Abstract
Activated microglia and astroglia are known to be involved in a variety of neurodegenerative diseases, including prion diseases. In the present experiments, we studied activation of astroglia and microglia after intraocular scrapie infection in transgenic mice expressing prion protein (PrP) in multiple cell types (tg7 mice) or in neurons only (tgNSE mice). In this model, scrapie infection and protease-resistant PrP deposition occurs in the retinas of both strains of mice, but retinal degeneration is observed only in tg7 mice. Our results showed that the retinas of tg7 and tgNSE mice both had astroglial activation with increased chemokine expression during the course of infection. However, only tg7 retinas exhibited strong microglial activation compared to tgNSE retinas, which showed little microglial activation by biochemical or morphological criteria. Therefore, microglial PrP expression might be required for scrapie-induced retinal microglial activation and damage. Furthermore, microglial activation preceded retinal neurodegeneration in tg7 mice, suggesting that activated microglia might contribute to the degenerative process, rather than being a response to the damage. Surprisingly, brain differed from retina in that an altered profile of microglial activation markers was upregulated, and the profiles in the two mouse strains were indistinguishable. Microglial activation in the brain was associated with severe brain vacuolation and neurodegeneration, leading to death. Thus, retinal and brain microglia appeared to differ in their requirements for activation, suggesting that different activation pathways occur in the two tissues.
Transmissible spongiform encephalopathies, or prion diseases, are a group of fatal degenerative brain diseases that occur naturally in primates and ruminants. They include scrapie in sheep, bovine spongiform encephalopathy in cattle, and several human diseases, such as Creutzfeldt-Jakob disease, kuru, and Gerstmann-Sträussler-Scheinker syndrome. The normal host prion protein (PrP) is required for propagation of the scrapie agent and for development of clinical disease (7, 9), and conversion of normal protease-sensitive PrP (PrP-sen) to an aggregated partially protease-resistant structure (PrP-res) is a critical event in the disease process. PrP is attached to the cell surface by a glycolipid anchor and is normally expressed in most cells and tissues, including neurons (28), astrocytes (28, 35, 50), lymphocytes (11), follicular dendritic cells (34), and tumor cell lines of various lineages (12).
Both neurons and astrocytes have been emphasized as major sites of agent replication and PrP-res accumulation (3, 31, 44, 45). In several models of scrapie infection, the initial pathological changes identified have been the deposition and accumulation of PrP-res within a particular brain area simultaneously with or closely followed by astroglial activation, and later by neuropil vacuolation and, in some cases, neuronal loss (6, 19, 51). However, microglia are known to be required for some models of in vitro PrP peptide-induced toxicity (4, 5, 39), and there is a growing body of evidence that implicates microglial cells as potential mediators of in vivo neurodegeneration in transmissible spongiform encephalopathy disease (2, 5, 8, 16, 19, 46, 53). There is also evidence to support the hypothesis that microglia are activated early by exposure to infection or PrP-res and that these activated microglia may contribute to neuronal damage and clinical disease (23, 33, 51).
As an extension of the central nervous system (CNS), the retina has often been featured in pathological studies because of its relatively simple architecture and well-described physiological properties. Direct intraocular (i.o.) infection has the advantage of initiating a localized infection in the retina, in addition to determining pathogenic effects in the brain by providing the ability to follow the infection through well-defined visual tracts. Other mouse scrapie experiments have successfully utilized i.o. inoculation with the mouse scrapie strain ME7 (17, 37, 47, 48). Furthermore, recent work using scrapie infection of sheep has validated the importance of this model in a natural infection and indicated the retina and brain visual system as a prominent site of PrP-res accumulation and pathology (21).
Using transgenic mice that express hamster PrP (HaPrP) in multiple cell types (tg7 mice) (43) or mice that have HaPrP expression restricted to neurons (tgNSE mice) (44), we previously showed that tg7 retinas progressed to full degeneration following i.o. inoculation with hamster scrapie strain 263K, while tgNSE retinas exhibited few degenerative changes (25). In both strains of mice, the infection followed the visual tracts to the brain, and the mice succumbed to disease.
In the present study, we used this i.o. infection model to explore the relationship between the inflammatory response and neurodegeneration in both retina and brain by monitoring chemokines and astroglial and microglial activation markers. After retinal infection, astroglial markers and chemokines were upregulated in both tg7 and tgNSE mice. In contrast, microglial-activation markers were increased in the retinas of tg7 but not tgNSE mice. This activation was closely followed by severe retinal degeneration. These results suggested that scrapie infection of tg7 mice expressing PrP-sen on both neuronal and nonneuronal cell types is strongly associated with microglial activation, which could contribute to the subsequent retinal degeneration process.
MATERIALS AND METHODS
Infection of mice.
All mice were bred and raised at the Rocky Mountain Laboratories and were handled according to the policies of the Rocky Mountain Laboratories Animal Care and Use Committee and all applicable federal guidelines. Adult (6- to 8-week-old) animals were used for all experiments. The transgenic animals used for this study have been described previously (43,44). For i.o. inoculations, mice were deeply anesthetized by intramuscular injection of a combination anesthetic cocktail containing ketamine, xylazine, and acepromazine. Approximately 2 × 105 the intracranial 50% infectious dose (ID50) of hamster 263K scrapie infectivity in 2 μl of phosphate-buffered saline (PBS) (pH 7.2) supplemented with 2% fetal bovine serum (HyClone, Logan, UT) was injected unilaterally into the vitreous cavity using a 32-gauge needle attached to a 10-μl Hamilton syringe. After injection, the needle was left in the vitreal chamber for 1 min to minimize leaking of the inoculum. All mice were observed several times each week for clinical signs of scrapie, which included weight loss, kyphosis, ataxia, and an exaggerated high-stepping gait most noticeable in the hind limbs (43). Mice exhibiting short incubation periods (less than 100 days) died within 1 to 4 days after the appearance of clinical symptoms, whereas mice exhibiting longer incubation periods had more prolonged clinical disease that lasted 10 to 14 days. Brain samples from mice in each group with clinical evidence of scrapie were analyzed for PrP-res by Western blotting to confirm the clinical diagnosis (44).
Pathology and immunohistochemistry.
Mice used for histopathological analysis were lightly anesthetized, and then transcardially perfused with 30 ml of PBS. The eyes were removed and placed in Davidson's fixative (3 parts 100% ethanol, 2 parts 37 to 40% formaldehyde, and 1 part glacial acetic acid) for 24 h before dehydration and embedding in paraffin. Whole brains were removed and placed in 3.7% phosphate-buffered formalin for 3 to 5 days and then cut into 2- to 3-mm coronal sections before dehydration and embedding in paraffin. Serial 4-μm sections were cut using a standard Leica microtome, placed on positively charged glass slides, and dried overnight at 56°C. All histopathological procedures were performed on brain and eye sections from both mock-infected and scrapie-infected animals, including appropriate immunohistochemical controls. Immunohistochemical staining was performed using the Ventana automated Nexus stainer (Ventana, Tucson, AZ). Slides were deparaffinized and rehydrated to Tris-HCl buffer, pH 7.5 (glial fibrillary acidic protein [GFAP] staining), or 0.1 mM citrate buffer, pH 6.0 (3F4 staining). Staining for GFAP utilized a standard avidin-biotin complex immunoperoxidase protocol using anti-GFAP at a dilution of 1:1,000 (Dako, Carpinteria, CA), biotinylated goat anti-rabbit immunoglobulin G (IgG) at a dilution of 1:250 (Vector Laboratories, Burlingame, CA), and amino-ethyl carbazole as a substrate (Ventana). For the detection of PrP-res, it is necessary to perform antigen retrieval by treating the sections with formic acid or subjecting them to autoclaving and/or proteinase K digestion (20, 27). The sections in this study were rehydrated and autoclaved for 20 min at 122°C and 22 lb/in2 in 0.1 mM citrate buffer (pH 6.0). PrP-res was then stained with mouse monoclonal antibody 3F4 cell culture supernatant (1:50) (24). Detection was performed with biotinylated horse anti-mouse IgG diluted 1:250 (Vector), followed by Supersensitive Streptavidin diluted 1:3 in Tris-HCl, pH 7.5, buffer (Biogenex) and amino-ethyl carbazole. For the Iba1 stain (22), slides were deparaffinized and rehydrated to 0.1 mM citrate buffer, pH 6.0. The slides were pretreated in citrate buffer for 30 seconds at 120°C using the Biocare decloaking chamber (Walnut Creek, CA) and then incubated with a standard avidin-biotin complex immunoperoxidase protocol using anti-Iba1 at a dilution of 1:1,000 (WAKO Chemicals, Richmond, VA), biotinylated goat anti-rabbit IgG at a dilution of 1:250 (Vector), and amino-ethyl carbazole as a substrate (Ventana).
Glial cell culture and staining.
Primary glial cells were prepared from the brains of both tg7 and tgNSE neonatal mice (12 to 36 h postbirth). The brains were dissected to remove the cerebella, olfactory bulbs, and midbrains. The remaining cerebral cortices were triturated under sterile conditions in Dulbecco's modified Eagle's medium/F12 medium (GIBCO, Carlsbad, CA) containing 10% fetal bovine serum (HyClone, Logan, UT), 100 μg/ml streptomycin, 100 U/ml penicillin, and 2 mM l-glutamine (GIBCO) and cultured in 25-cm2 flasks under standard conditions. The confluent (5- to 7-day-old) cultures were agitated overnight on a rotary shaker at 37°C and 250 rpm to remove the microglia. The separated microglia were transferred to Lab-Tek Chamber Slides (Nunc, Naperville, IL) and allowed to grow for 48 to 72 h to a subconfluent density prior to being stained. Astrocytes were removed from the 25-cm2 flasks by trypsinization and were similarly transferred to Lab-Tek slide chambers for culture and staining.
To detect PrP-sen expression, tg7 and tgNSE microglial and astroglial cell cultures were rinsed with PBS and then incubated for 30 min at 37°C with 1:500 monoclonal antibody 3F4 (24). The cells were rinsed with PBS and fixed for 15 min in 3.7% formaldehyde, followed by permeabilization in a solution of 0.1% Triton X-100 and 0.1% sodium citrate for 10 min. The slides were blocked for 30 min with 0.1 M glycine in PBS and for 30 min with 3% bovine serum albumin in PBS (Sigma, St. Louis, MO). To distinguish cell types, either rabbit anti-GFAP (Dako) or rabbit anti-Iba1 (WAKO) was applied (1:1,000) for 30 min at 37°C. Anti-3F4 was visualized by reaction with a secondary antibody, Alexa Fluor 488 goat anti-mouse IgG (Invitrogen, Carlsbad, CA), at 1:2,500 for 30 min at 37°C. Anti-GFAP or anti-Iba1 was visualized by reaction with Alexa Fluor 568 goat anti-rabbit IgG at 1:2,500 for 30 min at 37°C. The slides were then mounted with coverslips using ProLong Gold antifade reagent with DAPI (4′,6′-diamidino-2-phenylindole) (Invitrogen, Eugene, OR). The slides were examined on an Olympus BX51 microscope (Olympus America, Melville, NY), and images were captured with MicroSuite 5 imaging software (Soft Imaging System Corp., Lakewood, CO).
Infectivity titration.
tg7 and tgNSE mice were inoculated i.o. as described above. At several time points postinfection (p.i.), the eyes from three individual mice were removed and homogenized (10% [wt/vol]) in Tris buffer (pH 7.2). The homogenates were spun at 600 × g for 5 min to remove large cell debris and then sonicated for 1 min. Serial 10-fold dilutions (10−1 to 10−8) of each homogenate were made in PBS supplemented with 2% fetal calf serum, and then 50 μl of each dilution was inoculated intracranially into groups of four tg7 recipient mice. The mice were observed weekly and sacrificed upon the appearance of clinical symptoms or after >400 days. Infectivity titers/50 μl homogenate (ID50) were determined by the Spearman-Karber method (15).
Real-time quantitative RT-PCR.
Quantitative real-time reverse transcriptase (RT) PCR was performed as previously described, with some modifications (14). Brain and eye tissues were harvested and immediately placed in RNALater (QIAGEN, Valencia, CA). Total RNA was isolated from whole eye and brain tissues of individual transgenic mice using the RNeasy minikit (QIAGEN) plus DNase treatment. The left half of each brain was dissected into the thalamus, superior colliculus, and dorsal cortex, and each piece was processed individually for total RNA. Each RNA was reversed transcribed into cDNA using reverse transcription reagents with random hexamers (Applied Biosystems, Foster City, CA). The cDNAs were PCR amplified in triplicate by using TaqMan PCR Master Mix (Applied Biosystems) in a total reaction volume of 10 μl. All primer and probe sequences used were from gene expression assays (Applied Biosystems). The relative quantity of each gene amplified was normalized with mouse β-actin for each sample. The normalized relative expression values for each gene from scrapie-infected samples were compared with the gene expression levels from mock-infected samples to determine the differences.
Immunoblot analysis for PrP-res.
Eye tissues were analyzed for the presence of PrP using Western blotting techniques as previously described (42), with some modifications. For PrP-res, 10% (wt/vol) homogenates of eye tissue were made in modified RIPA buffer (5 mM Tris-HCl, pH 7.4, 0.5% Triton X-100, 0.5% sodium deoxycholate, 150 mM NaCl, and 5 mM EDTA). To remove nucleic acids, the homogenates were treated with 20 U Benzoase nuclease (Novagen, Madison, WI) for 30 min at 37°C, and debris was removed by centrifugation at 1,000 × g for 5 min at 4°C. The homogenates were treated with a final concentration of 50 μg/ml proteinase K for 1 hour at 37°C. Five-milligram tissue equivalents were diluted in sample buffer and run on 16% polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride membranes and probed with monoclonal antibody 3F4 as described previously, except that detection was with goat anti-mouse IgG-horseradish peroxidase (1:3,000) and standard enhanced-chemiluminescence procedures (GE Healthcare, Piscataway, NJ). Detection for enhanced chemifluorescence was performed with goat anti-mouse IgG-alkaline phosphatase (1:2,500). The blots were dried for 2 h, scanned with a Storm phosphorimager, and then analyzed with ImageQuaNT V5.2 software (Molecular Dynamics, Sunnyvale, CA).
RESULTS
PrP-sen expression in astroglia and microglia of tg7 and tgNSE mice.
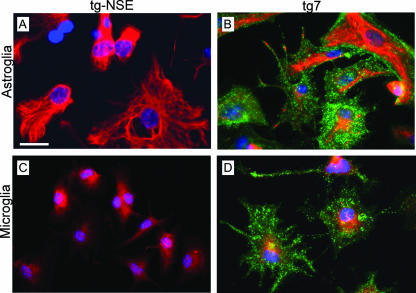
Expression of PrP-sen is required for susceptibility to prion disease infection. tg7 and tgNSE mice are both known to express PrP-sen in neurons (43, 44). Based on the broad cell type specificity of the PrP promoter (tg7) and the narrow “neuron-only” specificity of the NSE promoter (tgNSE), expression of PrP-sen in nonneuronal cells, including astroglia and microglia, was expected in tg7, but not in tgNSE, mice. Because of the possible importance of glial cells and glial cell-type-specific PrP expression in the current study, we examined PrP-sen expression directly in glial cultures from these mice. In these experiments, tgNSE astroglia and microglia had no detectable PrP-sen expression (Fig. 1A and C), whereas both astroglia and microglia from tg7 mice showed strong PrP-sen reactivity (Fig. 1B and D). Thus, the differences in PrP-sen expression in glial cells of tg7 and tgNSE mice might alter scrapie infection or stimulation of these cells by PrP-res, thereby influencing the pathogenic process.
FIG. 1.
Immunofluorescent staining of PrP-sen expressed by cultured astroglia and microglia from tg7 and tgNSE mice. All cells were stained live for HaPrP (green) with monoclonal antibody 3F4 and then fixed, permeabilized, and stained with either anti-GFAP (A and B) or anti-Iba1 (C and D, red). Primary antibodies were visualized with Alexa Fluor-conjugated secondary antibodies as described in Materials and Methods. Note the intense green PrP staining in both astroglial and microglial tg7 cells (B and D), but not in tgNSE cells (A and C). Scale bar, 25 μm. Original magnification, ×40.
Scrapie-induced pathogenesis in transgenic retinas.
We previously showed that after i.o. scrapie infection, both tg7 and tgNSE mice generated PrP-res in the retina by 6 to 8 weeks p.i. (25). Furthermore, both mouse strains had PrP-res deposition and astrogliosis in the retina, but only tg7 mice had retinal degeneration.
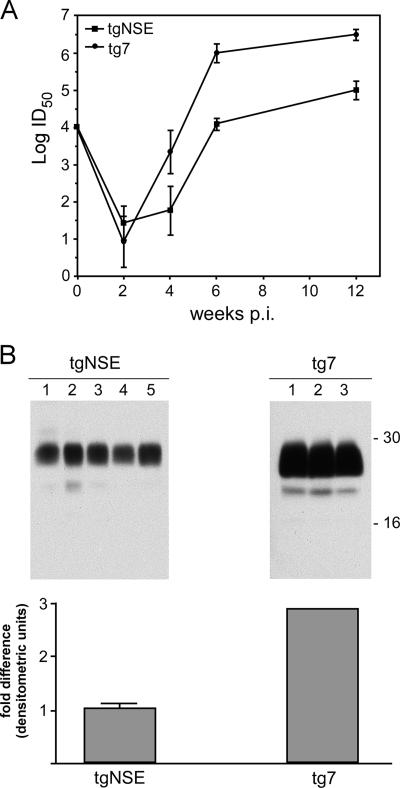
It was surprising that tgNSE retinas were not damaged by scrapie infection. To demonstrate ocular scrapie infection and replication in these mice, we studied scrapie infection using a quantitative end point dilution infectivity assay. After i.o. scrapie infection in both mouse strains, an initial drop in infectivity at 2 weeks p.i. was followed by a steady increase in titer (Fig. 2A). There was significant propagation of infectivity in both strains, but tg7 mice had 30- to 50-fold-higher levels than tgNSE mice at 4, 6, and 12 weeks p.i., possibly due to the presence of PrP-sen in a wider variety of retinal cell types in tg7 mice.
FIG. 2.
(A) Infectivity from eyes of scrapie-infected tg7 and tgNSE mice. Whole eyes from mice that were i.o. infected (three mice per time point) were homogenized and titered by end point dilution in tg7 indicator mice. Log10 ID50/50 μl 10% eye homogenate is plotted for various time points up to 12 weeks p.i. Time zero is the i.o. dose given (104 ID50 in 2 μl/eye). The error bars indicate standard deviations. (B) Western blot of PrP-res from clinically sick tg7 and tgNSE mice. Each 10% eye homogenate was treated with proteinase K as described in Materials and Methods, and PrP-res bands representing multiple glycosylation forms are indicated by approximate molecular mass markers (16 to 30 kDa). Lanes 1 to 3 represent proteins from three individual tg7 animals, and lanes 1 to 5 represent proteins from five individual tgNSE animals. The gels were loaded with equivalent amounts of protein (5 mg/10 μl tissue equivalents), and exposure times were equivalent. The blots were processed, and densitometric analysis was performed to generate average units per eye homogenate. The difference (n-fold) in densitometric units relative to the tgNSE eye is shown.
We have previously demonstrated equal retinal expression of the normal PrP, PrP-sen, in both strains (25). To quantitate the amount of PrP-res that accumulated in transgenic retinas, Western blotting of retinal extracts from scrapie-infected transgenic mice was performed at 12 weeks p.i., which was near the clinical time of disease. After i.o. infection, tg7 mice accumulated threefold-higher levels of PrP-res than tgNSE mice (Fig. 2B). This small difference in PrP-res accumulation did not seem sufficient to account for the major difference in retinal damage seen in the two mouse strains. However, it suggested that the differences in PrP-sen expression on nonneuronal cells in the two strains might contribute to the retinal-degeneration patterns observed.
Histopathology and biochemical studies of infected retinas.
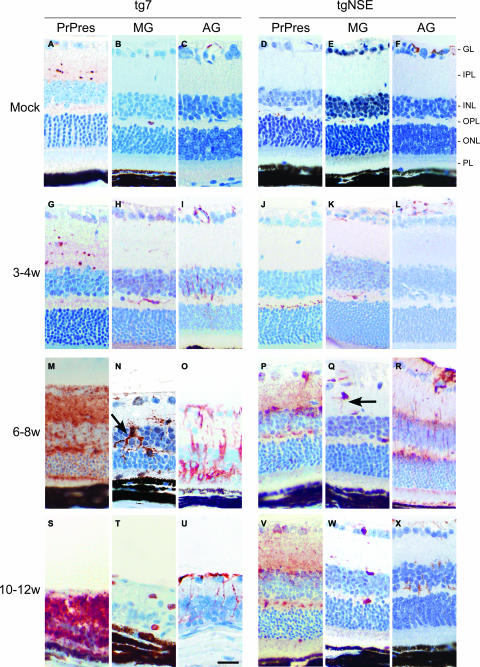
To study the kinetics of the pathogenic process in i.o. infected mice, immunohistochemistry was done on retinal sections from various times p.i. PrP-res deposition and the presence of GFAP-positive activated astrocytes were first detected in both mouse strains at 4 weeks p.i. The number of activated astrocytes increased and spread to multiple retinal layers at subsequent time points, until retinal degeneration at 12 weeks p.i. led to a decrease in all cell types seen (Fig. 3).
FIG. 3.
Immunohistochemical analysis of PrP-res, microglia, and activated astroglia in scrapie-infected tg7 and tgNSE retinas. Representative sections from uninfected (mock) or scrapie-infected mice sacrificed at 3 to 4 weeks (w), 6 to 8 weeks, or 10 to 12 weeks (clinical) p.i. are shown. Immunohistochemical staining was performed with anti-3F4 antibody (PrP-res), anti-Iba1 antibody for microglia (MG), or anti-GFAP antibody for activated astroglia (AG), as described in Materials and Methods. Compare activated microglia with thickened processes (arrow) in tg7 mice (N) to microglial cells with thin processes (arrow) in tgNSE mice (Q). Retinal layers are indicated as follows: PL, photoreceptor layer; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GL, ganglion layer. Scale bar, 20 μm (in panel U). Original magnification, ×40.
To observe microglia by immunohistochemistry, we used anti-Iba1, which detects both activated and nonactivated microglia and macrophages (22). In tg7 mice, an increase in the number of microglia was first detected at 3 to 4 weeks p.i., and by 6 to 8 weeks p.i., these cells were present in multiple retinal layers and showed thickened processes typically associated with activated microglia (Fig. 3N). These microglia were first detectable slightly preceding retinal degeneration, which began at 8 weeks p.i. and was severe at 10 to 12 weeks p.i. (Fig. 3T). Therefore, they might be contributing to the retinal degenerative process in tg7 mice. In contrast, tgNSE mice had only a slight increase in the number of microglia, mainly in the plexiform layers, at 6 to 8 weeks p.i., and these cells usually had thin processes characteristic of nonactivated microglia (Fig. 3Q). Furthermore, the number of these cells was not altered during the ensuing time up to 10 to 12 weeks p.i., which was near the end stage of disease (Fig. 3W).
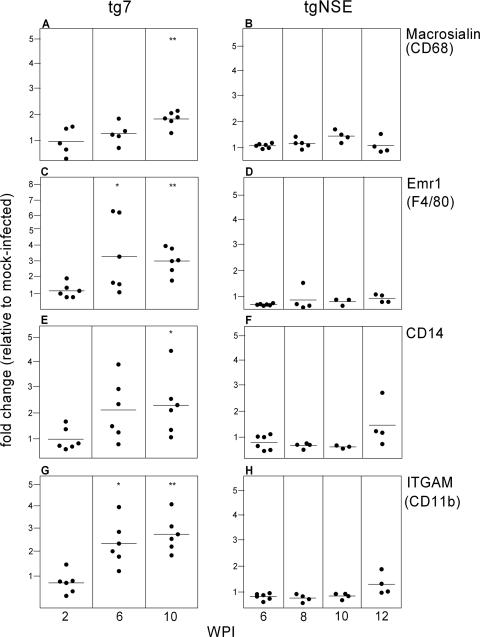
To determine more precisely whether the retinal microglia seen in tg7 and tgNSE mice were activated, we studied the expression of four different genes previously shown to be associated with activation of microglia (13, 46). Four transcripts were measured by RT-PCR in the infected retinas: macrosialin (CD68), a panmicroglia marker and a member of the Lamp family of receptors; Emr1 (F4/80), of the EGF/Tm7 family; CD14, a member of the class A scavenger receptors; and ITGAM (CD11b), of the integrin family. In the tg7 retinas, expression of all four microglial-activation markers was significantly increased at 6 and 10 weeks p.i. (Fig. 4). In contrast, in tgNSE mice, none of these markers were significantly upregulated at any time between 6 and 12 weeks p.i. Thus, these gene expression analyses indicated that activation of retinal microglia occurred following scrapie infection in tg7 mice but not in tgNSE mice.
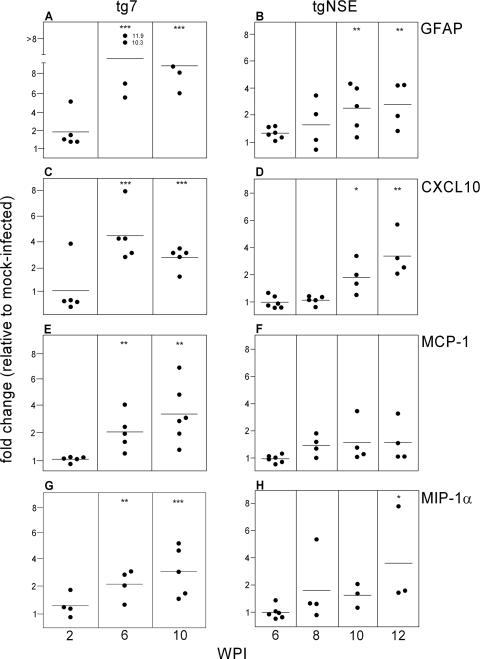
FIG. 4.
Real-time RT-PCR of tg7 and tgNSE eye RNAs for microglial-marker expression. The graphs show changes for individual eyes at 2, 6, and 10 weeks p.i. (tg7) and 6, 8, 10, and 12 weeks p.i. (tgNSE). The RNA was examined for macrosialin (CD68) (A and B), Emr1 (F4/80) (C and D), CD14 (E and F), and ITGAM (CD11b) (G and H). For each time point, four to six infected animals, as well as three or four mock-infected animals, were tested. The horizontal lines indicate the mean change, and the asterisks indicate statistically significant changes versus mock-infected controls (Mann-Whitney test; *, <0.05; **, <0.005).
Upregulation of chemokine RNAs in scrapie-infected retinas.
Several proinflammatory cytokines and chemokines have been shown previously to be associated with scrapie pathogenesis (10, 16, 30). To investigate whether such molecules were produced in scrapie-infected retinas from our transgenic mice, we used real-time quantitative RT-PCR to test mRNAs at several times p.i. We tested the expression levels of 15 different genes, many of which have been detected in various models of CNS degenerative, infectious, or inflammatory diseases (1, 10, 16, 26, 29, 32, 37, 49, 52, 54). Of the genes examined, the 11 unaltered genes were those for interleukin-1α, tumor necrosis factor alpha, CCL5 (RANTES), CCL4 (MIP-1β), CXCR3, CHOP-1, C1q-β, iNOS, eNOS, nNOS, and XCT-1. Only GFAP, an astrocyte activation marker, and the chemokines CCL3 (MIP-1α), CCL2 (MCP-1), and CXCL10 (IP-10) were found to be altered. In all four cases, upregulation was detected in both mouse strains, but the kinetics were slightly faster and the levels were higher in the tg7 mice (Fig. 5). Since all three of the upregulated chemokines have been shown to be produced by activated astrocytes, this result was consistent with our histopathological results showing astroglial activation. Possibly in tg7 mice activated microglia might also produce some of these chemokines, and this could account for the slightly increased levels seen in tg7 retinas.
FIG. 5.
Real-time RT-PCR of tg7 and tgNSE eye RNAs for chemokine expression. The graphs show changes for individual eyes at 2, 6, and 10 weeks p.i. (tg7) and 6, 8, 10, and 12 weeks p.i. (tgNSE). The RNA was examined for GFAP (A and B), CXCL10 (C and D), MCP-1 (E and F), and MIP-1α (G and H). For each time point, four to six infected animals and three or four mock-infected animals were tested. The horizontal lines indicate the mean change, and the asterisks indicate statistically significant changes versus mock-infected controls (Mann-Whitney test; *, <0.05; **, <0.005; ***, <0.0005).
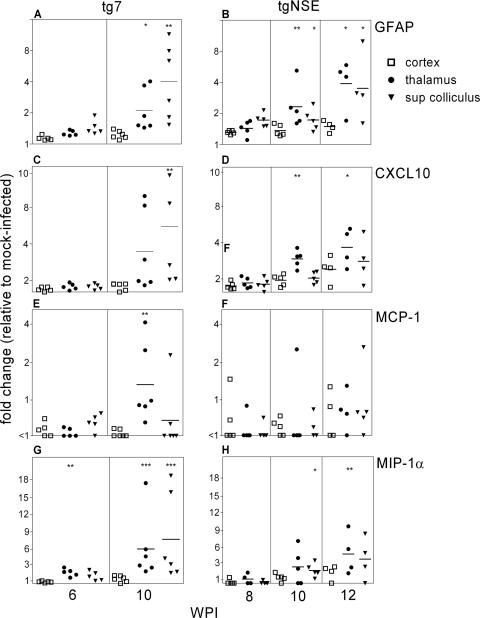
Astroglial and microglial activation in transgenic mouse brains.
Following ocular scrapie infection, PrP-res ascends via the visual tract from the eye to the visual areas of the brain (17, 25, 48). To study whether scrapie-induced activation of microglia and astroglia in the brain was similar to activation in the retina, we tested two areas of the visual system (the thalamus and superior colliculus) and one nonvisual region (the dorsal cortex) of the brains of tg7 and tgNSE mice for expression levels of the genes previously studied in the retina. Of this group, the 11 genes that were unaltered in the retina were also unaltered in the brain. The four upregulated genes seen in the retina, those for GFAP, CCL2, CCL3, and CXCL10, were also upregulated in visual areas of both infected tg7 and tgNSE mice (Fig. 6). These alterations were likely scrapie specific, as they were not seen in the dorsal cortex, which was only minimally infected at these times.
FIG. 6.
Real-time RT-PCR of tg7 and tgNSE brain RNAs for chemokine expression. The graphs show changes for individual thalamus, superior colliculus, and cortex brain regions at 6 and 10 weeks p.i. (tg7) and 8, 10, and 12 weeks p.i. (tgNSE). The RNA was examined for GFAP (A and B), CXCL10 (C and D), MCP-1 (E and F), and MIP-1α (G and H). For each time point, four to six infected animals and three or four mock-infected animals were tested. The horizontal lines indicate the mean change, and the asterisks indicate statistically significant changes versus mock-infected controls (Mann-Whitney test; *, <0.05; **, <0.005).
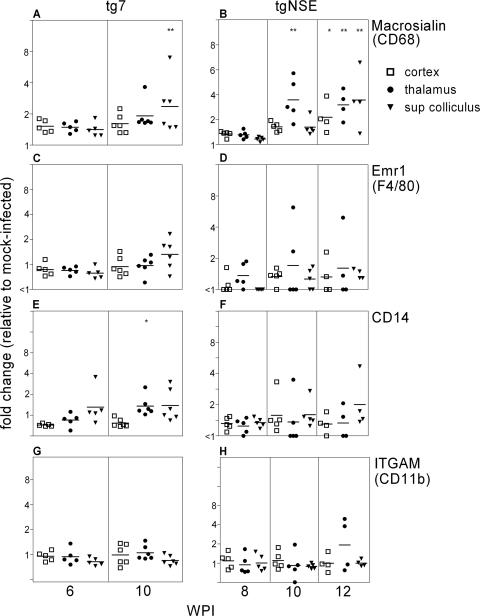
When we tested the microglial markers, only macrosialin (CD68) was increased in the visual areas, and surprisingly, this occurred in both tg7 and tgNSE mice (Fig. 7). The other three microglial activation markers, Emr1 (F4/80), CD14, and ITGAM (CD11b), were not increased significantly in the brain. Thus, the profile of activation markers expressed in brain microglia differed markedly from that in the retina, where all four microglial activation markers were upregulated. The timing of upregulation of macrosialin was somewhat delayed in the brain compared to the retina (6 weeks in the retina versus 10 weeks in the thalamus and superior colliculus), but this delay was likely due to the time required for PrP-res generation to progress from the retina to the brain via the optic nerve.
FIG. 7.
Real-time RT-PCR of tg7 and tgNSE brain RNAs for microglial-marker expression. The graphs show changes for individual thalamus, superior colliculus, and cortex brain regions at 6 and 10 weeks p.i. (tg7) and 8, 10, and 12 weeks p.i. (tgNSE). The RNA was examined for Macrosialin (CD68) (A and B), Emr1 (F4/80) (C and D), CD14 (E and F), and ITGAM (CD11b) (G and H). For each time point, four to six infected animals and three or four mock-infected animals were tested. The horizontal lines indicate the mean change, and the asterisks indicate statistically significant changes versus mock-infected controls (Mann-Whitney test; *, <0.05; **, <0.005).
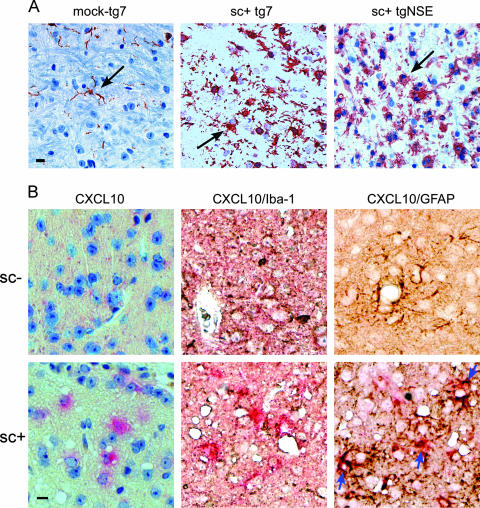
To confirm that these biochemical results truly were indicative of microglial activation, brain sections were immunostained with anti-Iba1. In scrapie-infected mice of both tg7 and tgNSE lines, there was obvious evidence of a very large increase in the number of Iba1-positive microglia in the thalamus (Fig. 8A), and many of these cells had plumb cell bodies and short stubby processes characteristic of activated microglia. Therefore, immunohistochemical analysis correlated with the RNA analysis, suggesting that activated microglia in both strains of mice were associated with areas of neurodegeneration. Furthermore, since there was extensive neurodegeneration and vacuolation leading to death in both tg7 and tgNSE mice, the presence of activated microglia in the brain correlated with neuronal damage in both strains.
FIG. 8.
(A) Comparison of Iba1-positive microglia in the thalamus of scrapie-infected (sc+) tg7 and tgNSE mice at the time of clinical disease versus an aged-matched tg7 mock-infected control. Immunohistochemical staining was performed with anti-Iba1 antibody as described in Materials and Methods. The arrows indicate fine microglial processes in the mock-infected brain compared to thickened processes and more numerous cells in the infected brain. Scale bar, 20 μm. (B) In situ hybridization and immunohistochemical analysis of CXCL10 expression in astrocytes of mock-infected (sc−) or scrapie-infected (sc+) tg7 mice. Representative sections from the superior colliculus were hybridized with digoxigenin-labeled antisense RNA for CXCL10 and developed using Fast Red stain (bright pink around blue nuclei). The sections were then incubated with anti-Iba1 or anti-GFAP and developed with diaminobenzidine (brown/black). All sections were counterstained with hematoxylin. The blue arrows indicate colocalization of CXCL10 expression with anti-GFAP stain. This colocalization was not seen with anti-Iba1. Scale bar = 20 μm. Original magnification, ×40.
DISCUSSION
In the present work, we studied the pathogenesis of retinal damage after i.o. hamster scrapie infection in two transgenic mouse strains that express HaPrP in differing cell types. In the retinas of both mouse lines, scrapie replication was detected, PrP-res was generated, and astroglial activation was observed; however, only tg7 mice developed microglial activation and retinal degeneration. In addition, since retinal gliosis and PrP-res accumulation slightly preceded retinal degeneration, activation of both astroglia and microglia appeared to be an early response to scrapie infection and PrP-res generation, rather than being a late response to retinal damage. Thus, activated microglia appeared to play a critical role in scrapie-induced retinal damage, in contrast to activated astroglia, which were not sufficient to induce retinal damage in scrapie-infected tgNSE mice.
Several possible mechanisms might explain these effects. First, since tg7 retinas had higher levels of scrapie infection and PrP-res than tgNSE retinas, these differences might account for the difference in retinal degeneration. Although PrP-res is likely to be the primary cause of initial retinal neuronal and glial stimulation, it seems unlikely that a threefold reduction in PrP-res levels in tgNSE mice would produce no detectable retinal damage during the 12 to 13 weeks before the mice succumbed to brain disease. Alternatively, it is possible that the 30- to 50-fold difference in scrapie infectivity is more important to the retinal damage than the level of PrP-res.
A second possible explanation for the lack of retinal degeneration in tgNSE mice is related to the lack of PrP-sen expression on tgNSE microglia (Fig. 1). Lack of PrP-sen expression might prevent microglial activation by PrP-res generated in retinal neurons. PrP-sen is a glycophosphatidylinositol-anchored protein known to interact with signal transduction proteins and has been shown to signal through the mitogen-activated protein kinase pathway following exposure to toxic PrP molecules (36, 40). Thus, tgNSE microglia could not be activated in this manner, since there is no microglial PrP-sen to initiate the process. Microglia can also be activated directly via their G-protein-coupled chemokine receptors; however, the chemokine signals coming from PrP-res-damaged neurons were insufficient to activate microglia in tgNSE retinas through this mechanism. Therefore, by either direct or indirect mechanisms, the PrP-negative microglia in tgNSE retinas appeared to be refractory to scrapie-induced activation.
Retinal microglia might also be activated indirectly by cytokines or other non-PrP molecules produced by activated astroglial cells. Chemokines CCL2, CCL3, and CXCL10 were detected in the retinas and brains of both mouse lines. Using in situ hybridization, CXCL10 was shown to be produced primarily by astroglia (Fig. 8B), and it is possible that the other chemokines are also produced by activated astrocytes, as this has been seen in other systems (38). However, in the present system, astrocyte activation in scrapie-infected tgNSE mice occurred in the absence of glial PrP-sen expression, and these activated astrocytes were apparently not sufficient to signal activation of retinal microglia or assist in the retinal neurodegenerative process in these mice.
A third possibility for the lack of retinal degeneration in tgNSE mice during scrapie infection could be the lack of microglial recruitment to the tgNSE retina. The lack of PrP-sen expression on tgNSE macrophages and monocytes might prevent these cells from being attracted to the retina by the generation of PrP-res during infection. In many chronic neurodegenerative diseases with different etiologies, such as Alzheimer's disease, multiple sclerosis, human immunodeficiency virus type 1 encephalitis, and Creutzfeldt-Jakob disease, accumulation and activation of microglia precede or are concomitant with neuronal and glial cell damage (1), indicating that neuronal and/or glial cell signals can both activate the resident glial cells and recruit the peripheral monocytes to the CNS. In scrapie retinal infection, microglia can be activated/recruited to the retina within days (33). In recent work by Priller et al. (41), peripheral scrapie infection facilitated brain engraftment by bone marrow-derived microglia, indicating a very efficient mechanism of CNS microglial recruitment associated with scrapie infection. However, in our own transplantation experiments, we did not observe a significant recruitment of microglia to the retina (data not shown). This could be due to retina-specific barriers or to the weaker chemokine signals from the tgNSE retina.
In the brain, microglial activation correlated with degeneration and was found in both tg7 and tgNSE mice. Therefore, the activation and function of tgNSE microglia are clearly different in the retina and brain. Possibly in tgNSE mice more PrP-res is produced by neurons in the brain than in the retina, and brain microglia might therefore be more readily activated. In addition, even in tg7 mice, resident brain microglia behaved differently than retinal microglia. For example, in the tg7 retina, upregulation of four different markers of microglial activation was seen (Fig. 3), whereas in the brain, only one marker (macrosialin) was significantly upregulated, and this was similar in tg7 and tgNSE thalamic and superior colliculus regions (Fig. 7). Macrosialin is a scavenger receptor found almost exclusively on macrophages, and therefore, these activated cells in the brain might function in part to phagocytose dead and/or dying cells. However, they probably do not represent perivascular macrophages that have migrated to the parenchyma, since that population of CNS phagocytes has recently been shown to lack expression of macrosialin (18). It is more likely that these cells represent a combination of resident microglia that became activated and infiltrating monocytes/macrophages from the blood. Possibly, the robust infection of the visual areas of the brain attracts infiltrating monocytes/macrophages from the blood, which could explain the similarly increased numbers of cells observed in the two strains of mice.
In summary, our experiments with transgenic mice indicated that activated microglia are strongly associated with neurodegeneration, but in the retina and brain, these cells might act by different mechanisms. The results in the tgNSE retinas are an example of limited damage, even in the presence of active PrP-res accumulation.
Acknowledgments
We thank Sue Priola, Sonja Best, and Kim Hasenkrug for careful reading of the manuscript and helpful suggestions.
Footnotes
Published ahead of print on 25 July 2007.
REFERENCES
- 1.Ambrosini, E., and F. Aloisi. 2004. Chemokines and glial cells: a complex network in the central nervous system Neurochem. Res. 29:1017-1038. [DOI] [PubMed] [Google Scholar]
- 2.Baker, C. A., D. Martin, and L. Manuelidis. 2002. Microglia from Creutzfeldt-Jakob disease-infected brains are infectious and show specific mRNA activation profiles J. Virol. 76:10905-10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barmada, S. J., and D. A. Harris. 2005. Visualization of prion infection in transgenic mice expressing green fluorescent protein-tagged prion protein. J. Neurosci. 25:5824-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bate, C., R. S. Boshuizen, J. P. Langeveld, and A. Williams. 2002. Temporal and spatial relationship between the death of PrP-damaged neurones and microglial activation. Neuroreport 13:1695-1700. [DOI] [PubMed] [Google Scholar]
- 5.Bate, C., S. Reid, and A. Williams. 2001. Killing of prion-damaged neurones by microglia. Neuroreport 12:2589-2594. [DOI] [PubMed] [Google Scholar]
- 6.Betmouni, S., and V. H. Perry. 1999. The acute inflammatory response in CNS following injection of prion brain homogenate or normal brain homogenate. Neuropathol. Appl. Neurobiol. 25:20-28. [DOI] [PubMed] [Google Scholar]
- 7.Brandner, S., A. Raeber, A. Sailer, T. Blattler, M. Fischer, C. Weissmann, and A. Aguzzi. 1996. Normal host prion protein (PrPC) is required for scrapie spread within the central nervous system. Proc. Natl. Acad. Sci. USA 93:13148-13151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, D. R., B. Schmidt, and H. A. Kretzschmar. 1996. Role of microglia and host prion protein in neurotoxicity of a prion protein fragment. Nature 380:345-347. [DOI] [PubMed] [Google Scholar]
- 9.Bueler, H., A. Aguzzi, A. Sailer, R. A. Greiner, P. Autenried, M. Aguet, and C. Weissmann. 1993. Mice devoid of PrP are resistant to scrapie. Cell 73:1339-1347. [DOI] [PubMed] [Google Scholar]
- 10.Burwinkel, M., C. Riemer, A. Schwarz, J. Schultz, S. Neidhold, T. Bamme, and M. Baier. 2004. Role of cytokines and chemokines in prion infections of the central nervous system. Int. J. Dev. Neurosci. 22:497-505. [DOI] [PubMed] [Google Scholar]
- 11.Cashman, N. R., R. Loertscher, J. Nalbantoglu, I. Shaw, R. J. Kascsak, D. C. Bolton, and P. E. Bendheim. 1990. Cellular isoform of the scrapie agent protein participates in lymphocyte activation. Cell 61:185-192. [DOI] [PubMed] [Google Scholar]
- 12.Caughey, B., R. E. Race, and B. Chesebro. 1988. Detection of prion protein mRNA in normal and scrapie-infected tissues and cell lines. J. Gen. Virol. 69:711-716. [DOI] [PubMed] [Google Scholar]
- 13.Chen, L., P. Yang, and A. Kijlstra. 2002. Distribution, markers, and functions of retinal microglia. Ocul. Immunol. Inflamm. 10:27-39. [DOI] [PubMed] [Google Scholar]
- 14.Dimcheff, D. E., S. Askovic, A. H. Baker, C. Johnson-Fowler, and J. L. Portis. 2003. Endoplasmic reticulum stress is a determinant of retrovirus-induced spongiform neurodegeneration. J. Virol. 77:12617-12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dougherty, R. M. 1964. Animal virus titration techniques, p. 183-186. In R. J. C. Harris (ed.), Techniques in experimental virology. Academic Press, San Diego, CA.
- 16.Eikelenboom, P., C. Bate, W. A. Van Gool, J. J. Hoozemans, J. M. Rozemuller, R. Veerhuis, and A. Williams. 2002. Neuroinflammation in Alzheimer's disease and prion. disease. Glia 40:232-239. [DOI] [PubMed] [Google Scholar]
- 17.Fraser, H., and A. G. Dickinson. 1985. Targeting of scrapie lesions and spread of agent via the retino-tectal projection. Brain Res. 346:32-41. [DOI] [PubMed] [Google Scholar]
- 18.Galea, I., K. Palin, T. A. Newman, R. N. Van, V. H. Perry, and D. Boche. 2005. Mannose receptor expression specifically reveals perivascular macrophages in normal, injured, and diseased mouse brain. Glia 49:375-384. [DOI] [PubMed] [Google Scholar]
- 19.Giese, A., D. R. Brown, M. H. Groschup, C. Feldmann, I. Haist, and H. A. Kretzschmar. 1998. Role of microglia in neuronal cell death in prion disease. Brain Pathol. 8:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardt, M., T. Baron, and M. H. Groschup. 2000. A comparative study of immunohistochemical methods for detecting abnormal prion protein with monoclonal and polyclonal antibodies. J. Comp. Pathol. 122:43-53. [DOI] [PubMed] [Google Scholar]
- 21.Hortells, P., M. Monzon, E. Monleon, C. Acin, A. Vargas, R. Bolea, L. Lujan, and J. J. Badiola. 2006. Pathological findings in retina and visual pathways associated to natural scrapie in sheep. Brain Res. 1108:188-194. [DOI] [PubMed] [Google Scholar]
- 22.Ito, D., Y. Imai, K. Ohsawa, K. Nakajima, Y. Fukuuchi, and S. Kohsaka. 1998. Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res. Mol. Brain Res. 57:1-9. [DOI] [PubMed] [Google Scholar]
- 23.Jeffrey, M., W. G. Halliday, J. Bell, A. R. Johnston, N. K. MacLeod, C. Ingham, A. R. Sayers, D. A. Brown, and J. R. Fraser. 2000. Synapse loss associated with abnormal PrP precedes neuronal degeneration in the scrapie-infected murine hippocampus. Neuropathol. Appl. Neurobiol. 26:41-54. [DOI] [PubMed] [Google Scholar]
- 24.Kascsak, R. J., R. Rubenstein, P. A. Merz, M. Tonna-DeMasi, R. Fersko, R. I. Carp, H. M. Wisniewski, and H. Diringer. 1987. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J. Virol. 61:3688-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kercher, L., C. Favara, C. C. Chan, R. Race, and B. Chesebro. 2004. Differences in scrapie-induced pathology of the retina and brain in transgenic mice that express hamster prion protein in neurons, astrocytes, or multiple cell types. Am. J. Pathol. 165:2055-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kielian, T. 2004. Microglia and chemokines in infectious diseases of the nervous system: views and reviews. Front. Biosci. 9:732-750. [DOI] [PubMed] [Google Scholar]
- 27.Kitamoto, T., J. Tateishi, T. Tashima, I. Takeshita, R. A. Barry, S. J. DeArmond, and S. B. Prusiner. 1986. Amyloid plaques in Creutzfeldt-Jakob disease stain with prion protein antibodies. Ann. Neurol. 20:204-208. [DOI] [PubMed] [Google Scholar]
- 28.Kretzschmar, H. A., S. B. Prusiner, L. E. Stowring, and S. J. DeArmond. 1986. Scrapie prion proteins are synthesized in neurons. Am. J. Pathol. 122:1-5. [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, H. P., Y. C. Jun, J. K. Choi, J. I. Kim, R. I. Carp, and Y. S. Kim. 2005. The expression of RANTES and chemokine receptors in the brains of scrapie-infected mice. J. Neuroimmunol. 158:26-33. [DOI] [PubMed] [Google Scholar]
- 30.Lewicki, H., A. Tishon, D. Homann, H. Mazarguil, F. Laval, V. C. Asensio, I. L. Campbell, S. DeArmond, B. Coon, C. Teng, J. E. Gairin, and M. B. Oldstone. 2003. T cells infiltrate the brain in murine and human transmissible spongiform encephalopathies. J. Virol. 77:3799-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallucci, G., A. Dickinson, J. Linehan, P. C. Klohn, S. Brandner, and J. Collinge. 2003. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science 302:871-874. [DOI] [PubMed] [Google Scholar]
- 32.Mander, P., and G. C. Brown. 2005. Activation of microglial NADPH oxidase is synergistic with glial iNOS expression in inducing neuronal death: a dual-key mechanism of inflammatory neurodegeneration J. Neuroinflamm. 2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marella, M., and J. Chabry. 2004. Neurons and astrocytes respond to prion infection by inducing microglia recruitment. J. Neurosci. 24:620-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McBride, P. A., P. Eikelenboom, G. Kraal, H. Fraser, and M. E. Bruce. 1992. PrP protein is associated with follicular dendritic cells of spleens and lymph nodes in uninfected and scrapie-infected mice. J. Pathol. 168:413-418. [DOI] [PubMed] [Google Scholar]
- 35.Moser, M., R. J. Colello, U. Pott, and B. Oesch. 1995. Developmental expression of the prion protein gene in glial cells Neuron 14:509-517. [DOI] [PubMed] [Google Scholar]
- 36.Mouillet-Richard, S., M. Ermonval, C. Chebassier, J. L. Laplanche, S. Lehmann, J. M. Launay, and O. Kellermann. 2000. Signal transduction through prion protein. Science 289:1925-1928. [DOI] [PubMed] [Google Scholar]
- 37.Perry, V. H., S. J. Bolton, D. C. Anthony, and S. Betmouni. 1998. The contribution of inflammation to acute and chronic neurodegeneration. Res. Immunol. 149:721-725. [DOI] [PubMed] [Google Scholar]
- 38.Peterson, K. E., J. S. Errett, T. Wei, D. E. Dimcheff, R. Ransohoff, W. A. Kuziel, L. Evans, and B. Chesebro. 2004. MCP-1 and CCR2 contribute to non-lymphocyte-mediated brain disease induced by Fr98 polytropic retrovirus infection in mice: role for astrocytes in retroviral neuropathogenesis. J. Virol. 78:6449-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peyrin, J. M., C. I. Lasmezas, S. Haik, F. Tagliavini, M. Salmona, A. Williams, D. Richie, J. P. Deslys, and D. Dormont. 1999. Microglial cells respond to amyloidogenic PrP peptide by the production of inflammatory cytokines. Neuroreport 10:723-729. [DOI] [PubMed] [Google Scholar]
- 40.Pietri, M., A. Caprini, S. Mouillet-Richard, E. Pradines, M. Ermonval, J. Grassi, O. Kellermann, and B. Schneider. 2006. Overstimulation of PrPC signaling pathways by prion peptide 106-126 causes oxidative injury of bioaminergic neuronal cells. J. Biol. Chem. 281:28470-28479. [DOI] [PubMed] [Google Scholar]
- 41.Priller, J., M. Prinz, M. Heikenwalder, N. Zeller, P. Schwarz, F. L. Heppner, and A. Aguzzi. 2006. Early and rapid engraftment of bone marrow-derived microglia in scrapie. J. Neurosci. 26:11753-11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Race, R., D. Ernst, A. Jenny, W. Taylor, D. Sutton, and B. Caughey. 1992. Diagnostic implications of detection of proteinase K-resistant protein in spleen, lymph nodes, and brain of sheep. Am. J. Vet. Res. 53:883-889. [PubMed] [Google Scholar]
- 43.Race, R., M. Oldstone, and B. Chesebro. 2000. Entry versus blockade of brain infection following oral or intraperitoneal scrapie administration: role of prion protein expression in peripheral nerves and spleen. J. Virol. 74:828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Race, R. E., S. A. Priola, R. A. Bessen, D. Ernst, J. Dockter, G. F. Rall, L. Mucke, B. Chesebro, and M. B. Oldstone. 1995. Neuron-specific expression of a hamster prion protein minigene in transgenic mice induces susceptibility to hamster scrapie agent. Neuron 15:1183-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raeber, A. J., R. E. Race, S. Brandner, S. A. Priola, A. Sailer, R. A. Bessen, L. Mucke, J. Manson, A. Aguzzi, M. B. Oldstone, C. Weissmann, and B. Chesebro. 1997. Astrocyte-specific expression of hamster prion protein (PrP) renders PrP knockout mice susceptible to hamster scrapie. EMBO J. 16:6057-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rock, R. B., G. Gekker, S. Hu, W. S. Sheng, M. Cheeran, J. R. Lokensgard, and P. K. Peterson. 2004. Role of microglia in central nervous system infections. Clin. Microbiol. Rev. 17:942-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russelakis-Carneiro, M., S. Betmouni, and V. H. Perry. 1999. Inflammatory response and retinal ganglion cell degeneration following intraocular injection of ME7. Neuropathol. Appl. Neurobiol. 25:196-206. [DOI] [PubMed] [Google Scholar]
- 48.Scott, J. R., and H. Fraser. 1989. Transport and targeting of scrapie infectivity and pathology in the optic nerve projections following intraocular infection. Prog. Clin. Biol. Res. 317:645-652. [PubMed] [Google Scholar]
- 49.Tsunoda, I., T. E. Lane, J. Blackett, and R. S. Fujinami. 2004. Distinct roles for IP-10/CXCL10 in three animal models, Theiler's virus infection, EAE, and MHV infection, for multiple sclerosis: implication of differing roles for IP-10. Mult. Scler. 10:26-34. [DOI] [PubMed] [Google Scholar]
- 50.van Keulen, L. J., B. E. Schreuder, R. H. Meloen, B. M. van den Poelen, G. Mooij-Harkes, M. E. Vromans, and J. P. Langeveld. 1995. Immunohistochemical detection and localization of prion protein in brain tissue of sheep with natural scrapie. Vet. Pathol. 32:299-308. [DOI] [PubMed] [Google Scholar]
- 51.Williams, A., P. J. Lucassen, D. Ritchie, and M. Bruce. 1997. PrP deposition, microglial activation, and neuronal apoptosis in murine scrapie. Exp. Neurol. 144:433-438. [DOI] [PubMed] [Google Scholar]
- 52.Williams, A. E., A. M. van Dam, A. H. W. Man, F. Berkenbosch, P. Eikelenboom, and H. Fraser. 1994. Cytokines, prostaglandins and lipocortin-1 are present in the brains of scrapie-infected mice. Brain Res. 654:200-206. [DOI] [PubMed] [Google Scholar]
- 53.Wojtera, M., B. Sikorska, T. Sobow, and P. P. Liberski. 2005. Microglial cells in neurodegenerative disorders. Folia Neuropathol. 43:311-321. [PubMed] [Google Scholar]
- 54.Xiang, W., O. Windl, G. Wunsch, M. Dugas, A. Kohlmann, N. Dierkes, I. M. Westner, and H. A. Kretzschmar. 2004. Identification of differentially expressed genes in scrapie-infected mouse brains by using global gene expression technology. J. Virol. 78:11051-11060. [DOI] [PMC free article] [PubMed] [Google Scholar]