Abstract
All herpesviruses contain a ubiquitin (Ub)-specific cysteine protease domain embedded within their large tegument protein, based on homology with the corresponding sequences of UL36 from herpes simplex virus type 1 and M48 from murine cytomegalovirus. This type of activity has yet to be demonstrated for cells infected with a gammaherpesvirus. By activity-based profiling, we show that the large tegument protein of murine gammaherpesvirus (MHV-68) ORF64 (273 kDa) is a functional deubiquitinating protease, as assessed by tandem mass spectrometry of adducts in extracts from MHV-68-infected cells that had been labeled with ubiquitin vinylmethylester, a ubiquitin-based active site-directed probe. The recombinantly expressed amino-terminal segment of ORF64 displays deubiquitinating activity toward Ub C-terminal 7-amido-4-methylcoumarin in vitro. The findings reported here for MHV-68 ORF64 extend those made for the alpha- and betaherpesvirus families and are consistent with an important, conserved enzymatic function of the tegument protein.
Herpesviruses have large genomes that encode both structural proteins and proteins necessary for the virus’ replicative success. The reliance on host functions for the generation of new virus particles has forced herpesviruses to acquire, in the course of their evolution, many specialized genes that regulate not only the host's cell cycle, but also protein synthesis and protein turnover. Presumably the removal of host proteins that interfere with the virus’ replicative strategies is carefully controlled and exploits the host cell's degradative apparatus.
In eukaryotic cells, the ubiquitin (Ub)-proteasome system controls cytosolic proteolysis. Unwanted proteins are tagged with Ub and then targeted to the proteasome (reviewed in reference 10). Any of a number of Ub-specific proteases (USPs) can revise a protein substrate's modification with Ub (1). The human mammalian genome encodes about 100 such proteases (19), and the functions of the vast majority are not known. The presence of USPs extends to pathogens, although these pathogens do not themselves possess functional Ub genes: bacteria such as Salmonella (21), Yersinia (20, 27), and Chlamydia (18) possess proteases that act on Ub conjugates or on proteins decorated with Ub-like modifiers. Manipulation of the Ub-proteasome pathway thus contributes to replicative success of the pathogen.
We have characterized in herpes simplex virus type 1 (HSV-1) a new type of USP (14). Its discovery relied on the use of a Ub-derived activity-based probe, which selectively targets USPs (6, 7). Such probes are equipped with an epitope tag and yield covalent adducts with their target enzymes. These adducts are then purified and the enzyme portion identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and liquid chromatography/tandem mass spectrometry (LC/MS/MS) (6, 7). In this fashion, we have now labeled and identified 40-odd distinct mammalian USPs. The HSV-1 USP, which was discovered by application of these probes to HSV-1-infected human foreskin fibroblasts, stands out because it lacks any detectable sequence similarity to mammalian USPs. The USP activity of HSV-1 is in the N-terminal portion of the large tegument protein UL36 and apparently released as a protein fragment in the course of infection (14).
All herpesviruses possess a large tegument protein homologous to UL36, although the extent of sequence similarity in the area to which we attribute enzymatic activity is low, on the order of 15% between HSV-1 and Epstein-Barr virus (EBV) (22). The residues implicated in the formation of the typical cysteine protease active site, Cys, His, Asp, and Gln, are among those shared by all herpesvirus homologs of UL36.
We have cloned and expressed the putative catalytic domain from murine cytomegalovirus (MCMV) and from EBV and have shown that both, when expressed recombinantly in Escherichia coli, possess enzymatic activity (22), as confirmed by crystallographic analysis of the MCMV M48 product in a complex with ubiquitin-vinylmethylester (UbVME) (23). The M48 protein (pUL48) from human cytomegalovirus (HCMV) likewise possesses enzymatic activity, as inferred from the ability of an electrophilic Ub derivative to label intact M48. This labeling requires the presence of the conserved cysteine residue at the presumptive catalytic center (26). For HCMV, there is no evidence that a smaller M48-derived fragment is generated in the course of infection (26).
Murine gammaherpesvirus 68 (MHV-68), also referred to as γ MHV68, is a natural pathogen of wild rodents (5). Its genome has been sequenced and reveals a close relationship with Kaposi's sarcoma herpesvirus and EBV (25). The functions of some of the MHV-68 gene products are similar to those of the corresponding gene products of human gammaherpesviruses. The MHV-68 model is therefore widely used to study the pathogenesis of gammaherpesviruses. Although catalytic activity was demonstrated for the recombinant amino acid 1 to 205 fragment of EBV (22), there is currently no information on the expression of USP activity for any of the gammaherpesviruses in the course of a virus infection.
Here, we investigate the presence and expression of the tegument gene-associated USP activity in MHV-68. We show that the MHV-68 large tegument protein (encoded by ORF64, referred to as ORF64) indeed displays the properties of a deubiquitinating enzyme (DUB). We further show that the proposed catalytic domain, as predicted based on the MCMV M48USP structure, shows enzymatic activity. This is the first report of USP activity associated with the tegument protein of a gammaherpesvirus in the course of a productive infection.
MATERIALS AND METHODS
Cells and viruses.
3T12 cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM glutamine at 37°C. MHV-68 was obtained from the American Type Culture Collection (VR1465). Virus stocks of MHV-68 were prepared in 3T12 cells. Plaque assays on virus stocks were performed as described elsewhere (8).
Probe labeling and detection.
Cells were infected with MHV-68 at a multiplicity of infection of 5 or 10 and harvested by trypsinization at the indicated times after infection. The cells were washed in 1× PBS and pelleted by centrifugation. The cells were lysed in NP-40 lysis buffer (50 mM Tris, pH 7.4, 0.5% NP-40, 5 mM MgCl2, 150 mM NaCl, 1 mM dithiothreitol, 1 mM phenylmethylsulfonylfluoride) for 20 min at 4°C. The cell lysates were centrifuged at 14,000 rpm in a microcentrifuge for 20 min at 4°C. The protein concentrations of the supernatants were determined by Bradford assay. For each labeling reaction, 20 μg of protein was used and was diluted with 1 volume of 50 mM Tris-HCl, pH 8, 100 mM NaCl and reacted with 0.2 μg hemagglutinin (HA)-UbVME (6, 7) for 1 h at room temperature. To block reactive sulfhydryl groups, the lysates were preincubated with 10 mM N-ethylmaleimide (NEM) for 20 min at room temperature. For labeling ORF64USP with HA-UbVME, the samples were incubated with an estimated twofold molar excess of probe in the absence or presence of NEM.
Samples were boiled in reducing sample buffer and separated by SDS-PAGE. Proteins were transferred to polyvinylidene difluoride membranes, blocked in 5% (wt/vol) milk, and immunoblotted with a monoclonal rat anti-HA-horseradish peroxidase antibody (3F10; Roche, Mannheim, Germany) for 45 min. Following three washes in PBS-Tween 20 (0.1%), the blots were developed by using Western Lighting Chemiluminescence Reagent (Perkin-Elmer, Boston, MA).
Identification of labeled DUB using MS.
3T12 cells were infected with MHV-68 at a multiplicity of infection of 10. The cells were lysed and processed as described above after 48 h of infection. A total of 1 × 108 cells were used for the immunoprecipitation; the supernatants were diluted 1:2 in 50 mM Tris-HCl, pH 8, 100 mM NaCl and incubated with 40 μg of probe for 2 h at room temperature. SDS was added to 0.4% (wt/vol), followed by vigorous mixing, and samples were diluted to 0.1% SDS with 50 mM Tris-HCl, pH 8, 100 mM NaCl. The samples were incubated with anti-HA beads (3F10) overnight at 4°C. The beads were extensively washed with NET buffer (50 mM Tris, pH 7.4, 0.5% NP-40, 150 mM NaCl, and 5 mM EDTA). Samples were boiled and subjected to SDS-PAGE, followed by silver staining. Proteins were excised from the gel and digested in situ with trypsin. The resulting peptides were analyzed by LC/MS/MS, and the data were correlated against the MHV-68 and NCBI mouse databases using SEQUEST. All proteins reported were identified by the presence of two or more peptides.
Plasmid constructs and purification of proteins.
The 1-to-235 and 10-to-235 segments of MHV-68 ORF64 were PCR cloned into pET28 as described previously (22), using MHV-68 bacterial artificial chromosome DNA (a kind gift of Ulrich Koszinowski) as the template. Briefly, proteins were expressed in E. coli Rosetta cells, and the fragments, equipped with an N-terminal His tag, were purified using Ni-nitrilotriacetic acid resin (QIAGEN) and subjected to gel filtration to obtain purified constructs.
Ub-AMC hydrolysis assay.
Ub-AMC (Ub C-terminal 7-amido-4-methylcoumarin) hydrolysis assays were performed as described previously (23). Briefly, the hydrolysis reaction was performed in reaction buffer supplemented with 0.1 mg/ml of bovine serum albumin (Roche) by incubating the enzyme ORF64USP (500 pM) with an excess of deconjugating enzyme substrate (100 nM). Ub-AMC, SUMO-1-AMC, NEDD8-AMC, and ISG15-AMC (Boston Biochem, Boston, MA) were used as substrates. For inhibition experiments, NEM (10 mM), UbVME, Nedd8-VME, or Sumo-VS (11) was added to the enzyme ORF64USP 30 min prior to the addition of substrate (Ub-AMC; 500 nM). All experiments were performed in triplicate.
Phylogenetic analysis of the DUB domain of MHV-68 ORF64.
We searched the nonredundant database of NCBI for sequences similar to the UL36 DUB-like domain of MHV-68. The obtained sequences were aligned automatically with CLUSTALX and manually with GeneDoc (http://www.nrbsc.org/downloads/). To analyze the results from a phylogenetic perspective, we used the Phylip package version 3.6 (http://evolution.genetics.washington.edu/phylip/getme.html). First, we produced 100 bootstrapped alignments from the original alignment with the Bootstrap program. Then, we calculated the most parsimonious trees consistent with these alignments by using the Protpars program. Finally, we obtained the consensus tree with the Consense program, using the maximum likelihood algorithm. In the resulting tree, every node that is displayed was present in at least 80% of the bootstrapped trees.
RESULTS
DUB expression in MHV-68-infected fibroblasts.
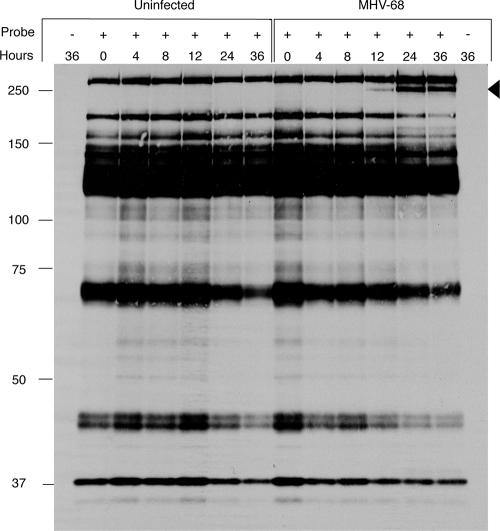
We began our investigations by infecting mouse fibroblasts with MHV-68. At various times after infection, cell lysates were prepared, which were incubated for 30 min at room temperature with HA-UbVME or left untreated. The extracts were then resolved by SDS-PAGE and immunoblotted for HA-UbVME-modified proteins using anti-HA antibodies. We observed a complex pattern of HA reactive species. In uninfected cells, these correspond to mouse USPs, as we have shown previously by MS/MS of immunopurified HA-UbVME-modified proteins (6, 7) (Table 1). In infected cells, we observed the corresponding set of modified host proteins, but after 12 h of infection, we detected an additional labeled polypeptide in the high-molecular-mass range (∼250 kDa) (Fig. 1). Its apparent mass was consistent with that predicted (273 kDa) for the tegument protein encoded by ORF64 as the labeled species. When we labeled extracts after preincubation with NEM, reactivity with the putative MHV-68 DUB and with all cellular DUBs was blocked, consistent with the involvement of a cysteine residue in the target proteins (data not shown). In some of the experiments we detected a virus-specific fragment with a smaller mass (∼30 kDa) (data not shown), as we also had seen for HSV-1-infected cells (14), but we did not observe its presence consistently.
TABLE 1.
Enzymes modified by the HA-UbVME probe
| Protein | Accession no. (NCBI) | Predicted mass (kDa) | Sequence coverage (%) |
|---|---|---|---|
| USP3a | 30580632 | 58.8 | 7.5 |
| USP4 | 2851531 | 108.2 | 32.7 |
| USP5 (isopeptidase T) | 3024764 | 95.8 | 40 |
| USP7 (herpesvirus-associated USP) (mHAUSP) | 81891295 | 128.4 | 34 |
| USP8 | 31981044 | 122.5 | 23 |
| USP FAF-X | 2501465 | 290.4 | 18.3 |
| USP10 | 32700079 | 87 | 14.4 |
| USP11 | 118572705 | 105.3 | 8.7 |
| USP12 | 81881643 | 42.9 | 9.2 |
| USP14 | 20178168 | 56 | 44.6 |
| USP15 | 28558361 | 112 | 35.1 |
| USP16 | 13195676 | 93.3 | 13.7 |
| USP19 | 47825366 | 150.4 | 12.9 |
| USP20a | 123857967 | 102.1 | 2.3 |
| USP22a | 78103329 | 60 | 8.4 |
| USP24 | 123858181 | 293.8 | 3.7 |
| mUSP25 | 46397896 | 121.1 | 20.2 |
| USP28 | 78103330 | 119.2 | 10.5 |
| USP30a | 85986575 | 58.2 | 5.6 |
| USP32a | 126032299 | 181.6 | 5.4 |
| USP38a | 38503388 | 116 | 8.9 |
| USP40a | 122066588 | 140 | 10.2 |
| USP46a | 49065851 | 42.4 | 14.2 |
| USP47a | 68566204 | 157.4 | 27.9 |
| USP48a | 115311888 | 120.6 | 12.9 |
| CYLD (DUB CYLD) | 51315948 | 107 | 3.6 |
| UCH-L1 | 18203410 | 24.8 | 10.8 |
| UCH-L3 | 17380334 | 26.1 | 47.8 |
| UCH-L4a | 19924308 | 26.4 | 4.7 |
| UCH-L5 | 18203574 | 37.6 | 48.3 |
| Otubain-2 (OTUB2)a | 44888287 | 27.3 | 11.1 |
| Zinc finger, A20 domain containing 1a | 71043959 | 91.9 | 22.7 |
USP not previously reported to react with UbVME.
FIG. 1.
Detection and expression profile of an MHV-68-induced DUB. 3T12 cells were either infected with MHV-68 or left uninfected, and the levels and activities of DUBs were assessed using an HA-UbVME probe. Cells were collected at different time points postinfection (0, 4, 8, 12, 24, and 36 h), lysed, and either left untreated or treated with HA-UbVME, followed by SDS-PAGE (8%) and immunoblotting with an anti-HA antibody.
Identification of the virus-encoded UbVME reactive protein.
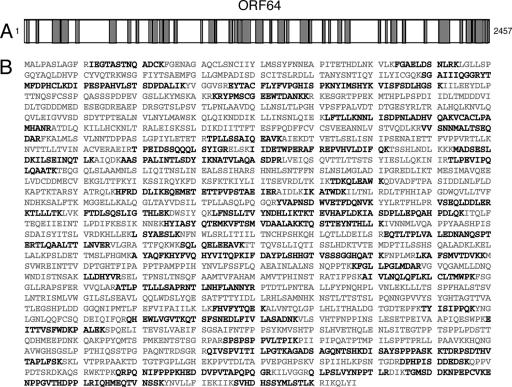
We accomplished the identification of the labeled species unique to virus-infected cells by large-scale immunopurification and resolution of purified materials by SDS-PAGE, followed by digestion in situ and resolution of the resultant peptides by LC/MS/MS. In this manner, we detected 115 peptides (35% sequence coverage) specific for the MHV-68 tegument protein ORF64 as the virus-specific high-molecular-weight (MW) USP. The level of sequence coverage and the sequences obtained unambiguously identified ORF64 as the source of the labeled fragment (Fig. 2). Because we obtained fairly even peptide coverage throughout the ORF64 sequence, and in view of the apparent MW of the labeled ORF64 species, we concluded that it is in all likelihood the fully intact ORF64 that was responsible for the generation of the labeled polypeptide. For the smaller polypeptide observed in some, but not all, of the labeling experiments performed on MHV-68-infected cells, we recovered peptides exclusively from the N-terminal segment of ORF64. In addition to the tegument protein encoded by ORF64, we found peptides corresponding to 13 other MHV-68 proteins (Table 2) . These proteins are most likely retrieved because they associate with the large tegument protein. As expected, most of the other peptides were derived from mouse DUBs found in infected cells and in uninfected controls (Table 1). Of note, we identified 13 new USPs in mouse 3T12 cells modified by the HA-UbVME probe, in addition to those previously found in EL-4 cells (7).
FIG. 2.
Polypeptides identified by MS for the MHV-68 DUB. Lysates from MHV-68-infected 3T12 cells were reacted with HA-UbVME probe on a preparative scale, followed by SDS-PAGE and silver staining. The proteins on the gel were extracted and subjected to MS/MS. The tegument protein ORF64 of MHV-68 was identified as a DUB. (A) Polypeptide coverage of ORF64 identified by MS (polypeptides identified are shown in gray). (B) Polypeptides identified by MS for ORF64 are shown in boldface for the sequence of the protein.
TABLE 2.
MHV-68-encoded proteins retrieved from large-scale immunopurification with the HA-UbVME probe
| Protein | Accession no. (NCBI) | Predicted mass (kDa) | Sequence coverage (%) | Remarksa |
|---|---|---|---|---|
| Tegument protein | 2317959 | 273.4 | 35 | ORF64 |
| Tegument protein/FGARATc | 2318000 | 145.7 | 13.3 | ORF75c |
| Tegument protein/FGARAT | 2318001 | 141.9 | 10.6 | ORF75b |
| Tegument protein | 2317997 | 25.9 | 14.2 | ORF67 |
| Major capsid protein | 2317938 | 153.2 | 4 | ORF25 |
| Capsid protein | 2317972 | 58.9 | 7.2 | ORF17 |
| Assembly/DNA maturation | 2317994 | 42.7 | 13.4 | ORF62 |
| Unknown | 2317944 | 35.7 | 8.3 | ORF33 herpesvirus UL16/UL94 family. UL16 protein may play a role in capsid maturation, including DNA packaging/cleavage |
| DNA polymerase processivity subunit | 1850858 | 41.9 | 22.3 | BALF3 (provisional) positional homolog to HSV and EHV2 gene 59, EBV BMRF1 |
| Single-stranded-DNA binding protein | 2317927 | 123.2 | 9.4 | ORF6 |
| Thymidine kinase | 2317936 | 72.2 | 6.8 | ORF21 |
| Unknown | 2317932 | 42.5 | 16.2 | ORF11 herpesvirus dUTPase family |
| Unknown | 2317975 | 42.5 | 11 | ORF23 herpesvirus BTRF1 protein conserved region |
| Unknown | 2317970 | 44.2 | 14 | M3 family of viral chemokine binding proteins |
EHV2, equine herpesvirus 2.
ORF64 contains a bona fide herpesvirus tegument USP domain.
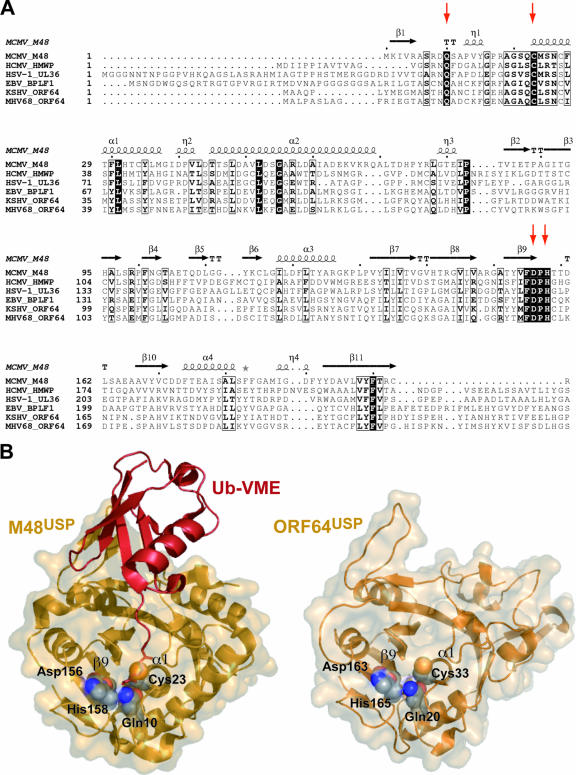
To assess whether the putative DUB activity can be faithfully mapped to the N-terminal portion of ORF64, we generated a sequence alignment based on representatives from all herpesvirus subfamilies and on MHV-68-encoded ORF64 itself. Overall, the large tegument proteins display a low degree of sequence identity (Fig. 3A). Nevertheless, residues that comprise the active site in the ORF64 homologue M48 (23) are strictly conserved in ORF64. To investigate the structural properties of the putative protease domain, we subjected the N-terminal portion of ORF64 to molecular modeling and obtained a three-dimensional model encompassing amino acids 10 to 247 (Fig. 3B). As judged by this hypothetical structure, ORF64 adopts an α-β-α sandwich fold that features a central catalytic cleft, ideally suited to accommodate the C-terminal stretch of Ub (Fig. 3B). Highly conserved residues that are likely involved in active-site formation (Fig. 3A) are located on the corresponding secondary-structure elements in both proteases (Fig. 3B), thus allowing the unambiguous assignment of Cys33, Asp163, and His165 as the catalytic triad. Moreover, Gln20 is ideally positioned to participate in oxyanion hole formation.
FIG. 3.
MHV-68 encoded ORF64 comprises an N-terminal domain with typical features of a herpesvirus tegument USP. (A) Sequence alignment of the N-terminal portions of herpesvirus tegument proteins. Residues that constitute the active site, also shown in panel B, are marked by red arrows. The secondary-structure depiction, shown on top, is deduced from the M48USP crystal structure (Protein Data Bank [PDB] code 2J7Q). (B) Structural comparison of MHV-68 and MCMV-encoded herpesvirus tegument USPs. Shown are ribbon representations of the M48USP structure in complex with Ub-VME (PDB code 2J7Q) (left) and an ORF64USP structure model (right) (modeled by SWISS-MODEL [15] using 2J7Q as a template in alignment mode). The solvent-accessible surface is depicted as semitransparent, and active site residues are shown as spheres. Secondary-structure elements harboring the active-site residues are marked as in panel A.
Taken together, these data suggest that the N-terminal portion of ORF64 constitutes a functional cysteine protease domain. We therefore cloned two fragments (residues 1 to 235 and 10 to 235) of ORF64 to allow a thorough biochemical characterization.
Substrate specificity of the active fragment of the MHV-68 DUB.
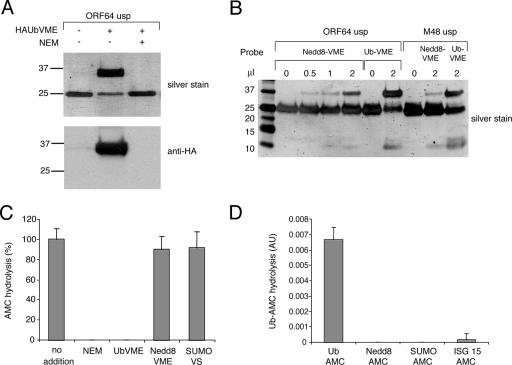
Two MHV-68 ORF64 fragments corresponding to those crystallized for MCMV M48 were expressed in E. coli in His-tagged form and purified to homogeneity. Upon incubation of the purified MHV-68 USP fragments with UbVME, we observed the formation of a covalent adduct, as judged from the shift in apparent MW and the appearance of a new, SDS-resistant complex at the expected MW (Fig. 4A). There was no difference in enzymatic activity between the two different ORF64USP fragments as measured by Ub-AMC hydrolysis (see below and data not shown). In all cases examined so far, the modification of candidate USPs by UbVME or similar Ub-based electrophiles has shown excellent correlation with enzymatic activity. As for MCMV M48USP, a smaller ORF64 fragment that contains the active-site residues is sufficient to confer both catalytic activity and substrate specificity (22). In view of the sequence divergence between ORF64USP and M48USP, we evaluated the substrate specificity of ORF64USP. We first performed a gel shift assay with increasing concentrations of the Nedd8-VME, in which M48USP was included as a negative control. In the case of ORF64USP, we observed a more pronounced formation of covalent adduct than with M48USP, as judged from the formation of an SDS-resistant complex of the expected MW. Even so, the Nedd8-VME adduct forms far less efficiently than the UbVME adduct (Fig. 4B). We also determined the hydrolytic activity of the purified ORF64USP fragment by its ability to hydrolyze the fluorogenic substrate Ub-AMC. We tested the abilities of several inhibitors, including Nedd8-VME, to block DUB activity in such Ub-AMC hydrolysis assays. The enzyme was preincubated for 30 min with a 100-fold molar excess of the electrophilic derivatives UbVME, Nedd8-VME, and SUMO-VS. UbVME completely inhibited AMC activity, whereas Nedd8-VME and SUMO-VS did not (Fig. 4C), showing that ORF64USP has affinity for Ub, but not for the other Ub-like molecules. Furthermore, ORF64USP hydrolyzed Ub-AMC in a manner sensitive to the inclusion of the alkylating agent NEM (Fig. 4C), again consistent with the involvement of the active-site cysteine. We next performed AMC assays with Ub-AMC, Nedd8-AMC, SUMO-1-AMC, and ISG15-AMC. The only substrate hydrolyzed by ORF64USP was Ub-AMC (Fig. 4D), again indicating that the formation of a covalent adduct of Nedd8-VME is unlikely to be of functional significance.
FIG. 4.
Evidence of deubiquitinating activity of ORF64. (A) Labeling of MHV-68 protease/tegument protein ORF64USP by HA-UbVME. ORF64USP was incubated with twofold molar excess of HA-UbVME in the absence or presence of NEM and subjected to SDS-PAGE. Silver stain was used for the upper gel, and anti-HA antibody directed against the probe was used in the lower gel. (B) ORF64USP and M48USP were incubated with increasing concentrations of the Nedd8-VME, as well as UbVME, for 30 min at 37°C and subjected to Tris-tricine SDS-PAGE, followed by silver staining. (C) The catalytic activity of ORF64USP was measured by an AMC assay. The enzyme was either untreated or incubated with a 100-fold molar excess of the indicated specific inhibitors (NEM, UbVME, Nedd8-VME, and SUMO-VS) for 30 min at room temperature prior to addition of Ub-AMC. Hydrolysis was measured over time by an increase in AMC fluorescence. The rate of Ub-AMC hydrolysis by the untreated construct was set to 100%. (D) AMC assays were performed for ORF64USP with Ub-AMC, Nedd8-AMC, SUMO-1-AMC, and ISG15-AMC. Hydrolysis was measured over time by an increase in AMC fluorescence. The data are presented as rates of hydrolysis in arbitrary units (AU). The error bars represent standard deviations.
Evolution of the herpesvirus DUBs.
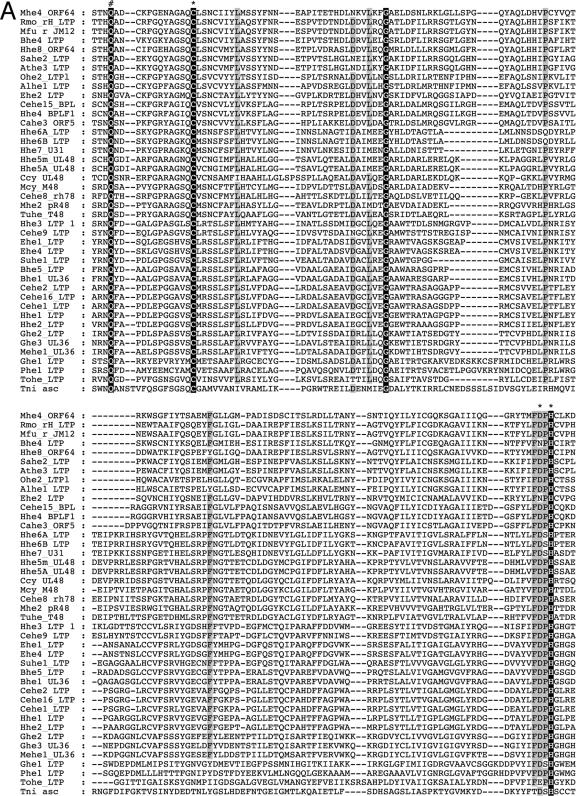
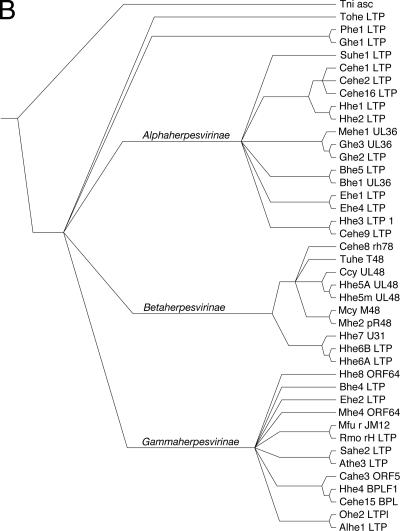
ORF64USP belongs to a recently discovered family of USPs, and we therefore characterized this novel protein domain phylogenetically. First, we looked for similar sequences in the NCBI databases, and since the overall similarity among the proteins that belong to this family is relatively low, we completed the search with an iterative position-specific BLAST. This strategy allowed us to find 40 similar sequences present in herpesviruses. Surprisingly, we also retrieved a protein from Trichoplusia ni ascovirus 2c, which was used as an outgroup to root the phylogenetic tree. The alignment of the 42 sequences obtained revealed several highly conserved motifs in this family (Fig. 5A). Contained in these motifs are the residues that conform to the catalytic triad of this cysteine protease, as well as the oxyanion hole (23). Notably, the sequences spaced between these motifs show considerable divergence in the different viruses. Next, we used the alignment of these protease domains to infer their phylogenetic relationships. The resulting phylogenetic tree places ORF64USP in the group Gammaherpesvirinae. In this tree, all three major herpesvirus groups are robustly clustered, with the exception of Gallid and Psittacid herpesvirus 1, which fail to cluster in the Alphaherpesvirinae group (Fig. 5B). Notably, if we construct the phylogenetic tree with less stringent parameters, these avian herpesvirus sequences, as well as the sequence from the unclassified Tortoise herpesvirus, do cluster with the Alphaherpesvirinae group (data not shown).
FIG. 5.
Phylogenetic analysis of herpesviral DUBs. (A) Amino acid alignment of viral UL36USP amino-terminal-like sequences. Catalytic residues are indicated with an asterisk. The oxyanion hole glutamine is indicated with a #. Absolutely conserved amino acids are highlighted in black, whereas positions with conservation higher than 80% are highlighted in gray. Alhe1, Alcelaphine herpesvirus 1; Athe3, Ateline herpesvirus 3; Bhe1, Bovine herpesvirus 1; Bhe4, Bovine herpesvirus 4; Bhe5, Bovine herpesvirus 5; Cahe3, Callitrichine herpesvirus 3; Ccy, Chimpanzee cytomegalovirus; Cehe1, Cercopithecine herpesvirus 1; Cehe15, Cercopithecine herpesvirus 15; Cehe16, Cercopithecine herpesvirus 16; Cehe2, Cercopithecine herpesvirus 2; Cehe8, Cercopithecine herpesvirus 8; Cehe9, Cercopithecine herpesvirus 9; Ehe1, Equid herpesvirus 1; Ehe2, Equid herpesvirus 2; Ehe4, Equid herpesvirus 4; Ghe1, Gallid herpesvirus 1; Ghe2, Gallid herpesvirus 2; Ghe3, Gallid herpesvirus 3; Hhe1, Human herpesvirus 1 (HSV-1); Hhe2, Human herpesvirus 2 (varicella zoster virus, VZV); Hhe3, Human herpesvirus 3; Hhe4, Human herpesvirus 4 (EBV); Hhe5A, Human herpesvirus 5 strain AD169 (HCMV); Hhe5m, Human herpesvirus 5 strain Merlin (HCMV); Hhe6A, Human herpesvirus 6A; Hhe6B, Human herpesvirus 6B; Hhe7, Human herpesvirus 7; Hhe8, Human herpesvirus 8 (KSHV); Mehe1, Meleagrid herpesvirus 1; Mfu, Macaca fuscata rhadinovirus; Mhe2, Murid herpesvirus 2 (RCMV); Mhe4, Murid herpesvirus 4 (MHV-68); Ohe2, Ovine herpesvirus 2; Phe1, Psittacid herpesvirus 1; Rmo, Rhesus monkey rhadinovirus H26-95; Sahe2, Saimiriine herpesvirus 2; Suhe1, Suid herpesvirus 1; Tni, Trichoplusia ni ascovirus 2c; Tohe, Tortoise herpesvirus; Tuhe, Tupaia herpesvirus. (B) Calculated phylogenetic relationships of viral UL36USP amino-terminal-like domains. A phylogenetic tree was calculated using the alignment displayed in panel A. The sequence from Trichoplusia ni ascovirus 2c was used to root the tree. The robustness of the tree, as measured by bootstrapping, is greater than 80%.
DISCUSSION
Based on homology with the corresponding primary and predicted secondary structures of UL36 from HSV-1 and M48 from MCMV, all herpesviruses contain a Ub-specific cysteine protease embedded within their large tegument proteins. The corresponding cloned fragments possess enzymatic activity. We have observed enzymatic activity in the course of active infection for an alphaherpesvirus (UL36 of HSV-1) (14) and for a betaherpesvirus (UL48 of HCMV) (26), but such activity has yet to be demonstrated for a gammaherpesvirus. Although we have shown that the cloned N-terminal segment of the gammaherpesvirus EBV displays deubiquitinating activity in vitro (22), it remained to be established whether this activity is manifest in infected cells, as well. Should enzymatic activity not be demonstrable, then the evolutionary conservation of the herpesvirus DUBs cannot be construed as an argument for their functional importance. We chose MHV-68 as a representative of the gammaherpesvirus family because of its ability to replicate to high titers in cultured cells and because there are no other data on deubiquitinating activity in MHV-68. We thus identified the first viral USP in cells infected with a gammaherpesvirus.
The HSV DUB (UL36) generates a smaller fragment that labels with UbVME. For cytomegalovirus, this does not appear to be the case, but in some of our labeling experiments, we also detected an ∼30-kDa polypeptide derived from ORF64. By MS, we recovered matching peptides only from the N terminus of ORF64 for the 30-kDa fragment. The observed fragment at lower mass could represent a degradation product, although its retrieval by our affinity purification clearly indicates involvement of the active site and retention of the covalent adduct with UbVME.
The M48USP structure defines a previously unknown class of DUBs termed herpesvirus tegument USPs (htUSPs) (23). The htUSP domains are generally located at the N termini of large tegument proteins. Sequence identities for these domains are in the range of 10 to 16% if one compares individual members from different subfamilies, yet secondary-structure predictions clearly suggest the conservation of the htUSP fold in all herpesvirus subfamilies (22). Moreover, residues that constitute the catalytic triad—strictly conserved throughout all members of the Herpesviridae—are located on identical secondary-structural elements according to these predictions and must adopt highly similar three-dimensional folds as judged from molecular modeling. The structural conservation of the htUSP fold is apparent from the modeled ORF64USP structure, in which all active-site residues are located at the corresponding positions compared to the M48USP crystal structure, notwithstanding a low level of sequence identity between ORF64 and M48 (16%). We have shown that the MCMV-encoded htUSP domain is necessary and sufficient to confer activity and specificity toward Ub-based substrates. While ORF64USP covalently binds UbVME and less efficiently binds Nedd8-VME as well, it hydrolyzes UbAMC, but not Nedd8-AMC. The covalent adducts of ORF64USP with Nedd8-VME are irreversible and thus tend to accumulate upon prolonged incubation. We conclude that the ability of ORF64USP to bind Nedd8 probably does not have functional significance.
The conservation of the tegument-associated USP activity in every member of the herpesvirus family examined to date suggests an important function for this activity. A recent publication showed that the carboxy-terminal sequence of VP1/2 tegument protein (encoded by UL36) in pseudorabies virus is an essential domain in this protein, whereas the amino-terminal DUB, as shown also for HCMV (26), is dispensable, at least in vitro, but its ablation results in poor propagation in vitro compared to the wild-type viruses (16). The consequences of ablation of USP activity for virus growth and persistence in vivo have yet to be explored in detail.
In addition to the MHV-68-encoded DUB, we recovered 32 cellular DUBs, 13 of which are new additions to those previously identified (7). As seen from the patterns of HA reactive species, there is little or no effect of MHV-68 infection on expression of cellular DUBs in 3T12 cells. This suggests the possibility that, in the context of the infected cell, the herpesvirus DUBs recognize distinct (sets of) substrates from those acted on by the endogenous host DUBs, whose activities appear not to be affected by herpesvirus infection. We further recovered peptides matching an additional 13 proteins encoded by MHV-68, mainly corresponding to other tegument proteins, capsid proteins, and proteins involved in transcription. The most likely explanation for the retrieval of these proteins is their interaction with the large tegument protein encoded by ORF64, leading to copurification with UbVME-tagged ORF64. None of the retrieved viral products showed obvious labeling with UbVME in infected cells and are therefore unlikely to be targets of covalent modification with UbVME.
The phylogenetic characterization of ORF64USP led us to the identification of the first putative member of the UL36USP family outside the herpesvirus family, namely, in the ascovirus Trichoplusia ni ascovirus 2c. The sequence of this novel domain displays low overall identity compared to the herpesvirus sequences. In contrast, those stretches presumably involved in catalytic activity show a high degree of similarity with herpesvirus proteases. Inspection of the alignment of UL36USP homologs suggests that the protease domain predates the ancestral herpesvirus and was inherited by at least herpesviruses and ascoviruses. While the paucity of ascoviral genomic information hampers a more detailed study of the origin of UL36USP homologs, the possibility of horizontal transfer between herpesviruses and ascoviruses is a distinct possibility. However, the ascoviral sequence can be effectively used as an outgroup of the herpesviral sequences, which supports a vertical-transmission hypothesis. When rooted with this ascoviral sequence, the inferred phylogenetic tree recapitulates the accepted taxonomic classification of herpesviruses. Thus, the tree shows three robustly predicted groups, corresponding to Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae. Only two avian Alphaherpesvirinae fail to cluster in their corresponding group.
Ubiquitination and deubiquitination regulate multiple essential cellular processes, such as protein degradation, cell signaling, DNA repair, and cell-cycle progression. Several viruses interfere with the ubiquitination system of the host through control of ubiquitination and/or deubiquitination. The γ-2 herpesviruses, to which MHV-68 and the human Kaposi's sarcoma virus belong, encode a Ub ligase, K3 in MHV-68 and K3 and K5 in Kaposi's sarcoma virus, responsible for down regulation of class I major histocompatibility complex expression in fibroblasts (24). In this case, ubiquitination is believed to serve as an immune evasion mechanism for γ-2 herpesviruses. Other viruses interact with the cullin RING Ub ligases, e.g., paramyxovirus and HIV-1 (4). Viruses presumably use these activities to create a host environment favorable for virus replication and dissemination. The first documented interaction of a herpesvirus protein and a DUB is that between the host-derived USP7 (also called HAUSP) and the ICP0 protein of HSV (9). USP7 also interacts with the EBNA1 protein of EBV (13). EBNA1 may interfere with the normal regulation of the mdm2-p53 pathway by USP7 (reviewed in reference 12). A number of other viruses also encode their own DUBs. Deubiquitinating activity is associated with an adenovirus protease (2), with the severe acute respiratory syndrome coronavirus papain-like protease (3, 17), and with the large tegument proteins of HSV-1 (14) and cytomegalovirus (26). We may now add MHV-68 to the list. Because MHV-68 encodes both a Ub ligase (K3) and a DUB (ORF64), the Ub pathway clearly is an important target for virulence factors carried by MHV-68. A more complete picture of the biological role of the herpesvirus USPs will require the identification of their natural substrates. No such substrates are known to date, but the ability to generate mutants of herpesviruses that lack this activity, as shown for HCMV (26), which can then be used in animal models of infectious disease may help attain this goal.
Acknowledgments
This work was carried out with financial support from the National Institutes of Health. S.G. is supported by a postdoctoral grant from the Wenner-Gren Foundations (Sweden). C.S. is supported by a EMBO long-term fellowship. V.Q. is supported by a postdoctoral grant from the Ministerio de Educación y Ciencia (Spain).
We thank Erica Young for technical assistance.
Footnotes
Published ahead of print on 18 July 2007.
REFERENCES
- 1.Amerik, A. Y., and M. Hochstrasser. 2004. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695:189-207. [DOI] [PubMed] [Google Scholar]
- 2.Balakirev, M. Y., M. Jaquinod, A. L. Haas, and J. Chroboczek. 2002. Deubiquitinating function of adenovirus proteinase. J. Virol. 76:6323-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barry, M., and K. Fruh. 2006. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci. STKE 2006:pe21. [DOI] [PubMed] [Google Scholar]
- 5.Blasdell, K., C. McCracken, A. Morris, A. A. Nash, M. Begon, M. Bennett, and J. P. Stewart. 2003. The wood mouse is a natural host for Murid herpesvirus 4. J. Gen. Virol. 84:111-113. [DOI] [PubMed] [Google Scholar]
- 6.Borodovsky, A., B. M. Kessler, R. Casagrande, H. S. Overkleeft, K. D. Wilkinson, and H. L. Ploegh. 2001. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 20:5187-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borodovsky, A., H. Ovaa, N. Kolli, T. Gan-Erdene, K. D. Wilkinson, H. L. Ploegh, and B. M. Kessler. 2002. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 9:1149-1159. [DOI] [PubMed] [Google Scholar]
- 8.Cardin, R. D., J. W. Brooks, S. R. Sarawar, and P. C. Doherty. 1996. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J. Exp. Med. 184:863-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 11.Hemelaar, J., A. Borodovsky, B. M. Kessler, D. Reverter, J. Cook, N. Kolli, T. Gan-Erdene, K. D. Wilkinson, G. Gill, C. D. Lima, H. L. Ploegh, and H. Ovaa. 2004. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol. Cell. Biol. 24:84-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holowaty, M. N., and L. Frappier. 2004. HAUSP/USP7 as an Epstein-Barr virus target. Biochem. Soc. Trans. 32:731-732. [DOI] [PubMed] [Google Scholar]
- 13.Holowaty, M. N., M. Zeghouf, H. Wu, J. Tellam, V. Athanasopoulos, J. Greenblatt, and L. Frappier. 2003. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 278:29987-29994. [DOI] [PubMed] [Google Scholar]
- 14.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547-557. [DOI] [PubMed] [Google Scholar]
- 15.Kopp, J., and T. Schwede. 2004. The SWISS-MODEL repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 32:D230-D234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, J. I., G. W. Luxton, and G. A. Smith. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80:12086-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindner, H. A., N. Fotouhi-Ardakani, V. Lytvyn, P. Lachance, T. Sulea, and R. Menard. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 79:15199-15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misaghi, S., Z. R. Balsara, A. Catic, E. Spooner, H. L. Ploegh, and M. N. Starnbach. 2006. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol. Microbiol. 61:142-150. [DOI] [PubMed] [Google Scholar]
- 19.Nijman, S. M., M. P. Luna-Vargas, A. Velds, T. R. Brummelkamp, A. M. Dirac, T. K. Sixma, and R. Bernards. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123:773-786. [DOI] [PubMed] [Google Scholar]
- 20.Orth, K., Z. Xu, M. B. Mudgett, Z. Q. Bao, L. E. Palmer, J. B. Bliska, W. F. Mangel, B. Staskawicz, and J. E. Dixon. 2000. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290:1594-1597. [DOI] [PubMed] [Google Scholar]
- 21.Rytkonen, A., J. Poh, J. Garmendia, C. Boyle, A. Thompson, M. Liu, P. Freemont, J. C. Hinton, and D. W. Holden. 2007. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc. Natl. Acad. Sci. USA 104:3502-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlieker, C., G. A. Korbel, L. M. Kattenhorn, and H. L. Ploegh. 2005. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J. Virol. 79:15582-15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlieker, C., W. A. Weihofen, E. Frijns, L. M. Kattenhorn, R. Gaudet, and H. L. Ploegh. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell 25:677-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevenson, P. G., S. Efstathiou, P. C. Doherty, and P. J. Lehner. 2000. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. USA 97:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Virgin, H. W. T., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 80:6003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou, H., D. M. Monack, N. Kayagaki, I. Wertz, J. Yin, B. Wolf, and V. M. Dixit. 2005. Yersinia virulence factor YopJ acts as a deubiquitinase to inhibit NF-kappa B activation. J. Exp. Med. 202:1327-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]