Abstract
Short interfering RNAs (siRNAs) targeting viral or cellular genes can efficiently inhibit human immunodeficiency virus type 1 (HIV-1) replication. Nevertheless, optimal HIV-1 gene silencing by siRNA requires precise complementarity with most of the target sequence. The emergence of mutations in the targeted gene could lead to rapid viral escape from the siRNA. In the present study, Escherichia coli endoribonuclease III (RNase III) or mammalian Dicer was used to cleave double-stranded RNA into endoribonuclease-prepared siRNA (esiRNA). esiRNAs generate a variety of siRNAs which can efficiently and specifically target multiple sites in the cognate RNA. esiRNAs targeting the region encoding the HIV-1 reverse transcriptase (RT) reduced viral replication by 90%. The inhibition was dose dependent and sequence specific because several irrelevant esiRNAs did not inhibit HIV-1 replication. Importantly, esiRNAs obtained from the prototypic RT sequence of the HXB2 strain and from highly mutated RT sequences showed similar degrees of viral inhibition, suggesting that the heterogeneous population of esiRNAs could overcome individual mismatches in the RT sequence. Finally, esiRNAs generated by Dicer cleavage were five times more potent than those generated by bacterial RNase III digestion. These results show that esiRNAs are potent HIV-1 inhibitors. Moreover, sequence targets do not need to be highly conserved to reach a high level of viral replication inhibition.
Double-stranded RNA (dsRNA) can induce the specific degradation of homologous mRNA species, a process termed RNA interference (RNAi) (14). dsRNAs are processed by the RNase Dicer, a member of the RNase III family of dsRNA-specific endonucleases, into ∼22-nucleotide fragments that bear 2-nucleotide 3′-end overhangs (2, 16, 50). These short interfering RNAs (siRNAs) are the effector molecules of this evolutionarily conserved mechanism. siRNAs are incorporated into the ∼500-kDa RNA-induced silencing complex (RISC) (16, 17, 50). One strand of the siRNA is used to target RISC to homologous mRNAs, which are cleaved and degraded. Transfection of 21-nucleotide siRNAs inhibits the expression of the target gene in a sequence-specific manner (13). siRNAs have become the method of choice for mammalian cell genetics as well as for sequence-specific therapeutic approaches (11, 12, 22, 24, 38, 39, 43). Several studies have reported the use of siRNAs to specifically inhibit human immunodeficiency virus type 1 (HIV-1) replication by targeting viral or cellular genes (4, 8, 9, 20, 29, 30, 33, 34, 36, 37, 40). These results suggest that RNAi represents an important new therapeutic approach for treating HIV-1 infection. However, a major problem of all antiretroviral therapies is the emergence of resistant variants. Recently, we showed that optimal HIV-1 gene silencing by siRNA requires precise complementarity with most of the target sequence and that substitutions at only a few positions at the 5′ and 3′ ends are partially tolerated (40). Not surprisingly, several studies have shown that HIV-1 promptly escapes previously effective siRNAs (4, 9, 46). Recent work with HIV-1 has also shown that tolerance to target sequence mismatches may depend on the sequence of the siRNA tested (30). This fact, coupled with the enormous genomic heterogeneity of HIV-1 quasispecies, may hinder the efficacy of single defined siRNAs. Coexpression of multiple siRNAs that target conserved RNA sequences could reduce the emergence of single-siRNA-resistant viruses, with an effect comparable to that achieved by three- or four-anti-HIV-drug combinations commonly known as highly active antiretroviral treatment. Recently, the use of multiple short hairpin RNAs (shRNAs) against HIV-1 has been shown to delay virus escape (45). Similarly, work with poliovirus has shown that targeting multiple viral sequences with a pool of siRNAs overcomes resistance mechanisms to RNAi and prevents viral escape (15).
In the present study, a mixed population of endoribonuclease-prepared siRNAs (esiRNAs) was generated to inhibit HIV-1 replication. esiRNAs produce a variety of siRNAs, which are able to efficiently and specifically silence target RNA (21, 25, 26, 28, 35, 44, 48, 49, 51). Escherichia coli RNase III or mammalian Dicer can efficiently digest dsRNA into short pieces with the same end structures as siRNAs (1, 50). Our data show that esiRNAs targeting the region encoding the HIV-1 reverse transcriptase (RT) may be a valid option for inhibiting viral replication and overcoming resistance to siRNAs.
MATERIALS AND METHODS
Generation of the esiRNA libraries.
DNA for in vitro transcription was generated by PCR using two oligonucleotides with the T7 promoter appended to the 5′ ends. The T7 promoter-containing PCR primers were used either in separate PCRs or in a single PCR to generate transcription templates for both strands of the dsRNA. The oligonucleotides for the amplification of the HIV-1 strain HXB2 plasmid DNA (AIDS Research and Reference Reagent Program, NIH, Bethesda, MD) were T7RT19B (sense) (5′-GCGTAATACGACTCACTATAGGGAGAGGACATAAAGCTATAGGTACAG-3′, HXB2 residues 2453 to 2475) and T7RT31486 (antisense) (5′-GCGTAATACGACTCACTATAGGGAGAGTTCTATGCTGCCCTATTTCTA-3′, HXB2 residues 3147 to 3127). The oligonucleotides for the amplification of the HIV-2 strain ROD plasmid DNA (Centralised Facility for AIDS Reagents, MRC, United Kingdom) were T7RT19HIV-2 (sense) (5′-GCGTAATACGACTCACTATAGGGAGATAATGACAGGCGACACCCCAA-3′, ROD residues 2306 to 2327) and T7RT3148HIV-2 (antisense) (5′-GCGTAATACGACTCACTATAGGGAGAAGTTCCTTGAGCTGCAGGA-3′, ROD residues 3004 to 2985). (The T7 RNA polymerase sequence is underlined.) For both amplifications, the PCR mixture contained 10 pmol of each oligonucleotide, a 200 μM concentration of each deoxyribonucleoside triphosphate, 2 mM MgSO4, 1× High-Fidelity PCR buffer (Invitrogen), and 0.5 U Taq DNA polymerase (Invitrogen) in a total reaction volume of 50 μl. Cycling parameters were one cycle of denaturation at 94°C for 2 min and then 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 30 s, and extension at 68°C for 30 s. This was followed by a 7-min incubation at 68°C.
DNA for in vitro transcription of samples obtained from HIV-1-infected individuals A1, B, C, D, G, and J was generated by nested RT-PCR. Plasma HIV-1 RNA was purified using the QIAamp viral RNA purification kit (QIAGEN). After viral RNA was isolated, 10 μl of resuspended RNA was PCR amplified using the Superscript one-step RT-PCR kit (Invitrogen). Oligonucleotides for RT-PCR were RT18deg (sense) [5′-GGA(AG)(AG)CC(AGC)A(AG)AAT(AG)ATAG(AG)GGGAATTGG(AG)GG-3′, HXB2 residues 2377 to 2407] and RT20deg (antisense) [5′-C(AT)GCCA(CT)(AG)TCTA(CT)(AG)TCTGCTTC(AG)T(AG)-3′, HXB2 residues 3462 to 3438]. Nested PCR was performed with oligonucleotides T7RT19deg (sense) [5′-GCGTAATACGACTCACTATAGGGAGAGGA(ACT)A(ACT)AA(AG)GCTA(CT)AGGTA-3′, HXB2 residues 2455 to 2472] and T7RT3148deg [5′-GCGTAATACGACTCACTATAGGGAGAATTTTT(CT)(AG)TCTATAG(AG)T(AC)C(AG)CTATT(AT)CTA-3′, HXB2 residues 3155 to 3127]. For the nested PCR, 1× High-Fidelity PCR buffer (Invitrogen) and 0.5 U Platinum Taq DNA polymerase (Invitrogen) were used under the conditions described above.
PCR oligonucleotides for generating the control esiRNA targeting the hepatitis C virus DNA (esiHCV) were T7Prot (sense) (5′-GCGTAATACGACTCACTATAGGGAGACCTATCACGGCCTACTCCCAA-3′, HCV-J reference strain residues 3401 to 3422) and T7Protrev (antisense) (5′-GCGTAATACGACTCACTATAGGGAGATCAAGACCGCATAGTAGTTTCCAT-3′, HCV-J reference strain residues 3954 to 3930). The HCV subgenomic replicon I389 was used as the template DNA (32). PCR oligonucleotides for generating the control esiRNA targeting the firefly luciferase RNA (esiLuc) were T7Luc (sense) (5′-GCGTAATACGACTCACTATAGGGAGCGCCAAAACATAAAGAAAGGC-3′, AB161988 reference sequence residues 9 to 21) and T7REVLuc (antisense) (5′-GCGTAATACGACTCACTATAGGGAGGGAACAACACTTAAAATCGCAGTATG-3′, AB161988 reference sequence residues 700 to 726).
Before in vitro transcription, PCR products were purified by electrophoresis in a 1% agarose gel, employing the QIAquick PCR purification kit (QIAGEN). Purified PCR products (1 to 2 μg) were in vitro transcribed in a final volume of 20 μl using the MEGAscript kit (Ambion, Austin, TX) according to the manufacturer's instructions. After incubation for 2 to 4 h at 37°C, the DNA template was digested with 0.5 U RNase-free DNase I (Ambion) for 15 min at 37°C. Complementary transcripts were annealed at 90°C for 1 min. The resulting dsRNAs were phenol extracted and ethanol precipitated. RNA integrity was determined by electrophoresis in a precast 0.8% agarose gel (Invitrogen). dsRNA was quantified photometrically.
The resulting long dsRNA was digested with bacterial RNase III (Ambion). The reaction conditions were 30 to 50 μg annealed template dsRNA, 1× RNase III buffer (Ambion), and 15 to 25 μl RNase III (Ambion) in a final volume of 50 μl, with incubation for 15 to 30 min at 37°C. Digestions with Dicer were performed using recombinant Dicer (Ambion). Briefly, 30 to 50 μg template dsRNA was digested for 16 to 18 h with 30 μl recombinant Dicer in a 100-μl volume at 37°C. The resulting siRNAs were purified by siRNA purification columns (Ambion), phenol-chloroform extracted, and ethanol precipitated. The pellet was washed with 70% ethanol, and purified siRNAs were resuspended in nuclease-free water (Ambion). RNA integrity was determined by electrophoresis in a precast 4% agarose gel (Invitrogen). siRNAs were quantified by measuring the absorbance at 260 nm.
esiRNA transfection and HIV-1 cotransfection and infection.
Human astroglioma U87-CD4 cells (1.5 × 105 cells) (AIDS Research and Reference Reagent Program, NIH) (10) were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, puromycin (100 μg/ml), and Geneticin (500 μg/ml). To examine silencing of virus production from plasmid templates, infectious clones (HXB2 or pROD) were mock transfected or transfected with different amounts of the appropriate esiRNA by using 2 μl Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. Cells at 70% confluence were transfected in 24-well plates for 4 h, and then the medium was replaced with fresh medium. When appropriate, 0.5 to 1 μg HIV-1 HXB2 infectious plasmid clone (AIDS Research and Reference Reagent Program, NIH) or HIV-2 ROD infectious plasmid clone (Centralised Facility for AIDS Reagents, MRC, United Kingdom) was included in the transfection mixture. The antiviral potency of esiRNAs was also assessed by transient transfection of U87-CD4 cells with esiRNA and infection 24 h later with 200 50% tissue culture infectious doses of wild-type HXB2 as described previously (40).
After HIV cotransfections or infections, the culture supernatants were collected and assayed for p24 antigen production by use of a commercial p24 antigen enzyme-linked immunosorbent assay (Innogenetics). To determine the inhibitory effects of the siRNAs, the level of p24 antigen in the control (mock-transfected) samples was set at 100%, and p24 antigen levels in test samples were expressed as percentages of the control level. siRT199 targeting the RT coding region comprising positions 199 to 227 of the HXB2 virus strain (GACAGTACTAAATGGAGAATT, HXB2 residues 2748 to 2766) (40) was also transfected using 2 μl Lipofectamine 2000 reagent (Invitrogen).
RESULTS
Escherichia coli RNase III-generated esiRNAs targeting the HIV-1 RT efficiently inhibit viral replication.
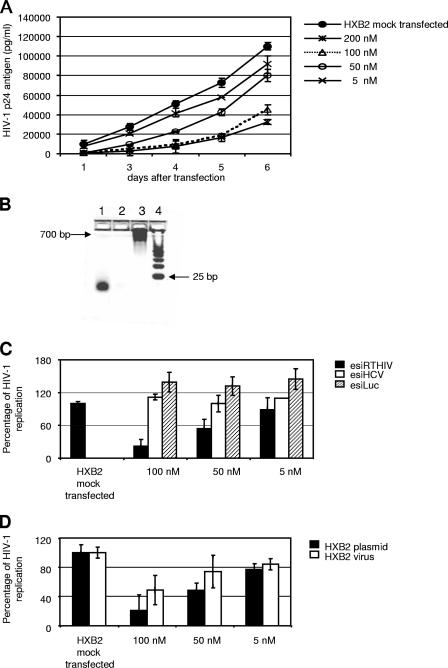
To test whether enzymatically prepared siRNAs have antiviral activity, we generated esiRNAs by bacterial RNase III digestion of a 0.7-kb HIV-1 RT dsRNA (esiRTHIV) (Fig. 1). The antiviral potency of esiRTHIV was assessed in U87-CD4 cells by cotransfecting an HIV-1 HXB2 infectious plasmid clone and different concentrations of esiRTHIV. As shown in Fig. 1A, HIV-1 replication was markedly inhibited by transfection of an esiRNA directed against the RT coding region. In particular, a 100 nM concentration of esiRTHIV suppressed viral replication at day 5 by nearly 90% compared with the control, demonstrating effective silencing activity. Cotransfection of different amounts of esiRTHIV demonstrated that the inhibition was dose dependent (Fig. 1A and C). The same inhibitory effect was obtained with an independently prepared batch of esiRNA (data not shown). As a control for specificity of inhibition, the antiviral activity of unrelated esiRNAs, namely, esiLuc and esiHCV, was assessed. Cells treated with either esiLuc or esiHCV did not exhibit an inhibitory effect on HIV-1 replication (Fig. 1C). These results demonstrated that HIV-1 replication can be efficiently inhibited in a dose-dependent manner by transfection with esiRTHIV and that this inhibition is sequence specific.
FIG. 1.
E. coli RNase III-generated esiRNAs targeting the HIV-1 RT coding region efficiently inhibit viral replication. (A) U87-CD4 cells were mock transfected or transfected with different concentrations (200, 100, 50, or 5 nM) of the esiRTHIV preparation. Cells were cotransfected with an HXB2 infectious clone. At days 1, 2, 3, 4, 5, and 6 after transfection, viral replication was monitored by determining the p24 antigen level in the culture supernatants. (B) Four percent agarose gel for determining esiRNA integrity. Line 1, 250 ng of of RNase III-digested esiRTHIV (HXB2); line 2, RNase III digestion reaction in the absence of dsRNA; line 3, 250 ng of undigested HIV-1 RT dsRNA (HXB2), line 4, size marker, 25-nucleotide DNA ladder. (C) U87-CD4 cells were mock transfected or transfected with different concentrations (100, 50, or 5 nM) of the esiRTHIV, esiHCV, and esiLuc preparations and the HXB2 infectious clone. Five days posttransfection, the supernatants of infected cells were assayed for p24 antigen levels. (D) U87-CD4 cells were mock transfected or transfected with different concentrations (100, 50, or 5 nM) of esiRTHIV. Cells were cotransfected with an HXB2 infectious clone or, alternatively, were infected 24 h after transfection with 200 50% tissue culture infectious doses of wild-type HIV-1 HXB2. Five days posttransfection or postinfection, the supernatants of infected cells were assayed for p24 antigen production. Values represent the means ± standard deviations from at least three independent experiments.
esiRTHIV was analyzed for its ability to inhibit infectious HIV-1. To this end, U87-CD4 cells were transfected with the concentration of esiRTHIV indicated below (Fig. 1D) and then infected with the wild-type HIV-1 HXB2 strain 24 h after transfection. As shown in Fig. 1D, these results paralleled those obtained with the cotransfected infectious viral plasmid. However, viral replication inhibition was lower than that obtained in cells cotransfected with the infectious HIV-1 clone. Recent findings indicate that the incoming HIV-1 RNA genome is not targeted by RNAi (47). Overall, these results demonstrated efficient inhibition of HIV-1 replication with esiRTHIV preparations.
esiRNAs generated from highly mutated RTs inhibit wild-type HIV-1 replication.
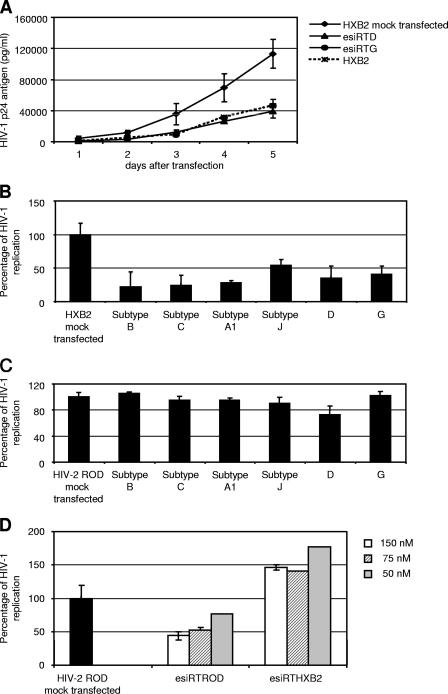
Our previous finding that a single mutation in the viral genome can facilitate escape from siRNA control prompted us to test whether the heterogeneous population of esiRNAs could efficiently inhibit the replication of viruses with mismatches in the target region. In other words, we questioned whether an esiRNA might overcome the limitations imposed by mutations in a region susceptible to nucleotide variation during antiretroviral therapy. To this end, the potency of esiRNA preparations generated from two subtype B viruses carrying several drug resistance-encoding substitutions within the RT target sequence was evaluated (Fig. 2A). These two viruses, D and G (7), differ by 3.5% and 5%, respectively, from the wild-type HXB2 RT target nucleotide sequence. Similar to the results described above, these two esiRNA preparations, esiRTHIVD and esiRTHIVG, efficiently inhibited the replication of the wild-type HXB2 isolate (Fig. 2A). This finding indicates that some nucleotide variation within a population of siRNA sequences obtained by digestion of dsRNA does not measurably affect the antiviral activity exerted by the esiRNA preparation. To test this hypothesis further, three additional esiRNAs were generated from different HIV-1 group M subtypes (A1, C, and J) (Fig. 2B) and tested against the wild-type HXB2 strain. The RT coding region of these three esiRNA preparations differed 10 to 12% in nucleotide sequence from the HXB2 strain. Notably, the replication of the wild-type HXB2 isolate could be inhibited by each of these three esiRNAs preparations (Fig. 2B). Moreover, the inhibition was sequence specific because HIV-2 ROD, which shares only 50% nucleotide identity with HIV-1 HXB2 in the targeted RT region, was not affected by any of the five esiRNA preparations tested (Fig. 2C). Instead, HIV-2 ROD replication was efficiently inhibited in a dose-dependent manner by its cognate esiRT HIV-2 preparation (Fig. 2D). These findings show that treatment with esiRNAs efficiently inhibits HIV-1 replication and that the heterogeneous population of esiRNAs circumvented potential mismatches within the target sequence.
FIG. 2.
esiRNAs generated from highly mutated RTs inhibit wild-type HIV-1 replication. (A) U87-CD4 cells were mock transfected or transfected with a 100 nM concentration of the esiRTHIV, esiRTHIVD, or esiRTHIVG preparation. Cells were cotransfected with an HXB2 infectious clone. At days 1, 2, 3, 4, and 5 after transfection, viral replication was monitored by determining the p24 antigen level in the culture supernatant. (B) U87-CD4 cells were mock transfected or transfected with 100 nM esiRTHIV subtype B, siRTHIV subtype C, siRTHIV subtype A1, siRTHIV subtype J, esiRTHIVD, or esiRTHIVG preparations. Cells were cotransfected with an HXB2 infectious clone. Five days posttransfection, the supernatants of transfected cells were analyzed for p24 antigen production. (C) U87-CD4 cells were mock transfected or transfected with 100 nM esiRTHIV subtype B, siRTHIV subtype C, siRTHIV subtype A1, siRTHIV subtype J, esiRTHIVD, or esiRTHIVG preparations. Cells were cotransfected with an HIV-2 (ROD strain) infectious clone. Five days posttransfection, the supernatants of infected cells were assayed for p24 antigen. (D) U87-CD4 cells were mock transfected or transfected with different concentrations (150, 75, or 50 nM) of esiRTROD or esiRTHXB2 preparation and the ROD or HXB2 infectious clone. Values represent the means ± standard deviations from at least three independent experiments.
esiRNAs generated by Dicer more efficiently inhibit HIV-1 replication.
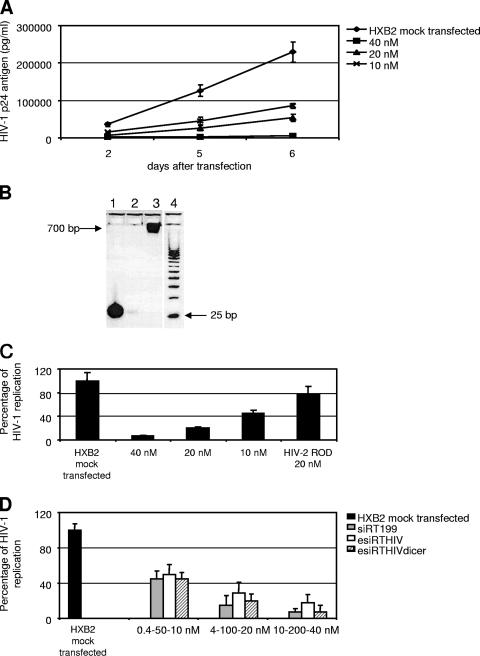
To establish an alternative method for generating esiRNA libraries, we explored the ability of the enzyme Dicer to substitute for the bacterial RNase III (Fig. 3). To our knowledge, there is no information regarding the efficacy of esiRNA preparations generated by Dicer compared with those obtained with bacterial RNase III. It was shown previously that recombinant Dicer efficiently converts large dsRNAs to siRNAs suitable for gene silencing (21, 35). These findings prompted us to use Dicer to generate esiRNA libraries from the same target HIV-1 RT sequence employed in the bacterial RNase III experiments. As expected, esiRTHIV preparations generated by Dicer (esiRTHIVdicer) elicited an effective silencing activity and were able to suppress HIV-1 replication (Fig. 3A). Importantly, lower concentrations of esiRTHIVdicer were able to reach the same level of silencing activity obtained with higher concentrations of esiRTHIV. Again, the observed silencing activity was specific because an esiRTHIV-2dicer preparation could not suppress HIV-1 replication (Fig. 3C). A comparison of the silencing efficiencies of esiRTHIV, esiRTHIVdicer, and a previously described siRNA (40) that effectively targets the HIV-1 RT region demonstrated that the esiRTHIVdicer preparation was five times more potent than esiRTHIV (Fig. 3D). Nevertheless, higher concentrations of esiRTHIV were necessary to reach the level of inhibition obtained with an effective siRNA (Fig. 3D). These results indicate the usefulness of Dicer-generated esiRNA preparations for inhibiting HIV-1 replication.
FIG. 3.
esiRNAs generated by Dicer more efficiently inhibit HIV-1 replication. (A) U87-CD4 cells were mock transfected or transfected with different concentrations (40, 20, or 10 nM) of the esiRTHIVdicer preparation. Cells were cotransfected with an HXB2 infectious clone. At days 2, 5, and 6 after transfection, viral replication was monitored by determining the p24 antigen level in the culture supernatants. (B) Four percent agarose gel for determining esiRNA integrity. Line 1, 250 ng of Dicer-digested esiRTHIV (HXB2); line 2, RNase III digestion reaction in the absence of dsRNA; line 3, 250 ng of undigested HIV-1 RT dsRNA (HXB2); line 4, size marker, 25-nucleotide DNA ladder. (C) U87-CD4 cells were mock transfected or transfected with different concentrations (40, 20, or 10 nM) of esiRTHIVdicer. Cells were cotransfected with an HXB2 infectious clone. At 5 days after transfection, the supernatants of infected cells were assayed for p24 antigen. In addition, 20 nM esiRTHIVdicer was cotransfected with an HIV-2 (ROD) infectious clone. (D) U87-CD4 cells were mock transfected or transfected with different concentrations of siRT199 (0.4, 4, or 10 nM), esiRTHIV (50, 100, or 200 nM), or esiRTHIVdicer (10, 20, or 40 nM). Cells were cotransfected with an HXB2 infectious clone. Five days posttransfection, the supernatants of infected cells were assayed for p24 antigen. Values represent the means ± standard deviations from at least three independent experiments.
DISCUSSION
With the development of new drugs and treatment strategies, therapeutic options for HIV continue to expand. RNAi provides a robust method for specifically inhibiting the expression of targeted cellular or viral genes, and it shows promise as a novel and broadly applicable approach to antiviral therapy. However, clinical applications of RNAi face several challenges, most notably the potential for viral escape (4, 9, 40). The present study investigated the silencing effects of esiRNAs targeted to a 0.7-kb genomic region encoding the HIV-1 RT. Taken together, our results show that treatment with esiRNAs provides an efficient approach for specific inhibition of HIV-1 replication.
Several studies have demonstrated the efficacy of specific siRNAs or shRNAs in inhibiting HIV-1 replication (4, 8, 9, 20, 27, 29, 30, 33, 34, 36, 37, 40, 45). However, some advantages of esiRNAs over siRNAs or shRNAs can be drawn from the present study. First, it seems extremely unlikely that a viral genome could accumulate enough point mutations to escape the antiviral activity of esiRNAs. Indeed, our results demonstrate that esiRNAs differing by >10% in the nucleotide target sequence can be used to efficiently block viral replication. Interestingly, it was noted recently that esiRNAs targeting a 1-kb sequence in the poliovirus RNA genome can prevent the generation of escape mutants (15). The results obtained here with HIV-2, which shares only 50% nucleotide identity with HIV-1 in the target sequence, suggest that resistance may be attained by variants approaching 50% nucleotide nonidentity in the target sequence. Second, the use of pooled siRNAs that target the same transcript can reduce off-target effects while maintaining efficient silencing of the specific target gene (6). Because the concentration of each siRNA in the mixture is relatively low and each siRNA has the same target but different off-targets, the use of pooled siRNAs may minimize the effects on unintended targets. Off-target effects for individual siRNAs have challenged the reliability of RNAi data (3, 5, 18, 19, 31, 41). Third, esiRNA is an efficient, specific, and adaptable tool that can be used to study different aspects of virus biology. As suggested previously (15), esiRNA may be used even when the exact sequence of the viral genome is unknown because the approach requires only specific PCR oligonucleotides for the amplification of the desired target sequence.
An interesting finding of the present study is the efficacy of esiRNA preparations generated by Dicer. Recently, siRNAs 25 to 30 nucleotides in length were found to be 100-fold more potent than the corresponding conventional 21-mer siRNAs (23, 42). The enhanced potency of the longer duplexes is attributed to the fact that they are substrates of the Dicer endonuclease, directly linking the production of siRNAs to incorporation into RISC. Similarly, the present results indicate that siRNAs generated by Dicer from long dsRNA can have enhanced efficacy for RNAi. As we have previously suggested, it may be interesting to test whether a significant increase in the potency of siRNAs weakens the ability of HIV-1 to escape RNAi inhibition (40). In conclusion, these findings show that esiRNAs are potent viral inhibitors and that sequence targets do not need to be highly conserved among different viral strains to reach high levels of viral replication inhibition.
Acknowledgments
This work was supported by the Spanish Minsiterio de Educación y Ciencia (MEC) project BFU2006-01066/BMC, Fondo de Investigación Sanitaria (FIS) project PI070098, and Fundación para la investigación y la prevención del SIDA en España (FIPSE 36549/06).
Footnotes
Published ahead of print on 25 July 2007.
REFERENCES
- 1.Amarasinghe, A. K., I. Calin-Jageman, A. Harmouch, W. Sun, and A. W. Nicholson. 2001. Escherichia coli ribonuclease III: affinity purification of hexahistidine-tagged enzyme and assays for substrate binding and cleavage. Methods Enzymol. 342:143-158. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 3.Birmingham, A., E. M. Anderson, A. Reynolds, D. Ilsley-Tyree, D. Leake, Y. Fedorov, S. Baskerville, E. Maksimova, K. Robinson, J. Karpilow, W. S. Marshall, and A. Khvorova. 2006. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3:199-204. [DOI] [PubMed] [Google Scholar]
- 4.Boden, D., O. Pusch, F. Lee, L. Tucker, and B. Ramratnam. 2003. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 77:11531-11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bridge, A. J., S. Pebernard, A. Ducraux, A. L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells Nat. Genet. 34:263-264. [DOI] [PubMed] [Google Scholar]
- 6.Buchholz, F., R. Kittler, M. Slabicki, and M. Theis. 2006. Enzymatically prepared RNAi libraries. Nat. Methods 3:696-700. [DOI] [PubMed] [Google Scholar]
- 7.Cabana, M., B. Clotet, and M. A. Martinez. 1999. Emergence and genetic evolution of HIV-1 variants with mutations conferring resistance to multiple reverse transcriptase and protease inhibitors. J. Med. Virol. 59:480-490. [DOI] [PubMed] [Google Scholar]
- 8.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, A. T., T. R. Brummelkamp, E. M. Westerhout, M. Vink, M. Madiredjo, R. Bernards, and B. Berkhout. 2004. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 78:2601-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 11.Dorsett, Y., and T. Tuschl. 2004. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 3:318-329. [DOI] [PubMed] [Google Scholar]
- 12.Dykxhoorn, D. M., and J. Lieberman. 2005. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annu. Rev. Med. 56:401-423. [DOI] [PubMed] [Google Scholar]
- 13.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 14.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 15.Gitlin, L., J. K. Stone, and R. Andino. 2005. Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches. J. Virol. 79:1027-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 17.Hammond, S. M., S. Boettcher, A. A. Caudy, R. Kobayashi, and G. J. Hannon. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146-1150. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, A. L., S. R. Bartz, J. Schelter, S. V. Kobayashi, J. Burchard, M. Mao, B. Li, G. Cavet, and P. S. Linsley. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21:635-637. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, A. L., J. Burchard, J. Schelter, B. N. Chau, M. Cleary, L. Lim, and P. S. Linsley. 2006. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12:1179-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawasaki, H., E. Suyama, M. Iyo, and K. Taira. 2003. siRNAs generated by recombinant human Dicer induce specific and significant but target site-independent gene silencing in human cells. Nucleic Acids Res. 31:981-987. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Ketzinel-Gilad, M., Y. Shaul, and E. Galun. 2006. RNA interference for antiviral therapy. J. Gene Med. 8:933-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, D. H., M. A. Behlke, S. D. Rose, M. S. Chang, S. Choi, and J. J. Rossi. 2005. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 23:222-226. [DOI] [PubMed] [Google Scholar]
- 24.Kim, D. H., and J. J. Rossi. 2007. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 8:173-184. [DOI] [PubMed] [Google Scholar]
- 25.Kittler, R., G. Putz, L. Pelletier, I. Poser, A. K. Heninger, D. Drechsel, S. Fischer, I. Konstantinova, B. Habermann, H. Grabner, M. L. Yaspo, H. Himmelbauer, B. Korn, K. Neugebauer, M. T. Pisabarro, and F. Buchholz. 2004. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 432:1036-1040. [DOI] [PubMed] [Google Scholar]
- 26.Kittler, R., V. Surendranath, A. K. Heninger, M. Slabicki, M. Theis, G. Putz, K. Franke, A. Caldarelli, H. Grabner, K. Kozak, J. Wagner, E. Rees, B. Korn, C. Frenzel, C. Sachse, B. Sonnichsen, J. Guo, J. Schelter, J. Burchard, P. S. Linsley, A. L. Jackson, B. Habermann, and F. Buchholz. 2007. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat. Methods 4:337-344. [DOI] [PubMed] [Google Scholar]
- 27.Konstantinova, P., W. de Vries, J. Haasnoot, O. ter Brake, P. de Haan, and B. Berkhout. 2006. Inhibition of human immunodeficiency virus type 1 by RNA interference using long-hairpin RNA. Gene Ther. 13:1403-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kronke, J., R. Kittler, F. Buchholz, M. P. Windisch, T. Pietschmann, R. Bartenschlager, and M. Frese. 2004. Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs. J. Virol. 78:3436-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 30.Lee, S. K., D. M. Dykxhoorn, P. Kumar, S. Ranjbar, E. Song, L. E. Maliszewski, V. Francois-Bongarcon, A. Goldfeld, M. N. Swamy, J. Lieberman, and P. Shankar. 14 April 2005. Lentiviral delivery of short hairpin RNAs protects CD4 T cells from multiple clades and primary isolates of HIV. Blood [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 31.Lin, X., X. Ruan, M. G. Anderson, J. A. McDowell, P. E. Kroeger, S. W. Fesik, and Y. Shen. 2005. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 33:4527-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 33.Martinez, M. A., B. Clotet, and J. A. Este. 2002. RNA interference of HIV replication. Trends Immunol. 23:559-561. [DOI] [PubMed] [Google Scholar]
- 34.Martinez, M. A., A. Gutierrez, M. Armand-Ugon, J. Blanco, M. Parera, J. Gomez, B. Clotet, and J. A. Este. 2002. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. AIDS 16:2385-2390. [DOI] [PubMed] [Google Scholar]
- 35.Myers, J. W., J. T. Jones, T. Meyer, and J. E. Ferrell, Jr. 2003. Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat. Biotechnol. 21:324-328. [DOI] [PubMed] [Google Scholar]
- 36.Nishitsuji, H., M. Kohara, M. Kannagi, and T. Masuda. 2006. Effective suppression of human immunodeficiency virus type 1 through a combination of short- or long-hairpin RNAs targeting essential sequences for retroviral integration. J. Virol. 80:7658-7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 38.Putral, L. N., W. Gu, and N. A. McMillan. 2006. RNA interference for the treatment of cancer. Drug News Perspect. 19:317-324. [DOI] [PubMed] [Google Scholar]
- 39.Rossi, J. J. 2006. RNAi as a treatment for HIV-1 infection. BioTechniques April 2006(Suppl.):25-29. [DOI] [PubMed]
- 40.Sabariegos, R., M. Gimenez-Barcons, N. Tapia, B. Clotet, and M. A. Martinez. 2006. Sequence homology required by human immunodeficiency virus type 1 to escape from short interfering RNAs. J. Virol. 80:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scacheri, P. C., O. Rozenblatt-Rosen, N. J. Caplen, T. G. Wolfsberg, L. Umayam, J. C. Lee, C. M. Hughes, K. S. Shanmugam, A. Bhattacharjee, M. Meyerson, and F. S. Collins. 2004. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells Proc. Natl. Acad. Sci. USA 101:1892-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siolas, D., C. Lerner, J. Burchard, W. Ge, P. S. Linsley, P. J. Paddison, G. J. Hannon, and M. A. Cleary. 2005. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 23:227-231. [DOI] [PubMed] [Google Scholar]
- 43.Stevenson, M. 2004. Therapeutic potential of RNA interference. N. Engl. J. Med. 351:1772-1777. [DOI] [PubMed] [Google Scholar]
- 44.Tan, C., B. Xuan, J. Hong, Z. Dai, R. Hao, Z. Li, and W. Huang. 2007. RNA interference against hepatitis B virus with endoribonuclease-prepared siRNA despite of the target sequence variations. Virus Res. 126:172-178. [DOI] [PubMed] [Google Scholar]
- 45.ter Brake, O., P. Konstantinova, M. Ceylan, and B. Berkhout. 2006. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol. Ther. 14:883-892. [DOI] [PubMed] [Google Scholar]
- 46.Westerhout, E. M., M. Ooms, M. Vink, A. T. Das, and B. Berkhout. 2005. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 33:796-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westerhout, E. M., O. ter Brake, and B. Berkhout. 2006. The virion-associated incoming HIV-1 RNA genome is not targeted by RNA interference. Retrovirology 3:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xuan, B., Z. Qian, J. Hong, and W. Huang. 2006. EsiRNAs inhibit hepatitis B virus replication in mice model more efficiently than synthesized siRNAs. Virus Res. 118:150-155. [DOI] [PubMed] [Google Scholar]
- 49.Yang, D., F. Buchholz, Z. Huang, A. Goga, C. Y. Chen, F. M. Brodsky, and J. M. Bishop. 2002. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA 99:9942-9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]
- 51.Zhu, C., J. Zhao, M. Bibikova, J. D. Leverson, E. Bossy-Wetzel, J. B. Fan, R. T. Abraham, and W. Jiang. 2005. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell 16:3187-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]