Abstract
Mammalian cells infected with human adenoviruses (Ads) undergo an apoptotic response as a result of expression of the viral E1A proteins, and this process is suppressed by the viral E1B-19K protein. The intermediary steps in the Ad-induced apoptosis pathway are not fully resolved. The apical step in the canonical mammalian apoptosis pathway involves functional activation of one or more of the BH3-only BCL-2 family proapoptotic proteins. Previous reports have suggested that Ad-induced apoptosis may be initiated at checkpoints downstream of the BH3-only proteins. Here, we undertook genetic and biochemical studies to determine the roles of BH3-only proteins in Ad-induced apoptosis. We examined the activities of the cellular antiapoptosis protein BCL-xL and its mutants expressed from the E1B region of the Ad5 genome. Our results showed efficient suppression of Ad-induced apoptosis by a BCL-xL mutant (mt1) deficient in interaction with multidomain proapoptotic proteins BAX and BAK but proficient in interaction with BH3-only proteins, suggesting a role for BH3-only proteins in the initiation of Ad-induced apoptosis. Further, the antiapoptotic activity of BCL-xL mt1 in Ad-infected cells was observed in spite of BAK activation as a consequence of MCL-1 degradation. Analysis of the mRNA levels of various BH3-only members by reverse transcription-PCR revealed prominent activation of the Bik gene. Further, the BIK protein was also modified into an apoptotically enhanced phosphorylated form during the viral infection. In addition to BIK, enhanced level of BIM was observed in Ad-infected cells. Between the two major E1A proteins coded by the 12S and 13S mRNAs, the 13S product appeared to contribute to the activation of these BH3-only members and apoptosis during viral infection. Depletion of BIK by the use of small interfering RNA reduced the level of Ad-induced apoptosis. Our results are consistent with a model that activation of the BH3-only members may initiate Ad-induced apoptosis.
Adenovirus (Ad) is a good model system to study the molecular mechanisms involved in cellular processes. During Ad replication, the infected quiescent cells enter into an S-phase-like state resulting in cell proliferation. This cellular environment facilitates the replication of viral DNA. The expression of viral immediate-early gene E1A is essential for the activation of cellular DNA synthesis and for facilitating viral DNA replication. The unscheduled DNA replication induced by E1A (i.e., cellular and viral) appears to contribute to the apoptotic response. It is well established that the apoptotic response induced during Ad infection is apparent in cells infected with viral mutants defective in E1B-19K (19,000-molecular-weight protein) (33, 37, 39, 45). These results have indicated that during normal Ad infection virus-induced apoptosis is suppressed by the E1B-19K protein. The cell death response in cells infected with an Ad recombinant virus that expresses the cellular antiapoptosis protein BCL-2 from the E1B region (i.e., in the absence of E1B-19K) is strongly suppressed (38). Similarly, the cytocidal effect of E1B-19K mutant virus is much reduced in cells that ectopically overexpress BCL-2 (7, 40). Thus, the apoptosis-like cell death induced by E1A can be suppressed by overexpression of the cellular BCL-2 protein.
The intermediate steps in the Ad-induced cell death pathway that lie between the actions of E1A and E1B-19K are not fully understood. In the canonical apoptotic cell death pathway, the various apoptotic stimuli activate functional expression of one or more members of a class of BCL-2 family proapoptotic proteins known as the BH3-only proteins. The BH3-only proteins (such as BIK, BIM, PUMA, etc.) function as the initiators of the apoptotic paradigm and activate a second class of proapoptotic proteins known as BH123 proteins (such as BAX and BAK) by unknown mechanisms. The BH123 proteins cause the cellular demise via mitochondrial dysfunction and ensuing activation of the caspases (reviewed in reference 12). Although E1B-19K efficiently inhibits the manifestation of the terminal apoptotic phenotypes, such as premature cell death and associated fragmentation of chromosomal DNA, it is unclear at what step in the apoptotic paradigm E1B-19K acts. Similarly, it is also unclear what proapoptotic initiators mediate Ad-induced apoptosis.
Several BH3-only members, such as BIM, PUMA, NOXA, and HRK, have been reported to be targets for the transcription factor E2F1 (2, 11, 15). Since E2F1 activation is one of the consequences of E1A expression (28), it is possible that some BH3-only members might be activated during Ad infection through the E2F pathway. However, results consistent with models that suggest initiation of Ad-induced apoptosis downstream of BH3-only proteins have been published. A study by Cuconati et al. has suggested direct functional activation of a BH123 protein, BAK, during Ad infection (10). These authors have observed that Ad infection resulted in a DNA damage response resulting in proteasomal degradation of the antiapoptotic protein MCL-1 and release of BAK from the MCL-1-BAK complex. Since E1B-19K interacts efficiently with BAK in immunoprecipitation studies, it was suggested that E1B-19K suppresses Ad-induced apoptosis primarily by sequestering BAK (10). A different study has reported that the expression of E1A resulted in transcriptional activation of genes coding for different initiator and effector caspases of the caspase cascade (26). It has been shown that transcription of these caspases is activated by the transcription factor E2F1 that is activated in cells transduced with E1A. Here, we report results of our studies designed to identify the critical early steps in Ad-induced apoptosis. Our results provide genetic and biochemical evidence that BH3-only BCL-2 family members induced during viral infection may initiate Ad-induced apoptosis.
MATERIALS AND METHODS
Cells and viruses.
Human A549, 293, HNK, and baby rat kidney cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. To construct Ad vectors that express wild-type (wt) BCL-xL and its mutants (4) under the transcriptional control of the cytomegalovirus (CMV) promoter, the PCR-amplified DNA fragments were cloned between HindIII and BamHI sites of an Ad transfer vector, pLendE1ACMV. This transfer plasmid vector contains Ad5 sequences from nucleotide (nt) 1 to 1710 (E1A region) and nt 3328 to 5788 (3′ end of E1B region) and the CMV promoter (in the 5′ region of E1B, deleting nt 1711 to 3327) (see Fig. 1A). The resultant plasmids were cotransfected with an Ad genomic plasmid pacAd5 9.2-100 (1) onto human 293 cells and incubated with growth medium containing 2% fetal bovine serum until the appearance of visible cytopathic effect. The recombinant viruses were purified by plaque assay. The isolated recombinants were screened for the expression of BCL-xL by Western blot analysis. Positive clones were further purified through a second round of plaque purification, amplified, and titrated on 293 cells. A control E1B-deleted Ad mutant designated Ad5ΔE1B was also constructed by cotransfection of the pLendE1ACMV vector with the adenovirus plasmid pacAd5 9.2-100.
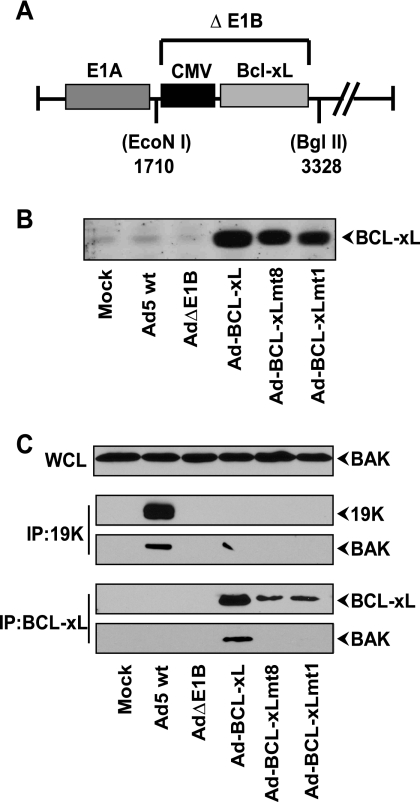
FIG. 1.
Ad-BCL-xL recombinants and protein expression. (A) Map of the E1B region of Ad-BCL-xL recombinant. The portion between nucleotide position 1710 (EcoNI) to 3328 (BglII) in ΔE1B is deleted. The deleted region is substituted with wt or mutant BCL-xL coding sequences under the transcriptional control of the CMV immediate-early promoter. (B) Expression of BCL-xL and mutant proteins. A549 cells were infected with the indicated viruses, and the expression of BCL-xL protein was analyzed by Western blotting. (C) Interaction of BCL-xL and E1B-19K with BAK. The whole-cell lysates (WCL) were either immunoprecipitated (IP) with the 19K antibody or BCL-xL antibody, and the blots were probed with the BAK antibody (Upstate, NY).
Cell death assay.
A549 cells (1 × 106 cells/60-mm dish) were infected with various viruses at 50 PFU/cell. Forty-eight hours after infection, adherent cells were collected by treatment with trypsin and combined with the floating cells collected from the culture medium. The cell suspensions were mixed with equal volumes of trypan blue, and the viable cells that excluded the dye were microscopically counted.
Analysis of DNA fragmentation.
A549 cells (1 × 106) were infected with various viruses at 50 PFU/cell. Twenty-four hours after infection, both adherent and floating cells were collected and lysed, and the low-molecular-weight DNA was prepared by Hirt extraction (38), treated with RNase, and analyzed by electrophoresis on a 1.5% agarose gel. The low-molecular-weight DNA was quantified with the aid of a phosphorimager, and the relative fragmentation was calculated.
Antibodies, immunoprecipitation, and Western blot analyses.
Rabbit polyclonal antibodies against BCL-xL and BFL-1/A1 and goat polyclonal antibody against actin were purchased from Santa Cruz Biotechnology (San Diego, CA). Mouse monoclonal BCL-xL antibody was purchased from Chemicon Int., Temecula, CA. MCL-1 (rabbit polyclonal), caspase-3 (rabbit polyclonal), and caspase-9 (mouse monoclonal) antibodies were purchased from Stressgen (British Columbia, Canada). Rabbit polyclonal antibodies to BAK and BAX were purchased from Upstate Biotechnology (Charlottesville, VA). Mouse monoclonal antibodies specific to BAK and poly(ADP-ribose) polymerase 1 (PARP-1) were purchased from Oncogene, Inc. (La Jolla, CA). The mouse monoclonal BAX antibody (6A7) was purchased from BD Pharmingen, San Jose, CA. The E1B-19K antipeptide antibody (13) was a gift from Maurice Green. A second 19K antibody raised against the same peptide target (prepared in our laboratory) was also used in some experiments. The following antibodies specific for BH3-only proteins were obtained from various commercial sources: rabbit polyclonal BIM (Pharmingen), NOXA, BIK, BID, and BNIP3-L (Santa Cruz Biotechnology), UREB1/LASU1/MULE (MCL1 ubiquitin E3 ligase) (Bethyl Lab), and PUMA (Sigma) antibodies, goat polyclonal HRK (Santa Cruz Biotechnology) antibody, and mouse monoclonal BNIP1 (BD Pharmingen) and BNIP3 (Sigma) antibodies. The monoclonal antibodies for E2F1 and p53 used in chromatin immunoprecipitation (ChIP) assays were purchased from Santa Cruz Biotechnology.
For coimmunoprecipitation studies, human A549 cells in 75-cm2 flasks were infected with various viruses at 50 PFU/cell and harvested after 24 h of infection. The cell pellets were washed with phosphate-buffered saline and suspended in 1.0 ml of lysis buffer (50 mM Tris-Cl, pH 7.6, 150 mM NaCl, 2 mM EDTA, 10% glycerol, and protease inhibitor cocktail) containing 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS). Cell lysis was carried out at 4°C for 2 h using a rotator, and the lysates were diluted 1:3 with a buffer containing 50 mM Tris-Cl, pH 7.6, and 150 mM NaCl. The coimmunoprecipitation studies were carried out as described previously (21) with the indicated antibodies. For Western blot analyses, both the floating and adherent cells were collected 24 h after infection and resuspended in 0.5 ml of sample buffer. Fifty-microliter samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 12% gels. The separated proteins were electrotransferred onto nitrocellulose membranes and probed with a primary antibody followed by horseradish peroxidase-conjugated secondary antibody and analyzed by using a chemiluminescence detection system (Roche Applied Science, Indianapolis, IN) according to the manufacturer's specifications.
RNA isolation and quantitative reverse transcription-PCR (RT-PCR) analysis.
A549 cells were infected with 50 PFU of wt Ad5. Twelve hours after infection, cells were lysed, and mRNA was extracted using Quick Prep micro mRNA purification kit (Amersham Biosciences) according to the manufacturer's instructions. cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (Invitrogen) and oligo(dT) primers (Ambion). Dilutions of the cDNA templates (1:10, 1:50, and 1:250) were amplified by PCR using Taq polymerase (Promega) and primers specific for the various BH3-only genes. PCR products were analyzed on 6% acrylamide gels stained with Vistra Green, and band intensities were imaged and quantified with STROM840 ImageQuant 5.2 software (Molecular Dynamics, Amersham Pharmacia Biotech). The intensity values obtained were normalized to the values obtained for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The gene-specific primers were designed using Primer3 software (35) (http://primer3.sourceforge.net/) so that they would hybridize solely within the coding region of mRNA sequences (GenBank database, NCBI) and would span at least two exons. The absence of an amplicon of the appropriate size from genomic DNA was verified for all primers.
ChIP.
The ChIP assay was performed using a commercial kit (EZ ChIP kit; Upstate) with modifications. A549 cells were either mock infected or infected with adenovirus type 5 (Ad5) in 100-mm dishes. Twenty-four hours after infection, cells were fixed with 1% formaldehyde, collected, and suspended in immunoprecipitation (IP) buffer (50 mM Tris, pH 8.0, 0.1% SDS, and protease inhibitor cocktail) containing 2 mM EDTA (IP-EDTA). After sonication, chromatin was prepared and precleared with protein G-agarose beads. The chromatin preparations were immunoprecipitated with antibodies specific to p53 or E2F1 overnight at 4°C. The immunoprecipitates were washed twice with the IP-EDTA buffer, once with half-strength IP-EDTA buffer with 0.25 M LiCl, and twice with Tris-EDTA buffer. The DNA was recovered from the bound protein-DNA complexes after reversal of the cross-link. The DNA recovered was amplified by PCR using BIK-specific primers (forward primer, −236 to −211, and reverse primer, −86 to −109) (41). PCR products were electrophoresed on a 1.5% agarose gel and photographed in ChemiDox-XRS (Bio-Rad). Control ChIP analysis of the GAPDH promoter region was performed as recommended (EZ ChIP kit; Upstate).
GST pull-down assay.
Recombinant glutathione S-transferase (GST) and GST fusion proteins were generated using pGEX-3X and pGEX-2T-BCL-xL (3). GST fusion proteins were expressed in Escherichia coli BL21, induced with 1 mM isopropyl-β-d-thiogalactopyranoside, and purified by lysing the bacterial pellet in NTEN buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% NP-40 in the presence of one Complete mini, protease inhibitor cocktail tablet [Roche]/10 ml buffer). The fusion proteins were purified using glutathione-agarose beads (Sigma) as described previously (3) and quantified by SDS-PAGE and staining with Sypro Orange (Molecular Probes) using bovine serum albumin as the standard. One microgram of GST or GST-BCL-xL beads was incubated with 100 μg of total protein from A549 mock-infected or wt Ad5-infected cell lysate for 1 h at room temperature. After extensive washing, the interacting proteins were eluted from the beads by boiling in SDS sample buffer for 5 min and analyzed by 15% SDS-PAGE. To determine the phosphorylation status of BIK, the cell lysates and GST-BCL-xL pull-down beads were treated with lambda protein phosphatase in 50 mM Tris-HCl buffer, pH 7.5, containing 2 mM dithiothreitol, 0.1 mM EGTA, 100 mM NaCl, and 0.01% Brij 35. The beads were incubated with lambda protein phosphatase (NEB) at a concentration of 400 units/80 μg of protein and incubated at 30°C for 20 min. Proteins were eluted from the complex by boiling in SDS sample buffer and analyzed by Western blotting.
RESULTS
Effects of BCL-xL mutants on Ad-induced cell death.
The available experimental evidence suggests that Ad-induced apoptosis might be initiated at two different apoptosis checkpoints downstream of BH3-only proteins: one at the level of BAK activation (10) and the second at the level of transcriptional activation of caspases (26). We sought to determine whether Ad-induced apoptosis might be mechanistically similar to the canonical apoptosis paradigm induced by nonviral stimuli, in which the BH3-only proteins are apical activators. For this purpose, first we used a genetic approach. Cheng and colleagues have constructed and characterized a panel of interesting BCL-xL mutants (4). Some of these mutants were defective in interaction with BH123 proteins BAX and BAK but retained the antiapoptosis activity. These mutants have served as valuable reagents to dissect BH3/BH123 apoptotic checkpoints (5). The BCL-xL mutants deficient in interaction with BAX and BAK retain the ability to complex with BH3-only members, such as tBID, BAD, and BIM, and antagonize the proapoptotic activity of these BH3-only proteins (5). We constructed three different Ad5 recombinant viruses in which the E1B region (encompassing the sequences coding for the 19K and 55K proteins) was substituted with an expression cassette that expresses wt or mutant BCL-xL proteins (Fig. 1A). It should be noted that we chose to delete the sequences that code for both E1B proteins, since several studies have shown that such deletions did not influence the apoptotic phenotype compared to deletion of only the 19K-coding region. Among the various BCL-xL mutants that have been characterized by Cheng et al. (4), we chose mt1 and mt8. Of these two mutants, mt1 (F131V and D133A) is proficient for the antiapoptotic activity and deficient in interaction with BAX and BAK, while mt8 (G138E, R139L, and I140N) is defective in the antiapoptotic activity as well as in interaction with BAX and BAK (4). Like wt BCL-xL, mutant mt1 has been shown to complex with ectopically expressed BH3-only proteins and inhibit their proapoptotic activity, while the functionally defective mutant mt8 has been shown to be defective in interaction with both classes of proapoptotic proteins (5).
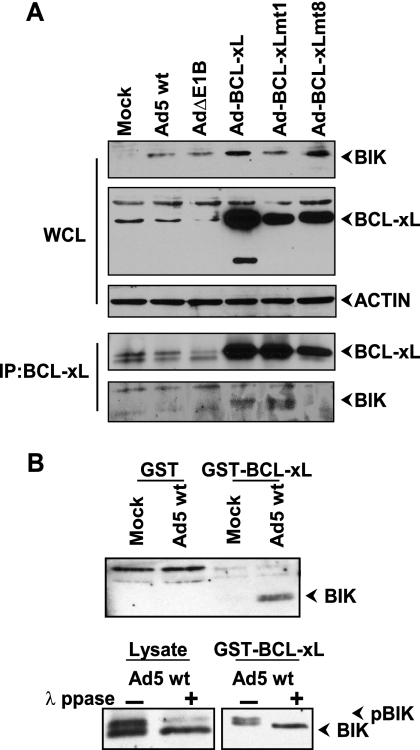
The expression of wt or mutant BCL-xL proteins from the recombinant viral genomes was determined by Western blot analysis of permissive human cells (A549) infected with various BCL-xL-expressing Ad recombinants. Cells infected with Ad-BCL-xL wt or mt1 or mt8 contained increased levels of BCL-xL protein compared to endogenous levels observed in mock-infected cells or in cells infected with wt Ad5 or Ad5ΔE1B (Fig. 1B). To determine the interaction of BH123 proteins with BCL-xL in Ad-infected cells, we carried out coimmunoprecipitation and Western blot analysis. The lysates of infected cells were immunoprecipitated with an antibody specific for BCL-xL and subjected to Western blot analysis. The blots were probed with the antibody specific for the BH123 protein BAK (Fig. 1C). As expected, BAK was coprecipitated with wt BCL-xL and not with mt1 and mt8. Similarly, BAK also readily coprecipitated with E1B-19K from the lysates of cells infected with wt Ad5 and not with the AdΔE1B mutant (lacking E1B-19K). Under the same conditions, there was no detectable coprecipitation of BAX with either BCL-xL or E1B-19K (not shown). The lack of direct interaction between BAX and E1B-19K in Ad-infected cells has previously been documented (9, 21). The reason for the lack of interaction between BCL-xL and BAX in Ad-infected cells is not known.
Effect of BCL-xL on Ad-induced apoptosis.
The effects of wt and mutant BCL-xL proteins on Ad-induced apoptosis were determined in comparison with wt Ad5 and AdΔE1B mutant. For this purpose, A549 cells were infected with various Ads, and the effect on cell viability was determined by trypan blue exclusion assay (Fig. 2A). The viabilities of cells infected with wt Ad-BCL-xL or Ad-BCL-xL mt1 mutant were more or less similar to that of cells infected with wt Ad5. In contrast, the viabilities of cells infected with AdΔE1B or Ad-BCL-xL mt8 were reduced. The extent of cell death observed in cells infected with Ad-BCL-xL mt8 was consistently higher than that of cells infected with AdΔE1B. It is possible that mt8 may have a proapoptotic activity (in addition to loss of the antiapoptotic activity) which might be additive with Ad-induced apoptosis. Then, we determined the effects of wt and mutant BCL-xL proteins on the DNA fragmentation (deg) phenotype conferred by the deletion of Ad E1B (Fig. 2B). The cells infected with AdΔE1B contained higher levels of DNA fragmentation than the cells infected with wt Ad5. Cells infected with Ad-BCL-xL or Ad-BCL-xL mt1 contained low levels of DNA fragmentation. These levels were generally lower than that observed in cells infected with wt Ad5, suggesting that wt and mt1 BCL-xL proteins suppress Ad-induced apoptosis more efficiently than the cognate viral antiapoptotic machinery. We also examined the effects of BCL-xL on certain prototypical signatures of apoptosis, such as processing of the initiator caspase procaspase-9, the effector caspase procaspase-3, and proteolytic cleavage of PARP (Fig. 2C). In cells infected with AdΔE1B, there was a decrease in the levels of procaspase-9 and procaspase-3, suggesting partial activation of caspase-9 and caspase-3. These results suggest that during Ad-induced apoptosis, in addition to caspase-9 and caspase-3, other initiator and effector caspases might be activated. In contrast to cells infected with AdΔE1B, in cells infected with Ad-BCL-xL mt8, strong effects on processing of procaspase-9 and procaspase-3 and cleavage of PARP were observed. The effect of BCL-xL mt8 is in agreement with the more potent apoptotic phenotype of the recombinant virus expressing this mutant. Consistent with the results on cell viability and DNA fragmentation, the expression of wt or mt1 BCL-xL inhibited the activation of caspases and processing of PARP. Thus, our results indicate that Ad-induced apoptosis can be efficiently suppressed by a BCL-xL mutant (mt1) that interacts with BH3-only proteins, but not with BH123 proteins.
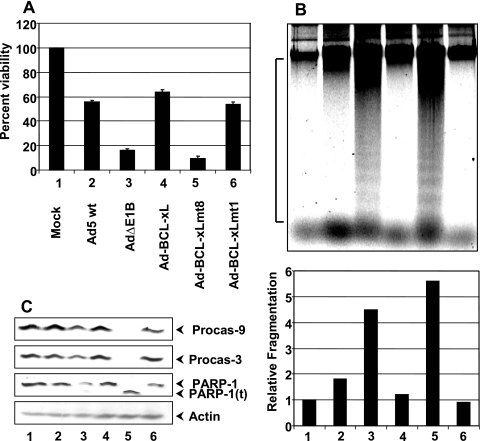
FIG. 2.
Effects of BCL-xL and mutants on Ad-induced cell death. (A) Cell viability assay. A549 cells were infected with the indicated viruses. Forty-eight hours after infection, the trypan blue-excluded live cells were counted microscopically and plotted against percent viability. The experiment was repeated four times in triplicate. (B) DNA fragmentation assay. The low-MW DNA was isolated from infected cells by the Hirt method and analyzed by agarose gel electrophoresis. The designations 1 to 6 represent different viruses as shown in panel A. The bottom panel shows the quantification of DNA fragmentation in relation to mock-infected cells. The area of the image used for quantification by the ImageQuant software is marked on the side of the panel. (C) Western blot analysis of apoptotic markers. The blots were probed with antibodies specific to caspase-9, caspase-3, PARP-1, and actin. Procas-9, procaspase-9; Procas-3, procaspase-3; PARP-1(t), truncated version of PARP-1.
Activation of BAX and BAK.
There is strong evidence that various apoptotic stimuli conformationally activate BAX by exposure of the N-terminal and C-terminal epitopes, resulting in translocation of BAX from the cytosol to mitochondria (16, 17, 48). Similarly, BAK has also been reported to be conformationally activated by exposure of an N-terminal epitope (14). The activation of these BH123 proteins leads to homo- and hetero-oligomerization of BAX and BAK. In Ad-infected cells, BAK has been reported to be activated by the release of free BAK from a complex with the antiapoptosis protein MCL-1 (10). We investigated whether MCL-1 was degraded in cells infected with Ad-BCl-xL or Ad-BCL-xL mt1. Western blot analysis (Fig. 3A) revealed degradation of MCL-1 in cells infected with wt Ad, AdΔE1B, or Ad-BCL-xL recombinants and not in cells infected with the E1A null mutant dl312. In contrast, there was no change in the levels of a different antiapoptosis protein BFL-1/A1. These results suggested that BCL-xL mt1 was able to suppress Ad-induced apoptosis in spite of release of BAK as a consequence of MCL-1 degradation.
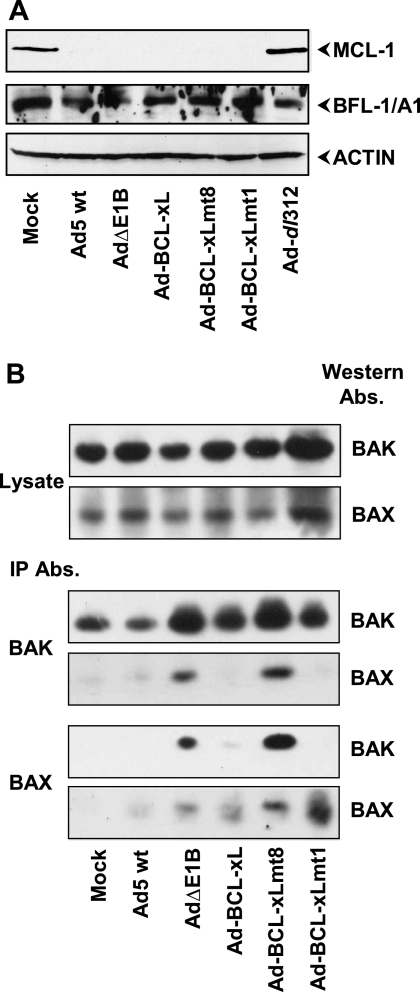
FIG. 3.
Activation of BH123 proteins during Ad-induced cell death. (A) Effects of Ad-BCL-xL recombinants on MCL-1. Lysates of A549 cells infected with the indicated Ads (24 h infection) were analyzed by Western blotting using antibodies specific for MCL-1, BFL-1/A1, or actin. (B) Conformational activation of BAX and BAK. The lysates prepared from Ad-infected A549 cells were immunoprecipitated (IP) with the conformation-specific BAX (6A7; BD Pharmingen) and BAK (AB-1) (AMO3; Oncogene) antibodies (Abs.). The Western blots were probed with the BAX (AB) (06-499; Upstate) and BAK (AB) (06-536; Upstate) antibodies.
We also investigated the conformational activation of BAX and BAK in cells infected with Ad-BCL-xL recombinants by immunoprecipitation and Western blot analysis (Fig. 3B). There were no significant differences in the levels of BAX and BAK between cells infected with various Ad mutants, when the blots were probed with the BAX or BAK antibodies that are not conformer specific (Fig. 3B, top two blots). When BAK was immunoprecipitated with the conformer-specific antibody, significant levels of BAK was immunoprecipitated in mock-infected cells as well as in cells infected with various Ad mutants (Fig. 3B, third blot from the top). However, relatively enhanced levels of BAK were immunoprecipitated from cells infected with AdΔE1B and Ad-BCL-xL mt8. It should be noted that cells infected with these two viruses are under apoptotic stress. These results suggest that a significant amount of BAK is present as an active conformer (i.e., with the N-terminal region exposed) in healthy mock-infected cells. However, in cells experiencing apoptotic stress, the level of BAK conformer was increased. When the immunoprecipitates obtained with the BAK antibody were probed for BAX, BAX was detected in extracts from cells infected with AdΔE1B and Ad-BCL-xL mt8. Similarly, when the extracts were immunoprecipitated with the conformer-specific BAX antibody (blot at the bottom of Fig. 3B) and probed for BAK (second blot from the bottom), BAK was detected only in cells infected with the proapoptotic mutants (AdΔE1B and Ad-BCL-xL mt8). These results suggest that the conformational activation of BH123 proteins and oligomerization was suppressed in cells expressing E1B-19K, wt BCL-xL and BCL-xL mt1. Thus, BCL-xL mt1 appears to prevent activation of BH123 proteins without direct interaction with them.
Activation of BH3-only members in Ad-infected cells.
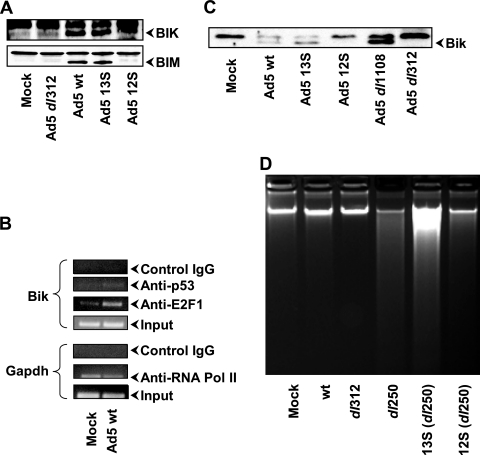
The suppression of Ad-induced apoptosis by BCL-xL mt1 suggests that some BH3-only members (that could be sequestered by it) might be important for the initiation of Ad-induced apoptosis. We then sought to determine whether any known BH3-only members are activated during Ad infection. First, we carried out RT-PCR analysis using primers specific for 15 different BH3-only members (Fig. 4A). These studies revealed that the transcript for BIK was present at higher levels in Ad-infected cells (about eightfold) than in mock-infected cells (Fig. 4B). Comparable results on BIK expression were also observed in HNK cells infected with Ad2 or a 19K mutant (dl250) (data not shown). Moderate enhancement in the levels of the transcripts for BIM and HRK (about three- to fourfold) was also observed in A549 cells.
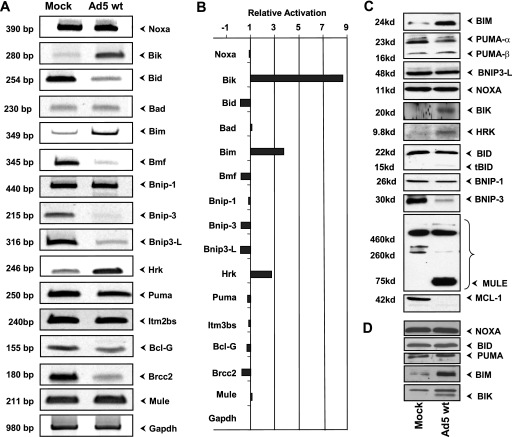
FIG. 4.
Effect of Ad infection on the expression of BH3-only family members. (A) RT-PCR analysis of various BH3-only members in A549 cells. The analysis was carried out using multiple dilutions of the first strand of the cDNAs, and representative images of the cDNA fragments were stained with Vistra Green. (B) Quantification of RT-PCR analysis. The relative intensities of the bands in panel A were quantified by using ImageQuant software. (C) Western blots of BH3-only proteins in mock-infected and wt Ad5-infected A549 cells. The effect on MCL-1 (BH1234) is shown in the bottom blot. (D) Western blots of BH3-only proteins in mock-infected and wt Ad5-infected HNK cells.
We then determined the expression of several BH3-only proteins by Western blot analysis (Fig. 4C). The expression of BIK, BIM, and HRK proteins was elevated. However, the extent of stable BIK protein expression was somewhat less than that of the mRNA levels. Part of the explanation for this might be that BIK is unstable. Previous reports have suggested that BIK protein levels were regulated by the proteasome pathways (23, 29). Among other BH3-only proteins examined, there was no remarkable difference between mock-infected and Ad-infected cells. The level of hypoxia-induced BH3-only member BNIP3 was generally lower in Ad-infected cells. Although there was no significant difference in the level of transcript of MULE (MCL1 ubiquitin E3 ligase) (43, 51), the protein blots probed with a MULE polyclonal antibody revealed a strong lower-molecular-weight [MW] band. The significance of the lower-MW band is not known. However, it correlated with the disappearance of MCL-1 in Ad-infected cells (bottom blot in Fig. 4C). Western blot analysis of Ad5-infected HNK cells also revealed enhanced expression of BIK and BIM (Fig. 4D).
Activation of BIK by E1A 13S product.
After establishing that BIK was the major BH3-only member activated during Ad infection in human cells, we then determined which of the two major E1A protein isoforms contributed to BIK activation. We carried out a Western blot analysis of A549 cells infected with Ad mutants individually expressing either the 12S or 13S mRNA product. Surprisingly, BIK was activated in cells infected with wt Ad5 and in cells infected with Ad5 13S (Fig. 5A). A similar pattern was also observed for BIM. A modest enhancement in BIK expression was also observed in cells infected with Ad5 12S. These results suggest that E1A 13S may be more efficient in activation of BIK and BIM than E1A 12S is. Then, we determined whether enhanced expression of BIK was mediated by p53 and/or E2F1 (two major cellular transcription factors activated by E1A) by ChIP assays (Fig. 5B). We found that the BIK promoter was occupied by E2F1. In contrast, the level of occupancy by p53 was low. Using the available E1A antibodies, we were unable to detect E1A at the BIK promoter. These results suggest that the cellular transcription factor E2F1 may mediate transcriptional activation of BIK as a consequence of E1A expression.
FIG. 5.
(A) Effect of E1A protein isoforms on BIK and BIM expression. A549 cells were infected with wt Ad5 or the indicated mutants, and expression of BIK and BIM was determined by Western blot analysis. (B) ChIP analysis of transcription factors associated with the Bik promoter. The results of ChIP analysis of E2F1 and p53 are shown. Immunoprecipitation with the antibody specific to RNA polymerase II (Pol II) and the PCR primers specific to the GAPDH promoter (Upstate) were used in the control ChIP assays. IgG, immunoglobulin G. (C) CR2-independent activation of BIK. BIK expression in A549 cells infected with the indicated Ad mutants was determined by Western blot analysis. (D) Roles of E1A protein isoforms on induction of apoptosis. A549 cells infected with the indicated viruses were maintained under 20 μg/ml of cytosine arabinoside for 36 h, and the low-MW intracellular DNA was analyzed as described in the legend to Fig. 2.
Since E2F1 activation by E1A is generally believed to be mediated by the CR2 region of E1A (28), we investigated the effect of an Ad CR2 (genomic E1A) deletion mutant dl1108 (18) on BIK expression. Western blot analysis (Fig. 5C) indicated efficient activation of BIK in A549 cells infected with dl1108. These results suggest that BIK expression in Ad-infected cells might be activated through a CR2-independent (E2F1-dependent) mechanism.
The results presented above revealed that among the proteins coded by the two major splice variants of E1A mRNAs, the 13S product activated BIK more efficiently than the 12S product. We then compared the apoptotic activity of Ad-19K (dl250) mutant (37) expressing either the 13S product or the 12S product in A549 cells. Since these mutants would be expected to have differential replication potential in A549 cells, the apoptotic activity (DNA fragmentation) was analyzed under conditions of suppression of viral DNA replication by the use of cytosine arabinoside. The results in Fig. 5D revealed that the 13S product induced an enhanced apoptotic response compared to the 12S product, while the effect of the genomic E1A (13S plus 12S) was more than that of 12S. Thus, our results suggest that the E1A 13S product may play a more important role in apoptosis induction during Ad infection.
Interaction of BIK with BCL-xL.
Since we observed significant activation of BIK expression in Ad-infected cells, it is possible that suppression of Ad-induced apoptosis by BCL-xL and BCL-xL mt1 might, at least partially, be attributed to sequestration of BIK by BCL-xL. To test this possibility, we carried out a coimmunoprecipitation analysis (Fig. 6A). As expected, expression of BIK was enhanced in all Ad-infected cells (top blot). In cells infected with Ad-BCL-xL recombinants, interaction between BIK and wt BCL-xL or BCL-xL mt1 was observed and not with BCL-xL mt8 that is deficient in the antiapoptotic activity.
FIG. 6.
Interaction of BIK with BCL-xL and mutants. (A) Western blot analysis of whole-cell lysates (WCL) and immunoprecipitate (IP). The blots were probed with the antibodies indicated to the right of the blots. (B) Interaction of phosphorylated BIK with BCL-xL. The lysates from mock-infected and Ad5-infected cells were incubated with GST and GST-BCL-xL and analyzed by Western blotting using BIK antibody (top blot). In the lower blots, the lysates and the proteins bound to the GST-BCL-xL affinity matrix were either untreated (−) or treated with lambda protein phosphatase (λ ppase) (+) and analyzed by Western blotting.
Since the apoptotic activity of BIK is enhanced by phosphorylation (42), we investigated the possibility that BIK might be phosphorylated in Ad-infected cells. We performed a GST pull-down assay using lysates from mock-infected or Ad-infected cells. The endogenous BIK protein present in Ad-infected cells interacted with GST-BCL-xL (Fig. 6B, top blot) in agreement with the immunoprecipitation data. After the GST-BCL-xL pull-down assay, the samples were then treated with λ protein phosphatase and analyzed by SDS-PAGE (Fig. 6B, bottom blot). This analysis indicated that BIK associated with GST-BCL-xL (not treated with phosphatase) moved to a position that corresponded to the phosphorylated form (often as a doublet) and that after treatment with phosphatase, it migrated faster. Thus, our results suggest that the expression of the BH3-only member BIK is transcriptionally activated in Ad-infected cells and the protein is modified by phosphorylation.
Effect of BIK in Ad-induced apoptotic cell death.
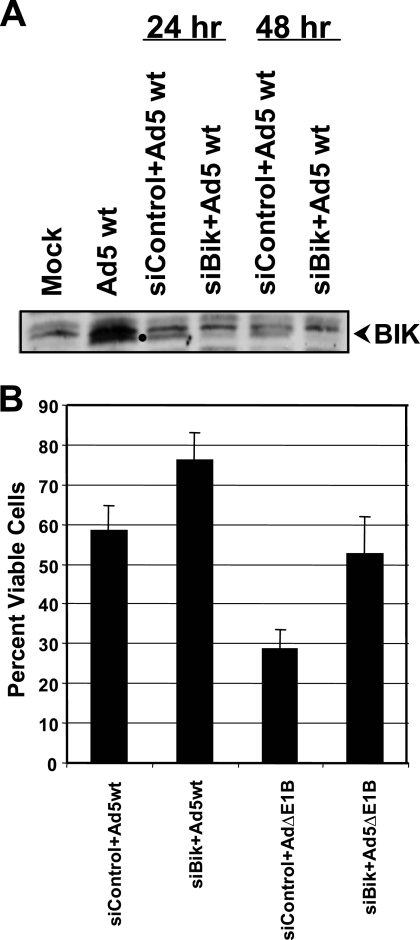
Since BIK was activated during Ad infection, we determined the effect of depletion of BIK on Ad-induced apoptotic cell death. A549 cells were transfected with control small interfering RNA (siRNA) or siRNA targeted against BIK. Twenty-four hours after transfection, cells were infected with wt Ad5. Western blot analysis revealed significant reduction in the level of BIK expression at 24 h after Ad5 infection (Fig. 7A). The viability of cells that were depleted for BIK and infected with wt Ad5 or AdΔE1B was determined 24 h after infection (Fig. 7B). There was an increase in the viability of cells infected with AdΔE1B of about 25%. Interestingly, depletion of BIK also enhanced the viability of cells infected with wt Ad5, suggesting modest levels of basal BIK-mediated cell death in the presence of E1B expression. These results suggest that BIK is a major contributor to Ad-induced apoptosis.
FIG. 7.
Effect of BIK depletion on Ad-induced cell death. A549 cells were transfected with control or BIK siRNA and the extent of BIK depletion was determined by Western blot analysis (A). The cells depleted for BIK were infected with Ad5 wt or AdΔE1B and cell viability was determined (B). The experiment was repeated twice in triplicate.
DISCUSSION
Our present study has provided genetic and biochemical evidence that BH3-only proapoptotic proteins might be the initiators of the Ad apoptosis paradigm. This conclusion is based on our results that Ad-induced apoptosis is efficiently suppressed by the expression of a BCL-xL mutant (mt1) that is proficient in interaction with BH3-only proteins and deficient in interaction with BH123 proteins (4, 5). Previous studies have identified that activation of the BH123 protein BAK (by liberation of BAK via degradation of MCL-1) might be a critical checkpoint for the onset of Ad-induced apoptosis (10). It is generally believed that activation of BH3-only proteins leads to activation of BAK and BAX (for a recent review, see reference 47). However, some evidence also suggests deviations of this general paradigm (i.e., BH3-only to BH123) during apoptosis induced by some stimuli. For example, it has been reported that the BH123 proteins can be directly activated (i.e., without the involvement of BH3-only proteins) by heat shock (31). Thus, it is possible that BAK activation may trigger the onset of Ad-induced apoptosis in the absence of BH3-only proteins. Although our data cannot rule out this possibility, the observation that BCL-xL mt1, which lacks the ability to complex with BAK, protects from Ad-induced apoptosis suggests that the activation of BH3-only proteins may be a critical step in the onset of Ad-induced apoptosis. We have also observed that BCL-xL mt1 suppressed Ad-induced apoptosis in spite of MCL-1 degradation (Fig. 3A).
A more downstream activation step at the level of transcriptional activation of caspases has also been proposed as a mechanism of E1A-induced apoptosis (26). Currently, there is no data to suggest that any of the BCL-2 family antiapoptotic proteins can directly antagonize the activities of caspases. Since Ad-induced apoptosis can be efficiently suppressed by the expression of E1B-19K (6, 44), BCL-2 (38), and BCL-xL (present study) from the viral genome, it appears that E1A-mediated transcriptional activation of the various caspase genes may not be a critical event in the onset of apoptosis during viral infection. Our results do not rule out initiation of apoptosis from BH123 activation and caspase activation checkpoints, under conditions of isolated E1A expression. Activation of caspases by E1A may enhance the overall apoptotic response during Ad infection.
Among the BH3-only members we have examined, BIK was prominently activated at the level of transcription in Ad5-infected A549 and HNK cells. Additionally, expression of BIM was also activated in both cell types. Since the list of BH3-only members continues to expand, the possibility that expression of certain other novel BH3-only members might also be activated in Ad-infected cells cannot be ruled out. Some of the BH3-only proteins are functionally redundant. For example, studies with mutant mice suggest that BIK and BIM are functionally redundant during mouse development and concomitant deletion of both genes is required for manifestation of certain mutant phenotypes (8). It is possible that activation of multiple BH3-only proteins might be a cellular defense strategy to limit viral multiplication. The spectrum of BH3-only proteins activated by Ad infection might also be influenced by the cell types. For example, we have observed modest activation of HRK in A549 cells and not in HNK cells.
We observed that BIK expression was enhanced to a higher degree in cells infected with Ad5-13S than in cells infected with Ad5-12S (Fig. 5A). These results correlate with the pattern of apoptotic response observed in cells infected with the Ad 19K mutant that either expresses the 13S or 12S product (Fig. 5D). In a previous report, human (HeLa) cells infected with Ad-13S have been shown to exhibit an apoptotic response comparable to wt Ad5, while the apoptotic response of Ad-12S was less pronounced and delayed (46). We note that Ad-12S has been shown to induce strong apoptotic response in NRK cells (25). However, the proficient apoptotic activity of Ad-12S was observed only under growth-inhibited conditions imposed by nutrient starvation or by contact inhibition.
We have observed that the Bik promoter was occupied by E2F1 and low levels of p53 in Ad-infected cells (Fig. 5B). In silico analysis of the human Bik promoter (41) suggests that it contains an E2F1 target site and a potential p53-binding site. Thus, it is possible that these cellular transcription factors may contribute to transcriptional activation of Bik in Ad-infected cells. Recently, it was reported that in certain cancer cells treated with the chemotherapeutic drug adriamycin, Bik was activated by E2F1 (34). It has previously been shown that expression of Bik was activated in cells infected with an Ad vector that expressed the p53 transgene from the E1 region, suggesting that Bik might be a transcriptional target for p53 (24). It is established that expression of E1A results in accumulation of p53 (22). Thus, increased accumulation of p53 may also contribute to enhanced expression of BIK in Ad-infected cells.
Our observation that E2F1 might contribute to transcriptional activation of Bik through the action of the E1A 13S mRNA product was rather surprising, considering that the well-known mechanism of E1A-mediated activation of E2F1 (i.e., the release of E2F1 from the pRb repression complex mediated by the E1A CR2 region) (27) is common to both E1A protein isoforms. We have further observed that an Ad5 (genomic E1A) mutant (dl1108) with a deletion in the CR2 region did not impair activation of BIK (Fig. 5C). It should be noted that Bim, another target for E2F1 (2, 15, 50) was also activated by E1A 13S (Fig. 5A). These observations suggest that the CR3 region of E1A might be important for the activation of Bik expression in Ad5-infected cells. Since the CR3 region is a potent trans-activation domain, it might play an indirect role through activation of E2F1 or other cellular transcription factors. In addition to the CR2-mediated E2F1 activation, a second mechanism has also been identified in Ad-infected cells. The latter mechanism depends on the E4-6/7 protein (30, 36). The E4 protein has been reported to complex with the E2F1/DP heterodimer and enhance the DNA binding activity of E2F1. It is possible that the activity of the basal level of “free” E2F1 present in Ad-infected A549 cells might be enhanced by the E4-6/7 protein. Since the E4 region is transcriptionally activated by the E1A CR3 region, the CR3 region might indirectly activate Bik through E4-mediated enhancement of E2F1 activity. A clear elucidation of the potential mechanism of transcriptional activation of Bik in Ad-infected cells would require an in-depth investigation using a combination of E1A and E4 mutants.
We have observed that BIK is phosphorylated in Ad-infected cells. We have previously reported that phosphorylation of BIK enhances its proapoptotic activity (42). Recently, the use of BIK mutants that mimic the phosphorylated form as a therapeutic agent against ovarian cancer is being advanced (20). The possibility that E1A might act synergistically with different anticancer drugs through activation of expression of apoptotically enhanced BIK would be of much interest with regards to the use of Ads as oncolytic agents. In this context, activation of BIK expression has also been observed in cells treated with certain anticancer drugs, such as doxorubicin (32) and adriamycin (34). The use of proteasomal inhibitors is on the rise for treatment of neoplastic diseases (19). Since proteasome inhibitors significantly enhance BIK accumulation (29, 49, 52), Ad oncolytic vectors in combination therapies with such inhibitors could also be envisioned.
Acknowledgments
We thank Marie Hardwick for providing BCL-xL mutants.
This work was supported by research grants CA-33616 and CA-73803.
Footnotes
Published ahead of print on 25 July 2007.
REFERENCES
- 1.Anderson, R. D., R. E. Haskell, H. Xia, B. J. Roessler, and B. L. Davidson. 2000. A simple method for the rapid generation of recombinant adenovirus vectors. Gene Ther. 7:1034-1038. [DOI] [PubMed] [Google Scholar]
- 2.Biswas, S. C., D. X. Liu, and L. A. Greene. 2005. Bim is a direct target of a neuronal E2F-dependent apoptotic pathway. J. Neurosci. 25:8349-8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, J. M., S. Malstrom, T. Subramanian, L. K. Venkatesh, U. Schaeper, B. Elangovan, C. D'Sa-Eipper, and G. Chinnadurai. 1994. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell 79:341-351. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, E. H., B. Levine, L. H. Boise, C. B. Thompson, and J. M. Hardwick. 1996. Bax-independent inhibition of apoptosis by Bcl-XL. Nature 379:554-556. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, E. H., M. C. Wei, S. Weiler, R. A. Flavell, T. W. Mak, T. Lindsten, and S. J. Korsmeyer. 2001. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell 8:705-711. [DOI] [PubMed] [Google Scholar]
- 6.Chinnadurai, G. 1998. Control of apoptosis by human adenovirus genes. Semin. Virol. 8:399-408. [Google Scholar]
- 7.Chiou, S. K., C. C. Tseng, L. Rao, and E. White. 1994. Functional complementation of the adenovirus E1B 19-kilodalton protein with Bcl-2 in the inhibition of apoptosis in infected cells. J. Virol. 68:6553-6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coultas, L., P. Bouillet, K. L. Loveland, S. Meachem, H. Perlman, J. M. Adams, and A. Strasser. 2005. Concomitant loss of proapoptotic BH3-only Bcl-2 antagonists Bik and Bim arrests spermatogenesis. EMBO J. 24:3963-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuconati, A., K. Degenhardt, R. Sundararajan, A. Anschel, and E. White. 2002. Bak and Bax function to limit adenovirus replication through apoptosis induction. J. Virol. 76:4547-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuconati, A., C. Mukherjee, D. Perez, and E. White. 2003. DNA damage response and MCL-1 destruction initiate apoptosis in adenovirus-infected cells. Genes Dev. 17:2922-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fortin, A., J. G. MacLaurin, N. Arbour, S. P. Cregan, N. Kushwaha, S. M. Callaghan, D. S. Park, P. R. Albert, and R. S. Slack. 2004. The proapoptotic gene SIVA is a direct transcriptional target for the tumor suppressors p53 and E2F1. J. Biol. Chem. 279:28706-28714. [DOI] [PubMed] [Google Scholar]
- 12.Green, D. R. 2005. Apoptotic pathways: ten minutes to dead. Cell 121:671-674. [DOI] [PubMed] [Google Scholar]
- 13.Green, M., K. H. Brackmann, L. A. Lucher, J. S. Symington, and T. A. Kramer. 1983. Human adenovirus 2 E1B-19K and E1B-53K tumor antigens: antipeptide antibodies targeted to the NH2 and COOH termini. J. Virol. 48:604-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths, G. J., L. Dubrez, C. P. Morgan, N. A. Jones, J. Whitehouse, B. M. Corfe, C. Dive, and J. A. Hickman. 1999. Cell damage-induced conformational changes of the pro-apoptotic protein Bak in vivo precede the onset of apoptosis. J. Cell Biol. 144:903-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershko, T., and D. Ginsberg. 2004. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J. Biol. Chem. 279:8627-8634. [DOI] [PubMed] [Google Scholar]
- 16.Hsu, Y. T., and R. J. Youle. 1998. Bax in murine thymus is a soluble monomeric protein that displays differential detergent-induced conformations. J. Biol. Chem. 273:10777-10783. [DOI] [PubMed] [Google Scholar]
- 17.Hsu, Y. T., and R. J. Youle. 1997. Nonionic detergents induce dimerization among members of the Bcl-2 family. J. Biol. Chem. 272:13829-13834. [DOI] [PubMed] [Google Scholar]
- 18.Jelsma, T. N., J. A. Howe, C. M. Evelegh, N. F. Cunniff, M. H. Skiadopoulos, M. R. Floroff, J. E. Denman, and S. T. Bayley. 1988. Use of deletion and point mutants spanning the coding region of the adenovirus 5 E1A gene to define a domain that is essential for transcriptional activation. Virology 163:494-502. [DOI] [PubMed] [Google Scholar]
- 19.Joazeiro, C. A., K. C. Anderson, and T. Hunter. 2006. Proteasome inhibitor drugs on the rise. Cancer Res. 66:7840-7842. [DOI] [PubMed] [Google Scholar]
- 20.Li, Y. M., Y. Wen, B. P. Zhou, H. P. Kuo, Q. Ding, and M. C. Hung. 2003. Enhancement of Bik antitumor effect by Bik mutants. Cancer Res. 63:7630-7633. [PubMed] [Google Scholar]
- 21.Lomonosova, E., T. Subramanian, and G. Chinnadurai. 2002. Requirement of BAX for efficient adenovirus-induced apoptosis. J. Virol. 76:11283-11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowe, S. W., and H. E. Ruley. 1993. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 7:535-545. [DOI] [PubMed] [Google Scholar]
- 23.Marshansky, V., X. Wang, R. Bertrand, H. Luo, W. Duguid, G. Chinnadurai, N. Kanaan, M. D. Vu, and J. Wu. 2001. Proteasomes modulate balance among proapoptotic and antiapoptotic Bcl-2 family members and compromise functioning of the electron transport chain in leukemic cells. J. Immunol. 166:3130-3142. [DOI] [PubMed] [Google Scholar]
- 24.Mathai, J. P., M. Germain, R. C. Marcellus, and G. C. Shore. 2002. Induction and endoplasmic reticulum location of BIK/NBK in response to apoptotic signaling by E1A and p53. Oncogene 21:2534-2544. [DOI] [PubMed] [Google Scholar]
- 25.Mymryk, J. S., K. Shire, and S. T. Bayley. 1994. Induction of apoptosis by adenovirus type 5 E1A in rat cells requires a proliferation block. Oncogene 9:1187-1193. [PubMed] [Google Scholar]
- 26.Nahle, Z., J. Polakoff, R. V. Davuluri, M. E. McCurrach, M. D. Jacobson, M. Narita, M. Q. Zhang, Y. Lazebnik, D. Bar-Sagi, and S. W. Lowe. 2002. Direct coupling of the cell cycle and cell death machinery by E2F. Nat. Cell Biol. 4:859-864. [DOI] [PubMed] [Google Scholar]
- 27.Nevins, J. R. 1995. Adenovirus E1A: transcription regulation and alteration of cell growth control. Curr. Top. Microbiol. Immunol. 199(Part 3):25-32. [DOI] [PubMed] [Google Scholar]
- 28.Nevins, J. R. 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10:699-703. [DOI] [PubMed] [Google Scholar]
- 29.Nikrad, M., T. Johnson, H. Puthalalath, L. Coultas, J. Adams, and A. S. Kraft. 2005. The proteasome inhibitor bortezomib sensitizes cells to killing by death receptor ligand TRAIL via BH3-only proteins Bik and Bim. Mol. Cancer Ther. 4:443-449. [DOI] [PubMed] [Google Scholar]
- 30.O'Connor, R. J., and P. Hearing. 1994. Mutually exclusive interaction of the adenovirus E4-6/7 protein and the retinoblastoma gene product with internal domains of E2F-1 and DP-1. J. Virol. 68:6848-6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pagliari, L. J., T. Kuwana, C. Bonzon, D. D. Newmeyer, S. Tu, H. M. Beere, and D. R. Green. 2005. The multidomain proapoptotic molecules Bax and Bak are directly activated by heat. Proc. Natl. Acad. Sci. USA 102:17975-17980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panaretakis, T., K. Pokrovskaja, M. C. Shoshan, and D. Grander. 2002. Activation of Bak, Bax and BH3-only proteins in the apoptotic response to doxorubicin. J. Biol. Chem. 277:44317-44326. [DOI] [PubMed] [Google Scholar]
- 33.Pilder, S., J. Logan, and T. Shenk. 1984. Deletion of the gene encoding the adenovirus 5 early region 1b 21,000-molecular-weight polypeptide leads to degradation of viral and host cell DNA. J. Virol. 52:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Real, P. J., C. Sanz, O. Gutierrez, C. Pipaon, A. M. Zubiaga, and J. L. Fernandez-Luna. 2006. Transcriptional activation of the proapoptotic bik gene by E2F proteins in cancer cells. FEBS Lett. 580:5905-5909. [DOI] [PubMed] [Google Scholar]
- 35.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol 132:365-386. [DOI] [PubMed] [Google Scholar]
- 36.Schaley, J., R. J. O'Connor, L. J. Taylor, D. Bar-Sagi, and P. Hearing. 2000. Induction of the cellular E2F-1 promoter by the adenovirus E4-6/7 protein. J. Virol. 74:2084-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian, T., M. Kuppuswamy, J. Gysbers, S. Mak, and G. Chinnadurai. 1984. 19-kDa tumor antigen coded by early region E1b of adenovirus 2 is required for efficient synthesis and for protection of viral DNA. J. Biol. Chem. 259:11777-11783. [PubMed] [Google Scholar]
- 38.Subramanian, T., B. Tarodi, and G. Chinnadurai. 1995. p53-independent apoptotic and necrotic cell deaths induced by adenovirus infection: suppression by E1B 19K and Bcl-2 proteins. Cell Growth Differ. 6:131-137. [PubMed] [Google Scholar]
- 39.Takemori, N., C. Cladaras, B. Bhat, A. J. Conley, and W. S. Wold. 1984. cyt gene of adenoviruses 2 and 5 is an oncogene for transforming function in early region E1B and encodes the E1B 19,000-molecular-weight polypeptide. J. Virol. 52:793-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarodi, B., T. Subramanian, and G. Chinnadurai. 1993. Functional similarity between adenovirus E1b 19k gene and Bcl2 oncogene: mutant complementation and suppression of cell death induced by DNA damaging agents. Int. J. Oncol. 3:467-472. [DOI] [PubMed] [Google Scholar]
- 41.Verma, S., M. L. Budarf, B. S. Emanuel, and G. Chinnadurai. 2000. Structural analysis of the human pro-apoptotic gene Bik: chromosomal localization, genomic organization and localization of promoter sequences. Gene 254:157-162. [DOI] [PubMed] [Google Scholar]
- 42.Verma, S., L. J. Zhao, and G. Chinnadurai. 2001. Phosphorylation of the pro-apoptotic protein BIK: mapping of phosphorylation sites and effect on apoptosis. J. Biol. Chem. 276:4671-4676. [DOI] [PubMed] [Google Scholar]
- 43.Warr, M. R., S. Acoca, Z. Liu, M. Germain, M. Watson, M. Blanchette, S. S. Wing, and G. C. Shore. 2005. BH3-ligand regulates access of MCL-1 to its E3 ligase. FEBS Lett. 579:5603-5608. [DOI] [PubMed] [Google Scholar]
- 44.White, E. 2001. Regulation of the cell cycle and apoptosis by the oncogenes of adenovirus. Oncogene 20:7836-7846. [DOI] [PubMed] [Google Scholar]
- 45.White, E., T. Grodzicker, and B. W. Stillman. 1984. Mutations in the gene encoding the adenovirus early region 1B 19,000-molecular-weight tumor antigen cause the degradation of chromosomal DNA. J. Virol. 52:410-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White, E., and B. Stillman. 1987. Expression of adenovirus E1B mutant phenotypes is dependent on the host cell and on synthesis of E1A proteins. J. Virol. 61:426-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willis, S. N., and J. M. Adams. 2005. Life in the balance: how BH3-only proteins induce apoptosis. Curr. Opin. Cell Biol. 17:617-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolter, K. G., Y. T. Hsu, C. L. Smith, A. Nechushtan, X. G. Xi, and R. J. Youle. 1997. Movement of Bax from the cytosol to mitochondria during apoptosis. J. Cell Biol. 139:1281-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeung, B. H., D. C. Huang, and F. A. Sinicrope. 2006. PS-341 (bortezomib) induces lysosomal cathepsin B release and a caspase-2-dependent mitochondrial permeabilization and apoptosis in human pancreatic cancer cells. J. Biol. Chem. 281:11923-11932. [DOI] [PubMed] [Google Scholar]
- 50.Zhao, Y., J. Tan, L. Zhuang, X. Jiang, E. T. Liu, and Q. Yu. 2005. Inhibitors of histone deacetylases target the Rb-E2F1 pathway for apoptosis induction through activation of proapoptotic protein Bim. Proc. Natl. Acad. Sci. USA 102:16090-16095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong, Q., W. Gao, F. Du, and X. Wang. 2005. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 121:1085-1095. [DOI] [PubMed] [Google Scholar]
- 52.Zhu, H., L. Zhang, F. Dong, W. Guo, S. Wu, F. Teraishi, J. J. Davis, P. J. Chiao, and B. Fang. 2005. Bik/NBK accumulation correlates with apoptosis-induction by bortezomib (PS-341, Velcade) and other proteasome inhibitors. Oncogene 24:4993-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]