible that the long acidic tail of gM was required for efficient delivery to the TGN and that the accumulation of the gM/gN complex in this compartment favored the subsequent transport to the AC. Thus, this long stretch of acidic amino acids could interact with PACS-1 and/or other cellular proteins to ensure a sufficient amount of gM/gN was available for transport to the AC and envelope morphogenesis. Interestingly, HCMV replication in cells in which PACS-1 expression was inhibited exhibited only a very modest decrease in virus yield (<2-fold), raising the possibility that a pathway involving retrieval of viral glycoproteins from the cell surface to the TGN in a PACS-1-dependent pathway could represent only a minor component of HCMV glycoprotein trafficking in infected cells (16). Importantly, these authors also noted that PACS-1 cycling between endosomes and the TGN could represent a dominant intracellular pathway for viral-glycoprotein transport to the TGN without a requirement for internalization and endocytosis of a minor population of cell surface viral glycoproteins (16). Thus, the importance of PACS-1 in HCMV virion envelope glycoprotein trafficking remains to be further defined.
The cytoplasmic domains of gM and gB also contain sequences that match consensus YXXØ tyrosine-based sorting signals that have been reported to have roles in ER export, endocytosis, lysosomal-endosomal sorting, and basolateral trafficking in polarized cells (1, 17, 34). A previous report suggested that the YXXØ in the cytoplasmic tail of HCMV gB served as an ER export signal (28). The YXXØ in the cytoplasmic domain of gM, however, does not appear to serve a similar function because mutation and/or deletion of this sequence did not prevent formation of the gM/gN complex or transport to the TGN. Thus, the YXXØ motif in gM likely served as a sorting signal that could potentially interact with several cellular adaptor molecules, including AP-1, -2, -3, and -4 and presumably GGA (for Golgi-localized, γ-ear containing, ADP ribosylation factor ARF binding) adaptor molecules (5, 25, 40, 50, 55, 62, 68). Although the YXXØ motif has been associated with internalization during endocytosis through interactions with AP-2, other studies have also shown that this sequence can interact with both AP-1 and AP-3 and served as a sorting signal for direct TGN-to-lysosomal-endosomal transport and basolateral trafficking in polarized cells (57, 70). We have previously observed that limited amounts of the gM/gN complex were expressed on the surfaces of infected and transfected cells (data not shown). We also used cells transfected with plasmids expressing gM mutant proteins and anti-gM/gN antibodies to estimate rates of endocytosis. In experiments carried out with transiently expressed gMΔYQAL/gN mutant-protein-containing complex, our results did not suggest a significant defect in retrieval from the cell surface, arguing that the YQAL motif of gM in the gM/gN complex was utilized primarily for intracellular trafficking and not for internalization during endocytosis (data not shown). Results from a similar experiment carried out with transiently expressed gMΔAC/gN mutant protein complex also failed to support a role for endocytosis of the gMΔAC/gN protein from the cell surface (data not shown). Thus, our findings argued that both the YQAL motif and the acidic cluster of gM were utilized for intracellular trafficking of the gM/gN complex and that endocytosis of gM/gN from the plasma membrane did not contribute significantly to the localization of this glycoprotein complex to the AC. These findings were also consistent with the lack of increased expression of the gM/gN on the surfaces of cells infected with either the gMΔYQAL or gMΔAC mutant virus. Additional support for our interpretation of the results is provided in the study by Jarvis et al., who noted that the total yield of HCMV during productive infection in virus-infected permissive cells was not influenced by the expression of the dominant-negative dynamin-I that blocked clathrin-dependent endocytosis (32). The conclusions of this study were that endocytosis played a minor role, if any, in the assembly and final envelopment of HCMV. Finally, our studies using FRAP to measuring the refilling of the AC indicated that significant differences existed between the wild-type gM/gN complex and the complexes containing the gM mutants, gMΔAC and gMΔYQAL. These experiments demonstrated that the half-life of refilling this compartment was on the order of 100 s (50% recovery) for wild-type gM/gN (Fig. 6). The time frame of this repopulation of the AC by the gM/gN complex appears too short to be accounted for by endocytic retrieval from the cell surface and transport to the AC. Although we cannot rule out a limited contribution of endocytosed cell surface gM/gN to the amount of gM/gN found in the AC, our results suggested that signals in the cytoplasmic domain of gM were of greater importance for the intracellular trafficking of this protein complex to the AC.
The replication-impaired phenotype of the gMΔAC and gMΔYQAL recombinant viruses was most consistent with a defect in virion assembly. Although several potential mechanisms could explain this defect in assembly, the delay in trafficking of the gMΔAC/gN and gMΔYQAL/gN complexes to the AC likely contributed to the replication defect exhibited by these viruses. The gMΔAC and gMΔYQAL mutations disrupted two well-described intracellular-trafficking motifs, both of which have been shown to direct the localization of cellular and viral proteins to the TGN through interactions with cellular adaptor proteins and clathrin-dependent pathways (5). In the transient-expression assays, mutations in either the acidic cluster (gMΔAC) or the YXXØ (gMΔYQAL) motifs of gM did not significantly alter the intracellular localization of the gM/gN complex from that observed for complexes containing wild-type gM/gN. However, these studies were done in transfected cells at steady state using static imaging and, more importantly, in the absence of virus-induced changes in the secretory pathway that were present in virus-infected cells. In addition, the kinetics of trafficking of these gM mutants to the TGN in transfected cells appeared to be similar to those of complexes containing wild-type gM. Thus, the findings that complexes containing mutant gM were delayed in localization to the AC in virus-infected cells were not predicted from studies using transient-expression assays and emphasize the importance of studies of protein trafficking in the environment of the virus-infected cell. Lastly, the quantitative relationship between the localization of gM/gN to the AC and virion assembly remains undefined, but the abundance of gM in the envelope of HCMV and the defect in virion assembly associated with the gMΔAC and gMΔYQAL mutant viruses argue that a delay in the delivery of the gM/gN complex to the AC could have resulted in a restriction in the cytoplasmic envelopment of the tegumented particle.
The role of gM in the envelopment of the particle is unknown. The abundance of this glycoprotein and its predicted seven membrane-spanning domains suggested that this multimembrane-spanning protein could function in the organization of the envelope of the virion. Although we do not have evidence for a direct role of gM in the morphogenesis of the HCMV envelope, our previous studies described defective cytoplasmic envelopment of a replication-impaired recombinant virus with a point mutation in gN (47). As noted above, electron microscopic analysis of cells infected with this gN mutant virus demonstrated large numbers of nonenveloped cytoplasmic particles in the cytoplasm of infected cells (47). This finding indicated that a loss of the function of the gM/gN complex resulted in a defect in envelopment and supported our hypothesis that gM has a major role in the envelopment of the HCMV particle. Thus, limiting amounts of the gM/gN complex in the AC secondary to delayed localization of gMΔAC/gN or gMΔYQAL/gN complexes in cells infected with the respective mutant virus could result in decreased assembly of infectious virions. In the case of the cytoplasmic domain deletion mutant gMΔCT virus, it is unclear whether gMΔCT/gN complexes localized to the AC or whether, once localized to the AC, the complex failed to function in virion envelopment. The decreased stability of the gMΔCT mutant protein could have contributed to a decrease in the concentration of the gMΔCT/gN complex in the AC and a subsequent loss of virion envelopment. Alternatively, the cytoplasmic domain could have a direct function in the process of envelopment, perhaps through interactions with tegument proteins, as described for the gM homolog of PRV (23).
Acknowledgments
This work was supported by grants from the HHS and NIH (AI49537 and AI50189 to W.J.B.) and from the Deutsche Forschungsgemeinschaft (grant MA929/6-5 to M.M.).
We thank Albert Tousson for his help with the FRAP assay.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Alconada, A., U. Bauer, B. Sodeik, and B. Hoflack. 1999. Intracellular traffic of herpes simplex virus glycoprotein gE: characterization of the sorting signals required for its trans-Golgi network localization. J. Virol. 73:377-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreoni, M., M. Faircloth, L. Vugler, and W. J. Britt. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods 23:157-167. [DOI] [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1993. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J. Virol. 67:1441-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billstrom, M. A., and W. J. Britt. 1995. Postoligomerization folding of human cytomegalovirus glycoprotein B: identification of folding intermediates and importance of disulfide bonding. J. Virol. 69:7015-7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonifacino, J. S., and L. M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395-447. [DOI] [PubMed] [Google Scholar]
- 6.Bowen, E. F., P. Wilson, M. Atkins, S. Madge, P. D. Griffiths, M. A. Johnson, and V. C. Emery. 1995. Natural history of untreated cytomegalovirus retinitis. Lancet 346:1671-1673. [DOI] [PubMed] [Google Scholar]
- 7.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britt, W., M. Jarvis, J. Y. Seo, D. Drummond, and J. Nelson. 2004. Rapid genetic engineering of human cytomegalovirus using a lambda phage-based linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J. Virol. 78:539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britt, W. J. 1984. Neutralizing antibodies detect a disulfide-linked glycoporotein complex within the envelope of human cytomegalovirus. Virology 135:369-378. [DOI] [PubMed] [Google Scholar]
- 10.Britt, W. J., and C. A. Alford. 1996. Cytomegalovirus, p. 2493-2523. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Raven Press, New York, NY.
- 11.Britt, W. J., and S. Boppana. 2004. Human cytomegalovirus virion proteins. Hum. Immunol. 65:395-402. [DOI] [PubMed] [Google Scholar]
- 12.Britt, W. J., M. A. Jarvis, D. D. Drummond, and M. Mach. 2005. Antigenic domain 1 is required for oligomerization of human cytomegalovirus glycoprotein B. J. Virol. 79:4066-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Browne, H., S. Bell, and T. Minson. 2004. Analysis of the requirement for glycoprotein M in herpes simplex virus type 1 morphogenesis. J. Virol. 78:1039-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-170. [DOI] [PubMed] [Google Scholar]
- 15.Compton, T. 1995. Towards a definition of the HCMV entry pathway. Scand. J. Infect. Dis. Suppl. 99:30-32. [PubMed] [Google Scholar]
- 16.Crump, C. M., C. H. Hung, L. Thomas, L. Wan, and G. Thomas. 2003. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J. Virol. 77:11105-11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danis, C., J. Deschambeault, S. Do Carmo, E. A. Cohen, E. Rassart, and G. Lemay. 2004. The tyrosine-based YXXO targeting motif of murine leukemia virus envelope glycoprotein affects pathogenesis. Virology 324:173-183. [DOI] [PubMed] [Google Scholar]
- 18.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. (Erratum, 84:1053.) [DOI] [PubMed] [Google Scholar]
- 19.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of the human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Favoreel, H. W., H. J. Nauwynck, H. M. Halewyck, P. Van Oostveldt, T. C. Mettenleiter, and M. B. Pensaert. 1999. Antibody-induced endocytosis of viral glycoproteins and major histocompatibility complex class I on pseudorabies virus-infected monocytes. J. Gen. Virol. 80:1283-1291. [DOI] [PubMed] [Google Scholar]
- 21.Feire, A. L., H. Koss, and T. Compton. 2004. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc. Natl. Acad. Sci. USA 101:15470-15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraile-Ramos, A., A. Pelchen-Matthews, T. N. Kledal, H. Browne, T. W. Schwartz, and M. Marsh. 2002. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3:218-232. [DOI] [PubMed] [Google Scholar]
- 23.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuchs, W., and T. C. Mettenleiter. 2005. The nonessential UL49.5 gene of infectious laryngotracheitis virus encodes an O-glycosylated protein which forms a complex with the non-glycosylated UL10 gene product. Virus Res. 112:108-114. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh, P., and S. Kornfeld. 2004. The GGA proteins: key players in protein sorting at the trans-Golgi network. Eur. J. Cell. Biol. 83:257-262. [DOI] [PubMed] [Google Scholar]
- 26.Gretch, D. R., B. Kari, L. Rasmussen, R. C. Gehrz, and M. F. Stinski. 1988. Identification and characterization of three distinct families of glycoprotein complexes present in the envelopes of human cytomegalovirus. J. Virol. 62:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahn, G., M. G. Revello, M. Patrone, E. Percivalle, G. Campanini, A. Sarasini, M. Wagner, A. Gallina, G. Milanesi, U. Koszinowski, F. Baldanti, and G. Gerna. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 78:10023-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heineman, T. C., P. Connolly, S. L. Hall, and D. Assefa. 2004. Conserved cytoplasmic domain sequences mediate the ER export of VZV, HSV-1, and HCMV gB. Virology 328:131-141. [DOI] [PubMed] [Google Scholar]
- 29.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Homman-Loudiyi, M., K. Hultenby, W. Britt, and C. Soderberg-Naucler. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-golgi network 46, and mannosidase II. J. Virol. 77:3191-3203. (Erratum, 77:8179.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber, M. T., and T. Compton. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J. Virol. 72:8191-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jarvis, M. A., K. N. Fish, C. Soderberg-Naucler, D. N. Streblow, H. L. Meyers, G. Thomas, and J. A. Nelson. 2002. Retrieval of human cytomegalovirus glycoprotein B from cell surface is not required for virus envelopment in astrocytoma cells. J. Virol. 76:5147-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarvis, M. A., T. R. Jones, D. D. Drummond, P. P. Smith, W. J. Britt, J. A. Nelson, and C. J. Baldick. 2004. Phosphorylation of human cytomegalovirus glycoprotein B (gB) at the acidic cluster casein kinase 2 site (Ser900) is required for localization of gB to the trans-Golgi network and efficient virus replication J. Virol. 78:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, A. O., M. A. Lampson, and T. E. McGraw. 2001. A di-leucine sequence and a cluster of acidic amino acids are required for dynamic retention in the endosomal recycling compartment of fibroblasts. Mol. Biol. Cell. 12:367-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jons, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jons, A., H. Granzow, R. Kuchling, and T. C. Mettenleiter. 1996. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J. Virol. 70:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kari, B., and R. Gehrz. 1993. Structure, composition and heparin binding properties of a human cytomegalovirus glycoprotein complex designated gC-II. J. Gen. Virol. 74:255-264. [DOI] [PubMed] [Google Scholar]
- 38.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kopp, M., H. Granzow, W. Fuchs, B. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kyttala, A., K. Yliannala, P. Schu, A. Jalanko, and J. P. Luzio. 2005. AP-1 and AP-3 facilitate lysosomal targeting of Batten disease protein CLN3 via its dileucine motif. J. Biol. Chem. 280:10277-10283. [DOI] [PubMed] [Google Scholar]
- 41.Lake, C. M., and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus that lacks glycoprotein gN is impaired in assembly and infection. J. Virol. 74:11162-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehner, R., H. Meyer, and M. Mach. 1989. Identification and characterization of a human cytomegalovirus gene coding for a membrane protein that is conserved among human herpesviruses. J. Virol. 63:3792-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ljungman, P., P. Griffiths, and C. Paya. 2002. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin. Infect. Dis. 34:1094-1097. [DOI] [PubMed] [Google Scholar]
- 44.Lopper, M., and T. Compton. 2004. Coiled-coil domains in glycoproteins B and H are involved in human cytomegalovirus membrane fusion. J. Virol. 78:8333-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mach, M., B. Kropff, P. Dal Monte, and W. Britt. 2000. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J. Virol. 74:11881-11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mach, M., B. Kropff, M. Krzyzaniac, and W. Britt. 2005. Complex formation by glycoproteins M and N of human cytomegalovirus: structural and functional aspects. J. Virol. 79:2160-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mach, M., K. Osinski, B. Kropff, U. Schloetzer-Schrehardt, M. Krzyzaniak, and W. Britt. 2007. The carboxy-terminal domain of glycoprotein N of human cytomegalovirus is required for virion morphogenesis. J. Virol. 81:5212-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacLean, C., L. Robertson, and F. Jamieson. 1993. Characterization of the UL10 gene product of herpes simplex virus type 1 and investigation of its role in vivo. J. Gen. Virol. 74:975-983. [DOI] [PubMed] [Google Scholar]
- 49.Masse, M. J., A. Jons, J. M. Dijkstra, T. C. Mettenleiter, and A. Flamand. 1999. Glycoproteins gM and gN of pseudorabies virus are dispensable for viral penetration and propagation in the nervous systems of adult mice. J. Virol. 73:10503-10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mattera, R., B. Ritter, S. S. Sidhu, P. S. McPherson, and J. S. Bonifacino. 2004. Definition of the consensus motif recognized by gamma-adaptin ear domains. J. Biol. Chem. 279:8018-8028. [DOI] [PubMed] [Google Scholar]
- 51.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9:423-429. [DOI] [PubMed] [Google Scholar]
- 53.Mocarski, E. S., and C. Tan Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 54.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakatsu, F., and H. Ohno. 2003. Adaptor protein complexes as the key regulators of protein sorting in the post-Golgi network. Cell Struct. Funct. 28:419-429. [DOI] [PubMed] [Google Scholar]
- 56.Netterwald, J. R., T. R. Jones, W. J. Britt, S. J. Yang, I. P. McCrone, and H. Zhu. 2004. Postattachment events associated with viral entry are necessary for induction of interferon-stimulated genes by human cytomegalovirus. J. Virol. 78:6688-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Odorizzi, G., C. R. Cowles, and S. D. Emr. 1998. The AP-3 complex: a coat of many colours. Trends Cell. Biol. 8:282-288. [DOI] [PubMed] [Google Scholar]
- 58.Ross, J., M. Williams, and J. I. Cohen. 1997. Disruption of the varicella-zoster virus dUTPase and the adjacent ORF9A gene results in impaired growth and reduced syncytia formation in vitro. Virology 234:186-195. [DOI] [PubMed] [Google Scholar]
- 59.Rubin, R. 2002. Clinical approach to infection in the compromised host, p. 573-679. In R. Rubin and L. S. Young (ed.), Infection in the organ transplant recipient. Kluwer Academic Press, New York, NY.
- 60.Rudolph, J., C. Seyboldt, H. Granzow, and N. Osterrieder. 2002. The gene 10 (UL49.5) product of equine herpesvirus 1 is necessary and sufficient for functional processing of glycoprotein M. J. Virol. 76:2952-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication. Characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scott, P. M., P. S. Bilodeau, O. Zhdankina, S. C. Winistorfer, M. J. Hauglund, M. M. Allaman, W. R. Kearney, A. D. Robertson, A. L. Boman, and R. C. Piper. 2004. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat. Cell. Biol. 6:252-259. [DOI] [PubMed] [Google Scholar]
- 63.Seo, J., and W. Britt. 2006. Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J. Virol. 80:5611-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seyboldt, C., H. Granzow, and N. Osterrieder. 2000. Equine herpesvirus 1 (EHV-1) glycoprotein M: effect of deletions of transmembrane domains. Virology 278:477-489. [DOI] [PubMed] [Google Scholar]
- 65.Shimamura, M., M. Mach, and W. J. Britt. 2006. Human cytomegalovirus infection elicits a glycoprotein M (gM)/gN-specific virus-neutralizing antibody response. J. Virol. 80:4591-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. USA 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stagno, S., and W. J. Britt. 2005. Cytomegalovirus, p. 389-424. In J. S. Remington and J. O. Klein (ed.), Infectious diseases of the fetus and newborn infant, 6th ed. W.B. Saunders, Philadelphia, PA.
- 68.Takatsu, H., Y. Katoh, Y. Shiba, and K. Nakayama. 2001. Golgi-localizing, gamma-adaptin ear homology domain, ADP-ribosylation factor-binding (GGA) proteins interact with acidic dileucine sequences within the cytoplasmic domains of sorting receptors through their Vps27p/Hrs/STAM (VHS) domains. J. Biol. Chem. 276:28541-28545. [DOI] [PubMed] [Google Scholar]
- 69.Tischer, B. K., D. Schumacher, M. Messerle, M. Wagner, and N. Osterrieder. 2002. The products of the UL10 (gM) and the UL49.5 genes of Marek's disease virus serotype 1 are essential for virus growth in cultured cells. J. Gen. Virol. 83:997-1003. [DOI] [PubMed] [Google Scholar]
- 70.Traub, L. M. 2003. Sorting it out: AP-2 and alternate clathrin adaptors in endocytic cargo selection. J. Cell. Biol. 163:203-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tugizov, S., E. Maidji, J. Xiao, and L. Pereira. 1999. An acidic cluster in the cytosolic domain of human cytomegalovirus glycoprotein B is a signal for endocytosis from the plasma membrane. J. Virol. 73:8677-8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp II, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. (Erratum, 78:13395.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, D., and T. Shenk. 2005. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J. Virol. 79:10330-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, D., and T. Shenk. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. USA 102:18153-18158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]