Abstract
Rous sarcoma virus (RSV) requires large amounts of unspliced RNA for replication. Splicing and polyadenylation are coupled in the cells they infect, which raises the question of how viral RNA is efficiently polyadenylated in the absence of splicing. Optimal RSV polyadenylation requires a far-upstream splicing control element, the negative regulator of splicing (NRS), that binds SR proteins and U1/U11 snRNPs and functions as a pseudo-5′ splice site that interacts with and sequesters 3′ splice sites. We investigated a link between NRS-mediated splicing inhibition and efficient polyadenylation. In vitro, the NRS alone activated a model RSV polyadenylation substrate, and while the effect did not require the snRNP-binding sites or a downstream 3′ splice site, SR proteins were sufficient to stimulate polyadenylation. Consistent with this, SELEX-binding sites for the SR proteins ASF/SF2, 9G8, and SRp20 were able to stimulate polyadenylation when placed upstream of the RSV poly(A) site. In vivo, however, the SELEX sites improved polyadenylation in proviral clones only when the NRS-3′ splice site complex could form. Deletions that positioned the SR protein-binding sites closer to the poly(A) site eliminated the requirement for the NRS-3′ splice site interaction. This indicates a novel role for SR proteins in promoting RSV polyadenylation in the context of the NRS-3′ splice site complex, which is thought to bridge the long distance between the NRS and poly(A) site. The results further suggest a more general role for SR proteins in polyadenylation of cellular mRNAs.
Generation of mature mRNA in eukaryotes generally requires multiple processing steps, including capping, splicing, and polyadenylation, that are coupled to ensure proper processing (reviewed in reference 31). Retroviruses utilize the host transcription/RNA processing machinery to generate viral RNA, but due to peculiarities of their replication scheme, they often utilize the RNA processing machinery in unique ways. In the simple avian retrovirus Rous sarcoma virus (RSV), the env and src mRNAs are generated by RNA splicing from a common 5′ splice site (ss) to one of two alternative 3′ ss (10). However, unlike most host genes, retrovirus replication requires that a substantial portion of the primary viral transcripts remain completely unspliced to serve as gag-pol mRNA and as genomic RNA for progeny virions. RSV employs several mechanisms to preserve the pool of unspliced RNA, including the maintenance of suboptimal 3′ ss (25, 59) and the action of splicing repressor elements within the gag gene (the negative regulator of splicing, or NRS) (3, 39, 49) and upstream of the src 3′ ss (the suppressor of src splicing) (1, 39, 50). Generation of functional unspliced viral mRNA poses problems for the coupling of the splicing and polyadenylation reactions.
The NRS has been well characterized and is thought to act as a pseudo-5′ ss that sequesters viral 3′ ss in a nonproductive splicing complex (reviewed in reference 9). An upstream region of the ∼230-nucleotide (nt) element binds SR proteins and hnRNP H (18, 35), whereas the 3′ region binds U1 and U11 snRNPs (presumably mutually exclusively, since the binding sites overlap) (7, 21, 23, 36). The SR proteins promote U1 snRNP binding to the NRS and an early interaction with a 3′ ss (11), which then matures into a noncatalytic splicing complex that prevents the 3′ ss from interacting with the authentic viral 5′ ss (20). U11 snRNP, the binding of which is mediated by hnRNP H interactions with downstream sites (37), antagonizes U1 binding and its inhibitory activity (23, 36, 44).
Use of the RSV poly(A) site is naturally inefficient, as ∼15% of viral RNAs represent read-through transcripts (22), and RSV polyadenylation substrates are very poorly used in vitro (56 and N. L. Maciolek and M. T. McNally, unpublished data). The first suggestion that the NRS might play a role in polyadenylation came from Miller and Stoltzfus (41), who observed that deletions that encompassed the NRS led to increased read-through transcription. More specific NRS mutations have confirmed a role for the NRS in polyadenylation control; mutations in the SR protein-binding region and/or in the U1/U11-binding sites led to decreased polyadenylation efficiency, which suggested that the ability of the NRS to control splicing was integral to its role in polyadenylation control (17, 44). It is known that 3′ ss stimulate polyadenylation (13, 14, 43), and several direct interactions between splicing and polyadenylation factors have been described that contribute to the coupling of splicing and polyadenylation. These include interactions between the 3′ ss-binding factor U2AF65 and the polyadenylation factors poly(A) polymerase and cleavage factor Im (CFIm), which acts early in poly(A) complex formation (42, 54), and between U2 snRNP components and cleavage and polyadenylation specificity factor (CPSF), which binds the poly(A) signal (27). Given the importance of 3′ ss in coupling splicing and polyadenylation, an attractive hypothesis was that the nonproductive splicing complex assembled on the NRS and viral 3′ ss stimulated polyadenylation in the absence of true splicing by stabilizing the binding of splicing factors to the weak viral 3′ ss (17).
In this study, in vitro and in vivo approaches were used to elucidate the mechanism of NRS-mediated polyadenylation control. The weak RSV poly(A) site could be activated in vitro by the NRS alone, indicating that the nonproductive splicing complex formed on the NRS and downstream 3′ ss is not necessary to stimulate RSV polyadenylation, and U1 and U11 snRNPs were dispensable for the effect. It was shown that SR protein binding to the NRS- or SELEX-binding sites was sufficient to stimulate polyadenylation in vitro. However, this was not true in vivo, in which case a requirement for the downstream NRS snRNP-binding region was demonstrated. SR protein-binding sites alone did promote polyadenylation in vivo when moved closer to the viral poly(A) site. The data suggest that SR proteins play a novel role in promoting RSV polyadenylation, but they do so only when they are positioned sufficiently close to the 3′ end of the RNA via an interaction of the NRS with a downstream viral 3′ ss.
MATERIALS AND METHODS
Plasmid constructs.
p3Z-SVL, to generate the positive control simian virus 40 (SV40) late (SVL) polyadenylation substrate was constructed by inserting an EcoRI fragment from pMXSVL (43) into the same sites of pGEM-3Z and then deleting the SacI-BamHI fragment encompassing the adenovirus type 2 major late splicing cassette. p3Z-RSVPvuI-PstI, to generate the RSV substrate, was made by inserting a 374-bp PvuI-PstI fragment (PvuI site blunted with Klenow) into the SmaI and PstI sites of pGEM-3Z. The PvuI-PstI fragment was obtained from pBSKS+RSVSa1I-SacII, which contains an 1,174-bp Sa1I-SacII fragment from pAPrC that surrounds the 5′ LTR. For p4Z-Ad3′-RSV, an oligonucleotide containing SacI, XhoI, BamHI, and EcoRI sites was inserted into the EcoRI and SacI sites downstream of the adenovirus (Ad) 3′ exon in the previously described pAd(KX)BB (12), generating pAdBB(XB). A PCR fragment (with appended BamHI and XhoI sites) containing 123 nt of upstream and 87 nt of downstream sequence relative to the RSV poly(A) site (nt 9185 to 9311; coordinates are as described by Schwartz et al. [46]) was inserted into the BamHI and XhoI sites of pAdBB(XB) to make pAdBB-RSV. This was then digested with HindIII and XbaI, blunted, and recircularized to delete the Ad 5′ exon and NRS. p3Z-src3′ was made by inserting an src PCR product containing 41 nt of src exon and 80 nt of upstream intron (nt 6975 to 7096) with appended XbaI and EcoRI sites into the same sites of pGEM-3Z. p4Zsrc3′-RSV was created by replacing the Ad splicing cassette and NRS in pAdBB-RSV with a HindIII-EcoRI src fragment from p3Z-src3′. A construct harboring the NRS directly upstream of the RSV poly(A) site (p4Z-BBΔ76-RSV) was made by replacing the original KpnI-XbaI NRS fragment in pAdBB-RSV with a 285-nt KpnI-XbaI fragment (nt 703 to 930) harboring a deletion from nt 799 to 874 [pAd(KX)BBΔ76] (34). The plasmid was digested with HindIII and KpnI, blunted, and recircularized to remove the Ad 5′ ss (to make p4ZBBΔ76-Ad3′-RSV), and finally the Ad 3′ ss was similarly removed by XbaI and XhoI digestion followed by recircularization. For the construct containing the NRS fused to the src 3′ ss (p4ZBBΔ76-src3′-RSV), the Ad 3′ ss in p4ZBBΔ76-Ad3′-RSV was replaced with the HindIII-EcoRI src 3′ fragment from p3Z-src3′.
To construct p4Z-RG11-RSV, p4Z-NRSmutH-RSV, pNRS5′-RSV, and pNRS3′-RSV, the wild-type NRS in pAdBB-RSV was replaced with NRS PCR fragments containing the RG11 or mutH mutation (see Fig. 3A) or the NRS 5′ and 3′ fragments (NRS5′ and NRS3′, respectively) (35). Subsequently, to remove the Ad 5′ exon, the plasmid was digested with HindIII and KpnI, blunted, and recircularized. The Ad 3′ exon was similarly removed by digestion with XbaI and XhoI as described above.
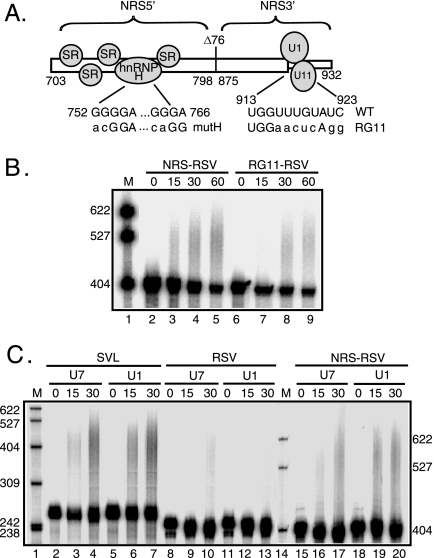
FIG. 3.
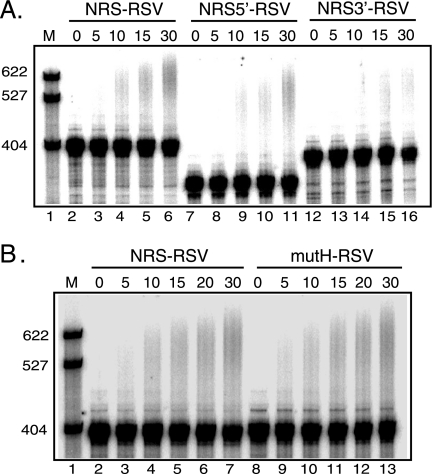
U1 and U11 snRNPs are not required for NRS-stimulated RSV polyadenylation. (A) Schematic of the NRS (nt 703 to 932) indicating the NRS5′- (nt 703 to 798), NRS3′- (nt 798 to 932), and NRS-binding factors. Shown are the binding of SR proteins and hnRNP H to NRS5′ and U1 and U11 snRNPs binding to NRS3′. Δ76 indicates a 76-nt deletion that does not markedly affect NRS function. The sequences of mutations that eliminate hnRNP H (mutH) and U1/U11 snRNP (RG11) binding are shown below the wild-type sequence. (B) RSV substrates harboring the wild-type NRS or the RG11 mutation were uniformly labeled with 32P, and polyadenylation was assessed in HeLa cell nuclear extract. (C) SVL, RSV, and NRS-RSV substrates were labeled as described for panel B, and polyadenylation was assayed in HeLa nuclear extract in which U1 or U7 snRNP was inactivated using 2′-O-methyl oligonucleotides. (B and C) Reaction mixtures were incubated for the times (in minutes) indicated above each lane and were subjected to electrophoresis on a 6% polyacrylamide gel that contains 8 M urea. (B) NRS-RSV samples were run ∼1 h longer to allow adequate separation of products. Polyadenylation appears as a slower-migrating smear. Images were obtained with a PhosphorImager and are representative of at least three independent experiments. M, 32P-end-labeled pBR322/MspI markers; WT, wild type. The results of quantitation of polyadenylation at 30 min are the following: (B) NRS-RSV, 9%; and RG11-RSV, 8%; (C) SVL/U7, 45%; SVL/U1, 31%; RSV/U7, 8%; RSV/U1, 5%; NRS RSV/U7, 20%; and NRS RSV/U1, 19%.
Proviral clones were generated in pJTM14 (41), which contains the Prague A strain provirus upstream of a chloramphenicol acetyltransferase (CAT) gene and the SV40 early poly(A) signal. Constructs containing NRS deletions (ΔNRS and Δ712-798) (NRS3′ in Fig. 7) or the RG11 mutation have been described previously (17). Proviruses in which the NRS was replaced with SELEX consensus SR protein-binding sites (see Fig. 6) (8, 29, 51) were made in two steps. Mutations were inserted into the SacII fragment of pBSKS-RSVSacII, and the SacII fragments were used to replace the wild-type sequence in pJTM14. Oligonucleotides harboring binding sites for ASF/SF2, 9G8, or a control were annealed and used to replace the MroI-SphI NRS fragment in pBSKS-RSVSacII (17). To replace just the NRS5′ (nt 703 to 798), PCR products containing control or SR protein-binding sites fused to the NRS 3′ end were inserted into the same sites of pBSKS-RSVSacII as those described above. For mutations in the downstream hnRNP H-binding sites, an MroI-SphI NRS fragment from pMS-mG1+2 (38) was inserted into the same sites of pBSKS+RSV SacII. The SacII fragments were then shuttled into the same sites of pJTM14 to generate pJTM14-ASF, pJTM14-9G8, pJTM14-control, pJTM14-ASF-NRS3′, pJTM14-9G8-NRS3′, pJTM14-control-NRS3′, and pJTM14-mG1+2. The proviral deletion clones that bring the ASF/SF2 SELEX and control sequences closer to the poly(A) site, pJTM14-ASFΔpol-env3′, pJTM114-968Δpol-env3′, and pJTM14-contΔpol-env3′, were made by deleting an FseI-NheI fragment from pJTM14-ASF, pJTM-968, and the pJTM14-control. Primer sequences are available upon request.
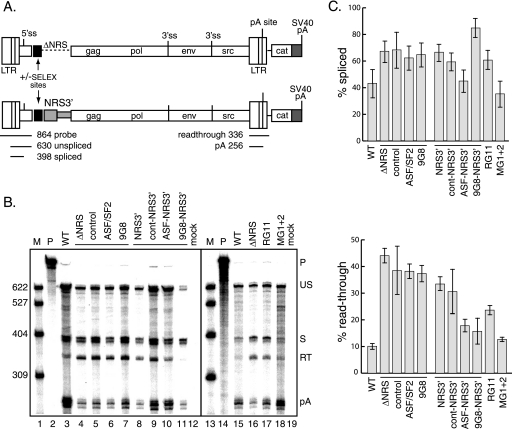
FIG. 7.
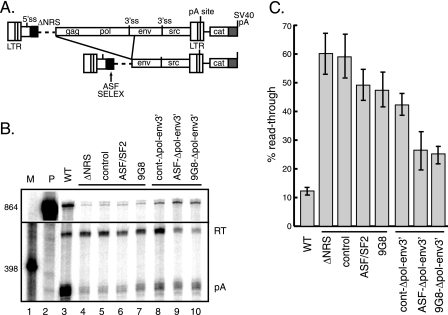
SR protein-binding sites stimulate polyadenylation of proviral clones in vivo. (A) Schematic of proviral constructs. Shown are the long terminal repeats (LTR); 5′ and 3′ ss; gag, pol, env, and src genes; poly(A) site; and downstream CAT gene and SVL poly(A) signal (shaded). At the top, the ASF/SF2 or 9G8 SELEX site (black box) replaced the entire NRS, while at the bottom, the SELEX sites were fused to NRS3′ (gray box). The positions and sizes of the RNase protection probe used for panel B and the protected products are shown below the lower schematic. (B) RNase protection assays were performed using RNA from CEFs transfected with the indicated proviral clones lacking the entire NRS (ΔNRS) or containing only NRS3′ or with constructs having insertions of control, ASF/SF2, or 9G8 SELEX sites. On the right are constructs with mutations that eliminate U1/U11 snRNP binding (RG11) or hnRNP H binding to the downstream sites (mG1+2). Products were subjected to electrophoresis on a 6% polyacrylamide gel that contains 8 M urea and visualized with a PhosphorImager. Protected products are labeled at the right. P, probe (864 nt); US, unspliced RNA (630 nt); S, spliced RNA (398 nt); RT, read-through RNA (336 nt); pA, polyadenylated product (256 nt); WT, wild type. M, 32P-end-labeled pBR322/MspI markers, the sizes of which are indicated at the left. (C) Quantitation of the data from three replicate experiments for the percentages of spliced (top) and read-through (bottom) RNA.
FIG. 6.
SR protein-binding sites stimulate RSV polyadenylation in vitro. (A) Schematic of RSV substrate and sequence of SR protein SELEX-binding sites inserted upstream of the RSV poly(A) site. Consensus sequences (upper case) used for ASF/SF2, 9G8, SRp20, and SRp40 are separated by a 7-nt spacer (lowercase letters) (8, 29, 51). The negative control contains three repeats of a sequence from the original ASF/SF2 SELEX pool (51). (B) SVL-, RSV-, and SELEX-containing substrates were uniformly labeled with 32P and incubated in HeLa nuclear extract for the times (in minutes) indicated above each lane. RNA was subjected to electrophoresis on a 6% polyacrylamide gel that contains 8 M urea. The image, representative of at least three independent experiments, was obtained with a PhosphorImager. Polyadenylation appears as an upward smear. M, 32P-end-labeled pBR322/MspI markers, the sizes of which are indicated at the left. The results of quantitation of polyadenylation are the following: SVL, 18%; RSV, 2%; cont-RSV, 4%; ASF/SF2-RSV, 10%; 9G8-RSV, 7%; SRp20-RSV, 9%; SRp40-RSV, 4%.
In vitro transcription of substrates.
To make RNA substrates by in vitro transcription (40), p3Z-RSV was linearized with BstEII, p3Z-SVL was linearized with HindIII, and NRS-containing substrates were linearized with BamHI. Alternatively, template DNA was generated by PCR using primers to the upstream T7 promoter and RSV or SVL substrate (these substrates were used for experiments shown in Fig. 2A, 3C, and 4). Templates for the SELEX site-containing RNAs (8, 29, 51) were obtained by PCR of p3Z-RSV with a primer for RSV as the antisense primer and template-specific primers containing a T7 promoter, the appropriate SR-binding sequence, and the RSV sequence. RNA was transcribed in vitro with T7 or SP6 polymerase and [32P]UTP in a capping reaction and was gel purified on a 6% polyacrylamide (19:1) gel that contains 8 M urea as described previously (35). Primer sequences are available upon request.
FIG. 2.
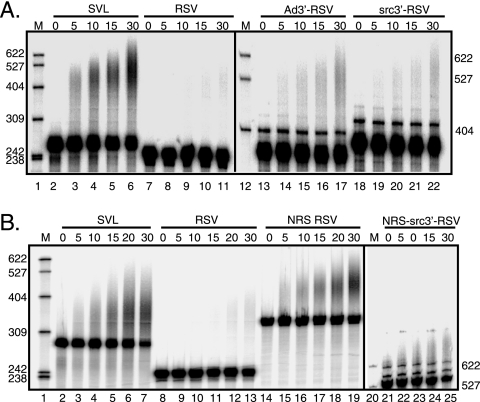
The NRS alone can stimulate RSV polyadenylation. The indicated RSV polyadenylation substrates (see Fig. 1) containing a 3′ ss (A) and/or the NRS (B) were uniformly labeled with 32P and incubated in HeLa nuclear extract for the times (in minutes) indicated above each lane. Polyadenylation appears as an upward smear. Samples were subjected to electrophoresis on a 6% 8 M urea polyacrylamide gel, and images were obtained with a PhosphorImager. Ad3′-RSV and src3′-RSV samples (A) and NRS-src3′-RSV samples (B) were run ∼1 h longer to allow adequate separation of products. The images are representative of at least three independent repeats. M, 32P-end-labeled pBR322/MspI markers, the sizes of which are indicated at the left and right. The results of quantitation of polyadenylation at 30 min are the following: (A) SVL, 32%; RSV, 2%; Ad3′-RSV, 8%; and src3′-RSV, 4%; (B) SVL, 47%; RSV, 6%; NRS-RSV, 63%; and NRS-src3′-RSV, 7%.
FIG. 4.
Polyadenylation stimulatory activity maps to NRS5′, but hnRNP H-binding sites are not required. (A) RNA substrates were uniformly labeled with 32P and incubated in HeLa nuclear extract for the times (in minutes) indicated above each lane. The NRS5′ and NRS3′ regions are the same as those bracketed in Fig. 3A, except that this NRS3′ version contained nt 801 to 932. RNA was subjected to electrophoresis on a 6% polyacrylamide gel that contains 8 M urea, and the image was obtained with a PhosphorImager; the results are representative of at least three independent experiments. Polyadenylation appears as a slower-migrating smear. The results of quantitation of polyadenylation at 30 min are the following: NRS-RSV, 13%; NRS5′-RSV, 12%; and NRS3′-RSV, 2%. (B) NRS-RSV and mutH-RSV, which contain the mutated hnRNP H-binding sites (Fig. 3A), were treated as described for panel A. The results of quantitation of polyadenylation at 30 min are the following: NRS-RSV, 13%; and NRSmutH-RSV, 13%. M, 32P-end-labeled pBR322/MspI markers, the sizes of which are indicated at left.
Polyadenylation assays.
Polyadenylation reactions were performed using 50,000 cpm of labeled substrate in 2.6% polyvinyl alcohol, 1 mM ATP, 20 μM creatine phosphate, 1 mM MgCl2, and 20 to 50% HeLa cell nuclear extract. Reaction mixtures were incubated at 30°C for the times specified in the figures. Samples were then proteinase K treated, phenol extracted, and ethanol precipitated, and RNA was resolved on a 6% polyacrylamide gel that contains 8 M urea. For U1 snRNP inactivation, HeLa nuclear extract was preincubated for 10 min at 30°C with 100 μM 2′-O-methyl RNA oligonucleotide U11-14 (47) or U73-20 (48). Polyadenylation reaction mixtures with S100 contained 34% HeLa S100 extract supplemented with 16% HeLa nuclear extract alone or with 1 μg purified SR proteins (58). Images were obtained using a STORM 820 PhosphorImager (Amersham Biosciences) and/or autoradiography and were quantitated with ImageQuant 5.2 software. Since substrates were subject to degradation over time, polyadenylation efficiency was quantitated as the phosphorimager units in the polyadenylated product divided by the units in the substrate at time zero.
Transfection and RNase protection assay.
Secondary chicken embryonic fibroblasts (CEFs) were grown in medium 199 (Invitrogen) supplemented with 2% tryptose phosphate broth, 1% bovine calf serum (HyClone), 1% chicken serum (GIBCO), 1× antibiotic-antimycotic (Invitrogen). Cells were transfected with 2 μg DNA in medium 199 containing 200 μg/ml DEAE-dextran, and after 4 h cells were subjected to a 2-min 10% dimethyl sulfoxide shock. Total RNA was harvested using the QIAGEN RNeasy kit 48 h later. RNase protection assays were carried out as previously described (17) using 2.5 μg of total RNA hybridized with 106 cpm of riboprobe transcribed from p5′XH1 (49) that spans the viral long terminal repeat and 5′ region of the gag gene (nt 218 to 630). Images were obtained by and quantitated with a STORM 820 PhosphorImager (Amersham Biosciences) using ImageQuant 5.2 software. Bands were normalized for uridine content.
RESULTS
3′ ss do not support efficient RSV polyadenylation in vitro.
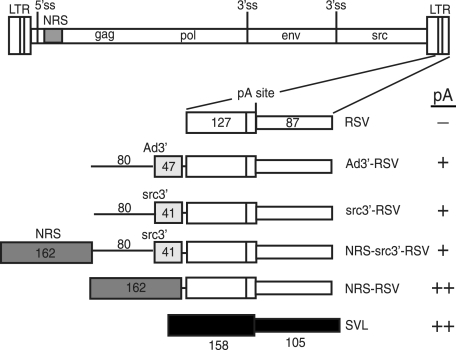
The poor in vitro polyadenylation efficiency observed for RSV polyadenylation substrates (57 and Maciolek and McNally, unpublished) led to the hypothesis that there is an additional element(s) within the viral genome that contributes to poly(A) site recognition in vivo. Because 3′ ss promote polyadenylation of cellular and RSV RNAs and can do so in the absence of a 5′ ss in vitro (41, 43, 53), we considered a role for the src 3′ ss in RSV polyadenylation control. The src 3′ ss was chosen because it is proximal to the viral poly(A) signal, and the NRS suppresses src splicing more efficiently than env, suggesting a stronger interaction with this ss (44). Substrates were designed with the RSV poly(A) site in the context of an Ad 3′ ss that is known to stimulate SV40 polyadenylation (43) or the weak src 3′ ss (Fig. 1). The RSV poly(A) signal alone served as a negative control, since it is poorly polyadenylated in vitro, while the positive control was the efficiently used SVL substrate. Substrates were uniformly radiolabeled and incubated in nuclear extract for the indicated times, and RNA was isolated and resolved in a denaturing polyacrylamide gel; polyadenylation manifests as an increase in size of the labeled substrate. After 30 min, the positive control SVL substrate was polyadenylated (30% in this experiment) (Fig. 2A, lanes 2 to 6) while, as observed previously (56 and Maciolek and McNally, unpublished), polyadenylation of the RSV substrate was barely detectable (∼1%) (lanes 7 to 11). Placement of the Ad 3′ ss upstream of the RSV poly(A) signal increased polyadenylation to 8% (lanes 13 to 17), whereas half this level was seen when the viral src 3′ ss was used (lanes 18 to 22). Thus, neither ss stimulated RSV polyadenylation to the level seen with SVL substrate, and the src 3′ ss was less effective than the Ad 3′ ss. It was also reported recently that the env 3′ ss does not stimulate efficient RSV polyadenylation (56). We conclude that the viral 3′ ss alone do not enable efficient RSV polyadenylation in vitro and that an additional element(s) is required.
FIG. 1.
Schematic of in vitro polyadenylation substrates. At the top is a diagram of the RSV provirus showing the long terminal repeats (LTR); 5′ ss; NRS (shaded); gag, pol, env, and src genes; and 3′ ss. Below is an expansion of the 3′ LTR and a schematic of the RSV substrates, which include the entire region downstream of the poly(A) site (87 nt) (thin open box) and 127 nt of upstream sequence (wide open box). Ad3′-RSV has 47 nt of Ad 3′ exon (lightly shaded box) with 80 nt of upstream intron (thin line). src3′-RSV has 41 nt of the src 3′ exon (shaded box) with 80 nt of upstream RSV intron. NRS-src3′-RSV and NRS-RSV have the 162-nt NRS BBΔ76 fragment (dark shaded box) inserted upstream of the RSV poly(A) signal, with or without the src 3′ ss region. The positive control SVL substrate (black boxes) contains 137 nt of upstream and 105 nt of downstream sequence relative to the SVL poly(A) site. A summary of polyadenylation activity is indicated at the far right (−, activity less than 10% of that of SVL; +, activity of up to 40% of that of SVL; ++, activity of 40% or more of that of SVL).
The NRS alone stimulates RSV polyadenylation in vitro.
Previous observations suggested that a functional NRS plays a role in RSV polyadenylation in vivo (17, 41, 44) and led to a model in which the nonproductive splicing complex assembled on the NRS and downstream 3′ ss promotes polyadenylation in the absence of true splicing. To explore this idea in vitro, substrates harboring the NRS alone or with the src 3′ ss (Fig. 1) were tested in polyadenylation reactions. A low level of activity was detected with the NRS-src3′-RSV polyadenylation substrate (7%) (Fig. 2B, lanes 21 to 25) that was only slightly better than that seen with src3′-RSV (Fig. 2A). Surprisingly, the NRS alone strongly stimulated polyadenylation (63%) (Fig. 2B, lanes 14 to 19). These data indicate that the nonproductive NRS-3′ ss complex is not required to stimulate RSV polyadenylation in vitro and that one or more NRS-binding factors are sufficient for this effect.
U1 and U11 snRNPs are not required for NRS-stimulated polyadenylation in vitro.
The NRS harbors binding sites for multiple factors, including U1 snRNP, U11 snRNP, hnRNP H, and SR proteins (Fig. 3A). A role for U1 snRNP or U1-associated proteins in SV40 polyadenylation has been suggested previously (30, 55), and the U1 site within the NRS is required for optimal RSV polyadenylation (17, 44). Therefore, the requirement of U1 and/or U11 snRNPs for NRS-stimulated polyadenylation was tested with a substrate (RG11) in which both binding sites were mutated (21, 36) (Fig. 3A). There does not appear to be an in vitro requirement for either snRNP, since there was no significant difference in polyadenylation activity between the wild-type (9%) and mutant NRS (8%) (Fig. 3B, lanes 2 to 5 and 6 to 9). To address the possibility that U1 might bind promiscuously to another site in the NRS-RSV substrate, polyadenylation assays were performed using nuclear extracts in which U1 snRNP was inactivated with a 2′-O-methyl RNA oligonucleotide that sequesters the 5′ end of the snRNA to prevent its interaction with substrates or with control extracts treated with a 2′-O-methyl oligonucleotide to U7 snRNA. Functional inactivation of U1 was evidenced by the inability of the extract to support splicing of an adenovirus splicing substrate (data not shown), but there was no consistent effect of oligonucleotide treatment on polyadenylation of any of the substrates (Fig. 3C). While evidence indicated that U1 was important for efficient polyadenylation of proviruses in vivo, this does not appear to be the case in vitro. Because U1 binds NRS3′ (nt 801 to 930), this observation suggested that the polyadenylation stimulatory activity resides in NRS5′ (nt 701 to 800). This proved to be the case; NRS5′ stimulated polyadenylation nearly as well as the full-length NRS, while NRS3′ was inactive (Fig. 4A). This suggested that SR proteins and/or hnRNP H, which bind NRS5′, promote polyadenylation.
hnRNP H is not required for in vitro NRS-stimulated polyadenylation.
hnRNP H can stimulate the use of viral and cellular poly(A) signals that harbor downstream G-rich tracts (2). The NRS has hnRNP H-binding sites within NRS5′ and also downstream of the U1/U11 sites (18, 37), which suggests that hnRNP H mediates NRS polyadenylation control. The minimal NRS used in this study does not include the downstream G tracts, but since NRS5′ was sufficient to stimulate polyadenylation, a role for hnRNP H in RSV polyadenylation was explored using point mutations that eliminate hnRNP H binding to NRS5′ (Fig. 3A) (17, 18). There was no difference in polyadenylation between the hnRNP H-binding mutant and the wild type (Fig. 4B). This suggests that hnRNP H is not required for NRS-stimulated polyadenylation, in agreement with previous data demonstrating that this hnRNP H-binding mutation had no effect on RSV polyadenylation in vivo (17).
SR proteins promote RSV polyadenylation in vitro.
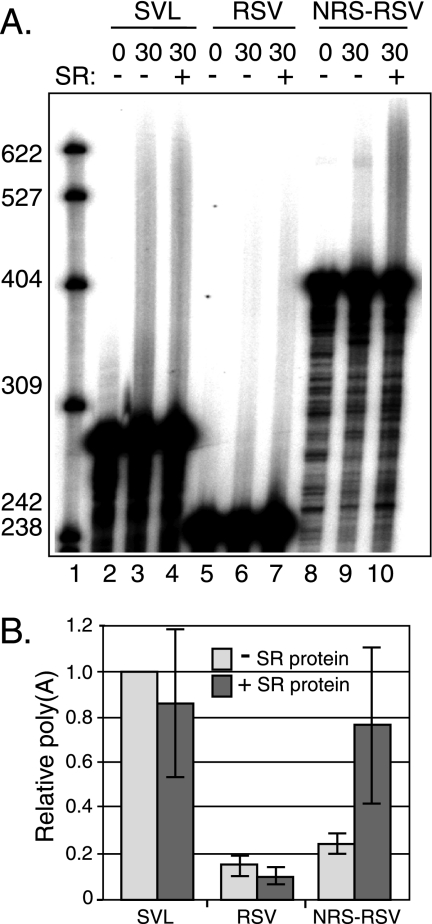
The 5′ region of the NRS contains a purine-rich region that binds SR proteins (35) and is required for optimal RSV polyadenylation (17). To test the hypothesis that SR proteins promote RSV polyadenylation, HeLa S100 extract that lacks SR proteins was used for polyadenylation assays, and purified total SR proteins were then added in an attempt to restore NRS-mediated polyadenylation activity. The S100 extract lacked a factor(s) required for polyadenylation, since it did not support polyadenylation of any substrate tested, including SVL substrate (data not shown). However, S100 was rendered polyadenylation competent when supplemented with a small amount of nuclear extract that, alone, was not active (data not shown). SVL polyadenylation was evident at 30 min (3% in this experiment), and addition of 1 μg of purified SR proteins had no effect (2%) (Fig. 5, lanes 3 and 4). RSV polyadenylation was barely detectable, and the addition of SR proteins also had no effect (∼0.5%) (lanes 6 and 7). Polyadenylation was detected with NRS-RSV (2%), but importantly, addition of purified SR proteins stimulated polyadenylation significantly (8%) (lanes 9 and 10). These data strongly support a role for the SR proteins in NRS-mediated RSV polyadenylation.
FIG. 5.
SR proteins are required for NRS-stimulated RSV polyadenylation in vitro. (A) SVL, RSV, and NRS-RSV substrates were labeled with 32P and incubated in HeLa cell S100 extract supplemented with 16% HeLa cell nuclear extract. Substrates were incubated with (+) or without (−) purified SR proteins for 30 min and were subjected to electrophoresis on a 6% polyacrylamide gel that contains 8 M urea. An image representative of three independent experiments was obtained with a PhosphorImager. Polyadenylation appears as a slower-migrating smear. M, 32P-end-labeled pBR322/MspI markers, the sizes of which are indicated at the left. (B) Quantitation of the data shown in panel A. The percent polyadenylation for SVL substrate in the absence of SR proteins was set at 1.0, and the relative levels for the RSV and NRS-RSV substrates were normalized to this value. Gray bars, without SR proteins; dark bars, with SR proteins. Error bars indicate standard deviations.
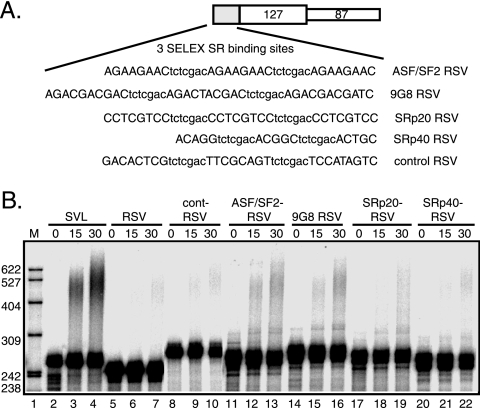
The NRS binds the SR proteins ASF/SF2, 9G8, SRp20, and SRp40 (17, 35), and to further demonstrate a role for individual SR proteins in RSV polyadenylation, three copies of SELEX consensus binding sites for ASF/SF2, 9G8, SRp20, and SRp40 (8, 29, 51) were fused to the RSV poly(A) signal (Fig. 6A), and polyadenylation activity was determined in nuclear extract. Compared to a control substrate, the activity of which was slightly elevated relative to that of RSV alone (Fig. 6B, compare lanes 5 to 7 and 8 to 10), the binding sites for ASF/SF2, 9G8, and SRp20 increased RSV polyadenylation (10, 7, and 9%, respectively) (lanes 11 to 19). The SRp40 sites were no more active than the control (lanes 20 to 22). One explanation is that the effect is specific to ASF/SF2, 9G8, and SRp20, or, alternatively, SRp40 is not as abundant as the other SR proteins in the nuclear extract. Regardless, the data support a role for the SR proteins ASF/SF2, 9G8, and SRp20 in NRS-stimulated RSV polyadenylation in vitro.
SR protein-binding sites stimulate RSV polyadenylation in vivo.
In light of the above evidence that SR proteins mediate NRS-stimulated RSV polyadenylation in vitro, it was important to demonstrate their importance in a proviral setting in vivo. To address whether SR proteins alone could perform this function, three SELEX consensus binding sites for ASF/SF2 and 9G8 or a random sequence was substituted for the NRS in a proviral clone containing the CAT gene and SV40 early poly(A) signal at the 3′ end of the genome (41). Inclusion of the downstream SV40 signal ensures the polyadenylation and stabilization of read-through transcripts. Since previous data suggested a role for the NRS U1-binding site in polyadenylation control (17, 44), proviral clones also were made in which the SELEX sites were fused to NRS3′ (Fig. 7A).
CEFs were transfected with the constructs, and splicing and polyadenylation were assessed in an RNase protection assay with a riboprobe complementary to the viral long terminal repeat and 5′ ss that allowed detection of unspliced, spliced, read-through, and polyadenylated products (17). RNA from wild-type provirus transfections showed ∼43% spliced RNA and ∼10% read-through transcripts (Fig. 7B, lane 3; quantitation of the data is shown in C), and as previously reported, deletion of the NRS resulted in increased splicing (∼67% spliced) and read-through RNA (∼44%) (lane 4), in accordance with its role in splicing repression and promoting polyadenylation (17, 44). Replacing the NRS with the SELEX sites did not rescue splicing inhibition, which was expected, since the U1 site is required for splicing inhibition, but polyadenylation control was not corrected either (lanes 5 to 7). Thus, in contrast to the in vitro results, SR protein-binding sites alone are not sufficient to promote polyadenylation in vivo.
To determine if 3′-end processing in vivo required SR proteins and the U1 site, constructs with SELEX sequences fused to NRS3′ were tested. Consistent with previous results, NRS3′ alone did not support efficient splicing inhibition (∼67% spliced) or polyadenylation (∼33% read-through) (Fig. 7B, lane 8). Fusing the ASF/SF2 or 9G8 binding sites to NRS3′ had variable effects on splicing inhibition but was sufficient to restore polyadenylation to near-wild-type levels (lanes 10 and 11). These effects were specific, since the level of splicing inhibition was unchanged by fusing the control sequence to NRS3′ (∼59% spliced), and polyadenylation was similar to that observed with the NRS3′ deletion (∼31% read-through) (lane 9). The ASF/SF2-binding site corrected the splicing defect when fused to NRS3′ (lane 10), but the 9G8 sites appeared to cause oversplicing (∼85% spliced) (lane 11). However, the level of unspliced 9G8 RNA appeared selectively reduced compared to that of other samples, such that the change in percent splicing may be due to destabilization of the unspliced RNA. Overexpression of SR proteins can result in reduced RNA stability (28, 60), so it is possible that 9G8 recruitment to the NRS3′-containing unspliced RNA may target it for degradation. Collectively, the results indicate that RSV polyadenylation control in vivo requires SR proteins and the integrity of the NRS-3′ ss inhibitory complex, perhaps to position the SR proteins nearer to the poly(A) site.
NRS3′ harbors binding sites for U1 snRNP, U11 snRNP, and hnRNP H, of which U1 has a demonstrated role in RSV polyadenylation (17). To determine if mutations in strong hnRNP H-binding sites in NRS3′ affect RSV 3′-end formation in vivo, proviral clones with mutations that eliminate hnRNP H binding were examined (37). As in previous studies, eliminating U1/U11 binding with the RG11 mutation compromised NRS function and caused increased splicing and read-through transcripts (Fig. 7B, lane 17; quantitation of the data is shown in C). No change in splicing or polyadenylation was observed with the hnRNP H mutant provirus (mG1+2) (lane 18). These results confirm a previous report, which showed that the snRNP-binding sites are required for RSV splicing and polyadenylation control (17), and suggest that the strong, downstream hnRNP H-binding sites are not required for either activity.
SR protein-binding sites can stimulate RSV polyadenylation in vivo independent of the NRS complex.
The observation that SR proteins are sufficient to stimulate RSV polyadenylation in vitro suggests that they perform the same function in vivo. While the requirement for the NRS-3′ ss complex for optimal RSV polyadenylation in vivo is consistent with positioning the SR proteins closer to the poly(A) site, it was also formally possible that an NRS-3′ ss complex factor other than SR proteins interfaces with the polyadenylation machinery (see Discussion). To determine if SR proteins alone could promote polyadenylation, a deletion was made in the ASF/SF2 and 9G8 SELEX and control proviral clones that moved the sites from ∼8,300 nt to ∼4,320 nt away from the poly(A) site (Fig. 8A); movement to this position is similar to what would occur if the NRS interacted with the env 3′ ss. As observed above, splicing was elevated with all the deletions (data not shown), and read-through transcription was elevated in the ΔNRS, control insert, and ASF/SF2 and 9G8 SELEX samples (Fig. 8B, lanes 4 to 7; quantitation of the data is shown in C), and moving the control sequence closer to the poly(A) site had no effect (Fig. 8B, lane 8). Significantly, repositioning the ASF/SF2- and 9G8-binding sites nearer to the poly(A) site decreased read-through transcription substantially (lanes 8 and 9). These results indicate that SR proteins influence NRS-mediated RSV polyadenylation, but they do so only when positioned an appropriate distance from the poly(A) site.
FIG. 8.
ASF/SF2 and 9G8 SELEX sites can activate RSV polyadenylation in vivo independently of the NRS-3′ ss complex. (A) Schematic of proviral constructs. Shown are the proviral long terminal repeats (LTRs); 5′ and 3′ ss; gag, pol, env, and src genes; poly(A) site; and downstream CAT gene and SVL poly(A) signal (shaded). The ASF/SF2 and 9G8 SELEX sites or control sequence (black box) replaced the entire NRS (ΔNRS; dashed line). The deletion (denoted by the lines) places the SELEX (or control) sequences nearer to the poly(A) site. (B) RNase protection assay (probe and products were the same as those used for Fig. 7) of total RNA isolated from CEFs transfected with the indicated proviral clones. Protected read-through and poly(A) products (designated on the right) were resolved on a 6% polyacrylamide gel that contains 8 M urea and visualized with a PhosphorImager. P, probe; RT, read-through RNA; pA, polyadenylated product; WT, wild type. M, 32P-end-labeled pBR322/MspI markers, the sizes of which are indicated at the left. Bands were quantitated using a PhosphorImager. (C) Quantitation of the three independent experiments.
DISCUSSION
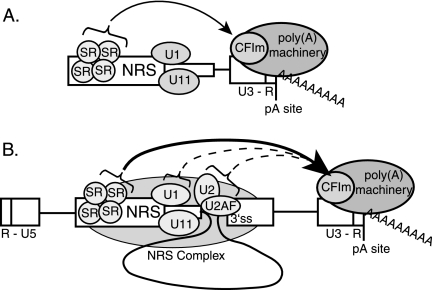
Previous work indicated that a functional NRS is required for optimal viral RSV polyadenylation (17, 44). In keeping with current models of coupled splicing and polyadenylation, it was proposed that the nonproductive splicing complex that forms on the NRS and a 3′ ss stimulate RSV polyadenylation by stabilizing the binding of splicing factors to the weak viral 3′ ss; these factors would then promote polyadenylation by conventional mechanisms (see below). In this model, NRS-binding factors are only indirectly involved in polyadenylation control. Surprisingly, we show here that the NRS alone promotes polyadenylation of an RSV substrate in vitro, and that U1 snRNP and a 3′ ss are not required for this effect. While this work was in progress, a report by Wilusz and Beemon (56) reached a similar conclusion. We further show that SR proteins mediate the polyadenylation stimulatory activity of the NRS in vitro, which has implications for potential polyadenylation control of cellular mRNAs. While the SR-related proteins have been shown to bind poly(A) factors and promote polyadenylation (32, 33, 42, 54), to our knowledge this is the first demonstration that SR proteins can promote polyadenylation. However, SR proteins alone were insufficient to stimulate polyadenylation in a proviral context in vivo. This is consistent with previous reports that the downstream snRNP-binding sites within the NRS are required for optimal viral polyadenylation (17, 44). One explanation for this discrepancy is that the distance between the SR protein-binding site and the poly(A) site is quite long in the provirus compared to those in the in vitro substrates, and there may be a distance constraint over which SR proteins can function. The NRS interaction with a viral 3′ ss would spatially bridge the long distance and position the SR proteins nearer to the poly(A) site (Fig. 9B).
FIG. 9.
Model for NRS-stimulated RSV polyadenylation in vitro and in vivo. (A) In vitro model. The NRS is shown with SR proteins and U1/U11 snRNPs bound. When in close proximity to the RSV polyadenylation site (pA site; at the end of the proviral U3-R region), as is the case with in vitro constructs, SR proteins recruit the polyadenylation machinery, possibly through an RS domain interaction with a similar domain in CFIm (arrow). (B) In vivo model. Shown are the 5′ end of viral RNA (R-U5) and the poly(A) site (pA site) at the 3′ end (U3-R). The NRS and associated factors are shown interacting with a 3′ ss to form the nonproductive NRS complex (shaded oval), which positions SR proteins at least 4,200 nt closer to the poly(A) site. SR proteins then recruit the polyadenylation machinery (arrow), possibly through CFIm. Potential interactions between the NRS-3′ ss complex factors U1, U2, and/or U2AF and the polyadenylation machinery also are shown (dotted arrows) (see the text for details). WT, wild type.
Interestingly, SR proteins bind an exonic splicing enhancer located just downstream from the env 3′ ss (25), but these sites do not appear to be sufficient for poly(A) control, since elevated levels of read-through RNA are detected when these sites are present and the NRS is deleted (41). These sites also are present in our ΔNRS proviruses, for which considerable poly(A) read-through was observed. Perhaps the env exonic splicing enhancer does not bind SR proteins as well as the NRS, which represents an extensive SR protein-binding platform (17, 35), or, alternatively, other components of the NRS-3′ ss complex contribute to polyadenylation (see below). It also was shown that deleting the region encompassing the env 3′ ss caused significant poly(A) read-through, but that a G-to-U mutation at the env 3′ ss was without effect (41). Our model would reconcile these observations; the env 3′ ss mutation would still allow the NRS-env complex to form, but deletion of the entire env 3′ ss region would disrupt the interaction with the NRS and repositioning of the SR proteins.
CFIm68, a large subunit of the CFIm polyadenylation factor, has a domain rich in RD-, RE-, and RS-dipeptide repeats that can interact with SR proteins, presumably through the RS domain that is known to mediate SR protein interactions with other RS-domain-containing proteins (16, 26, 45, 57). An attractive possibility to be tested in the future is that the NRS and associated SR proteins recruit or stabilize CFIm binding to the poorly utilized viral poly(A) site (Fig. 9A). A precedent for this idea comes from the observation that the RS domain within the 65-kDa subunit of U2AF stimulates polyadenylation in vitro via interaction with the alternating charged domain of CFIm59 (42).
Proviral deletions that moved the ASF/SF2 and 9G8 SELEX sites alone to a position similar to that which would occur from an NRS-env 3′ ss interaction partially restored polyadenylation, indicating that SR proteins can directly promote polyadenylation independent of the NRS complex. The partial rescue may reflect a decreased capacity of the SELEX site relative to NRS5′ to recruit SR proteins, or that other NRS complex components have a role in optimal RSV polyadenylation. For example, this could include U2AF65, whose binding to the viral 3′ ss might be stabilized within the NRS complex to recruit CFIm59 through their respective RS-like domains (42); U2AF65 also interacts with poly(A) polymerase (54). In another scenario, U2 snRNP binding to the suboptimal env branch point (19, 25) may be stabilized within the stalled NRS complex, and U2 might assist in RSV poly(A) site recognition. This is based on the recent demonstration that the SF3b components of U2 snRNP and subunits of the CPSF interact, and that this interaction mediates coupling of splicing and 3′-end formation (27). Finally, two observations suggest the possibility that U1 snRNP within the NRS complex contributes to RSV polyadenylation: U1A protein can interact with CPSF160 and stimulate polyadenylation (30), and U1 snRNP interacts with CFIm (4). Any of these interactions could collaborate with SR proteins to improve recognition of the RSV poly(A) site (Fig. 9B).
hnRNP H is an auxiliary polyadenylation factor for some viral and cellular poly(A) sites (2, 5), and hnRNP H/H′ sites have been found near numerous cellular poly(A) sites (24, 52). The NRS harbors two hnRNP H-binding regions, a weak upstream site that is embedded in the SR protein-binding region (17) and a strong downstream site(s) required to recruit U11 snRNP to the NRS (37). These observations suggested that hnRNP H might also contribute to RSV polyadenylation. This does not appear to be the case, since (i) a mutation in the upstream site had no effect on polyadenylation in vitro (the NRS substrate used in vitro lacked the downstream hnRNP H sites), (ii) mutation of the strong sites had no effect on RSV polyadenylation in vivo, and (iii) the ASF/SF2 SELEX sites alone promoted RSV polyadenylation in vivo. While evidence of a positive role for hnRNP H in RSV polyadenylation is lacking, a potential negative role was reported by Fogel et al. using proviral clones, but only when SR protein binding was compromised (17). This possibility was supported by a recent report in which, in vitro, sequestration of hnRNP H led to increased RSV polyadenylation, presumably by relieving competition with SR protein binding to NRS5′ (56). However, arguing against this idea is our finding that RSV polyadenylation was not increased in vivo when the strong hnRNP H sites were mutated. Additional in vivo experimentation will be required to clarify the natural influence of hnRNP H on RSV polyadenylation.
It has become clear that a large number of cellular poly(A) sites lack the canonical AAUAAA sequence, and these sites likely benefit from the action of a variety of auxiliary elements and binding factors (6, 24, 52). Our findings that SR proteins promote RSV polyadenylation suggest that SR proteins may be one such class of factors. Some SR proteins remain associated with spliced exons and may influence polyadenylation of cellular mRNAs more generally than has been appreciated. Alternatively, analogous to nonsplicing sites for U2AF that couple to polyadenylation (15), SR protein-binding sites that are distinct from those involved in splicing might act as auxiliary elements to influence recognition of suboptimal or alternative poly(A) sites. These elements would likely be in the 3′-terminal exon near the poly(A) site, unlike the distant sites in the NRS, which are thought to require the NRS-3′ ss complex for positioning SR proteins close to the poly(A) site.
Acknowledgments
We are grateful to members of the McNally laboratory for helpful comments on the manuscript.
This work was supported by Public Health Service grant R01 CA78709 from the National Cancer Institute to M.T.M.
Footnotes
Published ahead of print on 1 August 2007.
REFERENCES
- 1.Amendt, B. A., S. B. Simpson, and C. M. Stoltzfus. 1995. Inhibition of RNA splicing at the Rous sarcoma virus src 3′ splice site is mediated by an interaction between a negative cis element and a chicken embryo fibroblast nuclear factor. J. Virol. 69:5068-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arhin, G. K., M. Boots, P. S. Bagga, C. Milcarek, and J. Wilusz. 2002. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 30:1842-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrigo, S., and K. Beemon. 1988. Regulation of Rous sarcoma virus RNA splicing and stability. Mol. Cell. Biol. 8:4858-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awasthi, S., and J. C. Alwine. 2003. Association of polyadenylation cleavage factor I with U1 snRNP. RNA 9:1400-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagga, P. S., G. K. Arhin, and J. Wilusz. 1998. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 26:5343-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaudoing, E., S. Freier, J. R. Wyatt, J. M. Claverie, and D. Gautheret. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10:1001-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabello-Villegas, J., K. E. Giles, A. M. Soto, P. Yu, A. Mougin, K. L. Beemon, and Y. X. Wang. 2004. Solution structure of the pseudo-5′ splice site of a retroviral splicing suppressor. RNA 10:1388-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavaloc, Y., C. F. Bourgeois, L. Kister, and J. Stevenin. 1999. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA 5:468-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cochrane, A. W., M. T. McNally, and A. J. Mouland. 2006. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology 3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffin, J. M. 1996. Retroviridae: the viruses and their replication, p. 1767-1847. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Raven Press, New York, NY.
- 11.Cook, C. R., and M. T. McNally. 1999. Interaction between the negative regulator of splicing element and a 3′ splice site: requirement for U1 small nuclear ribonucleoprotein and the 3′ splice site branch point/pyrimidine tract. J. Virol. 73:2394-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook, C. R., and M. T. McNally. 1998. SR protein and snRNP requirements for assembly of the Rous sarcoma virus negative regulator of splicing complex in vitro. Virology 242:211-220. [DOI] [PubMed] [Google Scholar]
- 13.Cooke, C., and J. C. Alwine. 2002. Characterization of specific protein-RNA complexes associated with the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol. 22:4579-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooke, C., H. Hans, and J. C. Alwine. 1999. Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol. 19:4971-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danckwardt, S., I. Kaufmann, M. Gentzel, K. U. Foerstner, A. S. Gantzert, N. H. Gehring, G. Neu-Yilik, P. Bork, W. Keller, M. Wilm, M. W. Hentze, and A. E. Kulozik. 2007. Splicing factors stimulate polyadenylation via USEs at noncanonical 3′ end formation signals. EMBO J. 26:2658-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dettwiler, S., C. Aringhieri, S. Cardinale, W. Keller, and S. M. Barabino. 2004. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J. Biol. Chem. 279:35788-35797. [DOI] [PubMed] [Google Scholar]
- 17.Fogel, B. L., L. M. McNally, and M. T. McNally. 2002. Efficient polyadenylation of Rous sarcoma virus RNA requires the negative regulator of splicing element. Nucleic Acids Res. 30:810-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fogel, B. L., and M. T. McNally. 2000. A cellular protein, hnRNP H, binds to the negative regulator of splicing element from Rous sarcoma virus. J. Biol. Chem. 275:32371-32378. [DOI] [PubMed] [Google Scholar]
- 19.Fu, X.-D., R. A. Katz, A. M. Skalka, and T. Maniatis. 1991. The role of branchpoint and 3′ exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 5:211-220. [DOI] [PubMed] [Google Scholar]
- 20.Giles, K. E., and K. L. Beemon. 2005. Retroviral splicing suppressor sequesters a 3′ splice site in a 50S aberrant splicing complex. Mol. Cell. Biol. 25:4397-4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gontarek, R. R., M. T. McNally, and K. Beemon. 1993. Mutation of an RSV intronic element abolishes both U11/U12 snRNP binding and negative regulation of splicing. Genes Dev. 7:1926-1936. [DOI] [PubMed] [Google Scholar]
- 22.Herman, S. A., and J. M. Coffin. 1986. Differential transcription from the long terminal repeats of integrated avian leukosis virus DNA. J. Virol. 60:497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hibbert, C. S., R. R. Gontarek, and K. L. Beemon. 1999. The role of overlapping U1 and U11 5′ splice site sequences in a negative regulator of splicing. RNA 5:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu, J., C. S. Lutz, J. Wilusz, and B. Tian. 2005. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11:1485-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz, R. A., and A. M. Skalka. 1990. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol. Cell. Biol. 10:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohtz, J. D., S. F. Jamison, C. L. Will, P. Zuo, R. M. A. Garcia-Blanco, and J. L. Manley. 1994. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368:119-124. [DOI] [PubMed] [Google Scholar]
- 27.Kyburz, A., A. Friedlein, H. Langen, and W. Keller. 2006. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol. Cell 23:195-205. [DOI] [PubMed] [Google Scholar]
- 28.Lemaire, R., J. Prasad, T. Kashima, J. Gustafson, J. L. Manley, and R. Lafyatis. 2002. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. Genes Dev. 16:594-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, H. X., M. Zhang, and A. R. Krainer. 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 12:1998-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutz, C. S., K. G. Murthy, N. Schek, J. P. O'Connor, J. L. Manley, and J. C. Alwine. 1996. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 10:325-337. [DOI] [PubMed] [Google Scholar]
- 31.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 32.McCracken, S., M. Lambermon, and B. J. Blencowe. 2002. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol. Cell. Biol. 22:148-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCracken, S., D. Longman, I. L. Johnstone, J. F. Caceres, and B. J. Blencowe. 2003. An evolutionarily conserved role for SRm160 in 3′-end processing that functions independently of exon junction complex formation. J. Biol. Chem. 278:44153-44160. [DOI] [PubMed] [Google Scholar]
- 34.McNally, L. M., and M. T. McNally. 1998. An RNA splicing enhancer-like sequence is a component of a splicing inhibitor element from Rous sarcoma virus. Mol. Cell. Biol. 18:3103-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNally, L. M., and M. T. McNally. 1996. SR protein splicing factors interact with the Rous sarcoma virus negative regulator of splicing element. J. Virol. 70:1163-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNally, L. M., and M. T. McNally. 1999. U1 small nuclear ribonucleoprotein and splicing inhibition by the Rous sarcoma virus negative regulator of splicing element. J. Virol. 73:2385-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNally, L. M., L. Yee, and M. T. McNally. 2006. hnRNP H is required for optimal U11 snRNP binding to a retroviral RNA processing control element: implications for U12-dependent RNA splicing. J. Biol. Chem. 281:2478-2488. [DOI] [PubMed] [Google Scholar]
- 38.McNally, L. M., L. Yee, and M. T. McNally. 2004. Two regions promote U11 snRNP binding to a retroviral splicing inhibitor element (NRS). J. Biol. Chem. 279:38201-38208. [DOI] [PubMed] [Google Scholar]
- 39.McNally, M. T., and K. Beemon. 1992. Intronic sequences and 3′ splice sites control Rous sarcoma virus RNA splicing. J. Virol. 66:6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melton, D. A., P. A. Krieg, M. R. Rebagliati, T. Maniatis, K. Zinn, and M. R. Green. 1984. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12:7035-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, J. T., and C. M. Stoltzfus. 1992. Two distant upstream regions containing cis-acting signals regulating splicing facilitate 3′-end processing of avian sarcoma virus RNA. J. Virol. 66:4242-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Millevoi, S., C. Loulergue, S. Dettwiler, S. Z. Karaa, W. Keller, M. Antoniou, and S. Vagner. 2006. An interaction between U2AF 65 and CF I (m) links the splicing and 3′ end processing machineries. EMBO J. 25:4854-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niwa, M., S. D. Rose, and S. M. Berget. 1990. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 4:1552-1559. [DOI] [PubMed] [Google Scholar]
- 44.O'Sullivan, C. T., T. S. Polony, R. E. Paca, and K. L. Beemon. 2002. Rous sarcoma virus negative regulator of splicing selectively suppresses Src mRNA splicing and promotes polyadenylation. Virology 302:405-412. [DOI] [PubMed] [Google Scholar]
- 45.Rüegsegger, U., D. Blank, and W. Keller. 1998. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell 1:243-253. [DOI] [PubMed] [Google Scholar]
- 46.Schwartz, D. E., R. Tizard, and W. Gilbert. 1983. Nucleotide sequence of Rous sarcoma virus. Cell 32:853-869. [DOI] [PubMed] [Google Scholar]
- 47.Seiwert, S. D., and J. A. Steitz. 1993. Uncoupling two functions of the U1 small nuclear ribonucleoprotein particle during in vitro splicing. Mol. Cell. Biol. 13:3135-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, H. O., K. Tabiti, G. Schaffner, D. Soldati, U. Albrecht, and M. L. Birnstiel. 1991. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2′-O-methyl oligoribonucleotides. Proc. Natl. Acad. Sci. USA 88:9784-9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoltzfus, C. M., and S. J. Fogarty. 1989. Multiple regions in the Rous sarcoma virus src gene intron act in cis to affect the accumulation of unspliced RNA. J. Virol. 63:1669-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoltzfus, C. M., S. K. Lorenzen, and S. L. Berberich. 1987. Noncoding region between the env and src genes of Rous sarcoma virus influences splicing efficiency at the src gene 3′ splice site. J. Virol. 61:177-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tacke, R., and J. L. Manley. 1995. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 14:3540-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian, B., J. Hu, H. Zhang, and C. S. Lutz. 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33:201-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vagner, S., U. Ruegsegger, S. I. Gunderson, W. Keller, and I. W. Mattaj. 2000. Position-dependent inhibition of the cleavage step of pre-mRNA 3′-end processing by U1 snRNP. RNA 6:178-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vagner, S., C. Vagner, and I. W. Mattaj. 2000. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 14:403-413. [PMC free article] [PubMed] [Google Scholar]
- 55.Wassarman, K. M., and J. A. Steitz. 1993. Association with terminal exons in pre-mRNAs: a new role for the U1 snRNP? Genes Dev. 7:647-659. [DOI] [PubMed] [Google Scholar]
- 56.Wilusz, J. E., and K. L. Beemon. 2006. The negative regulator of splicing element of Rous sarcoma virus promotes polyadenylation. J. Virol. 80:9634-9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, J. Y., and T. Maniatis. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75:1061-1070. [DOI] [PubMed] [Google Scholar]
- 58.Zahler, A. M., W. S. Lane, J. A. Stolk, and M. B. Roth. 1992. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 6:837-847. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, L., and C. M. Stoltzfus. 1995. A suboptimal src 3′ splice site is necessary for efficient replication of Rous sarcoma virus. Virology 206:1099-1107. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, Z., and A. R. Krainer. 2004. Involvement of SR proteins in mRNA surveillance. Mol. Cell 16:597-607. [DOI] [PubMed] [Google Scholar]