Abstract
The interactions between herpes simplex virus gD and its nectin1 receptor or between gD, gB, and gH were analyzed by complementation of the N and C portions of split enhanced green fluorescent protein (EGFP) fused to the glycoproteins. The gDN-NectC complex was readily detected; the gDN-gCC complex was undetectable, highlighting the specificity of the assay. Split EGFP complementation was detected between proteins designated gDN+gHC, gDN+gBC, and gHN+gBC+wtgD (gB was deleted of endocytosis motifs), both in cells transfected with two-tree glycoproteins and in syncytia. The in situ assay provides evidence that gD interacts with gH and gB independently of each other and supports a model whereby gH and gB in complex exert their activities to gD.
The entry of herpes simples virus (HSV) into cells requires a multipartite fusion system made of a quartet of glycoproteins (3, 24, 28). The receptor-binding glycoprotein gD interacts with three alternative receptors, nectin1, herpesvirus entry mediator, and modified heparan sulfate (5, 11, 22, 27). gD also encodes a profusion domain at the ectodomain C terminus, which is required to trigger fusion (4). In the unliganded gD, the ectodomain C terminus folds around the N terminus. At receptor binding, the C terminus is displaced, gD adopts an open conformation, and fusion is triggered (8, 20). Three glycoproteins conserved across the Herpesviridae family, gB and gH·gL, execute fusion (2, 7, 25). The identity of the executor—whether it is gB, gH·gL, or the three glycoproteins together—remains unclear. Thus, gB exhibits a trimeric structure, properties typical of class I and II viral fusion proteins, and a candidate fusion loop (16, 17). On the other hand, gH exhibits elements typical of class I fusion glycoproteins, including two heptad repeats able to form a coiled coil and a candidate fusion peptide, besides having additional hydrophobic regions (9, 10, 12-15). Hemifusion (the fusion of the outer layers of the virion and cell membranes) requires gD and gH·gL; complete fusion (the mixing of both outer and inner lipid layers) additionally requires gB (29). Fusion between perinuclear virions and the outer nuclear membranes, which culminates in capsid release into the cytoplasm, requires gD plus either gB or gH·gL, implying that, under particular conditions, either gB or gH·gL suffice for fusion execution (6).
A key question in HSV entry/fusion centers on how gD signals the encounter with its receptor to the downstream glycoproteins and thus triggers fusion. The working model investigated in this laboratory envisions that the receptor-bound gD forms complexes with the downstream glycoproteins or with a subset of them (3). Indeed, by coimmunoprecipitation, gD was shown to be in a complex with gH (23).
The aim of this work was to investigate, by means of a protein complementation assay (CA) (19, 21), in intact cells, the molecular interactions that take place between gD and nectin1 and between the four glycoproteins. In the CA, proteins like enhanced green fluorescent protein (EGFP) are split into two portions that, if brought to an 8- to 10-Å proximity of each other, refold together and emit fluorescence (19, 21). In the current adaptation to membrane proteins, the EGFP N terminus (N; amino acids [aa] 1 to 157) was fused to the endodomain of HSV type 1 gD or gH, and the C terminus (C) was fused to the endodomain of gB, gH, gC, or nectin1. The glycoproteins under examination interact, or are likely to interact through their ectodomains; the EGFP portions were located in the endodomains. The assumption was made that any specific interaction occurring between the ectodomains would result in refolding and fluorescence emission of the endodomain-located EGFP fragments. Fluorescence intensity from the complemented EGFP was reported to be about 10% of that produced by the unsplit protein, essentially because only a subset of the fragments have a likelihood of associating with each other (see reference 19). This drawback is balanced by the fact that complementation is driven by specific interactions that occur between the proteins under investigation, to which the EGFP portions are fused. Moreover, the refolded EGFP adopts an irreversible conformation that contributes to stabilizing the complex and thus enables the detection of transient and weak complexes (18, 19).
Plasmid construction.
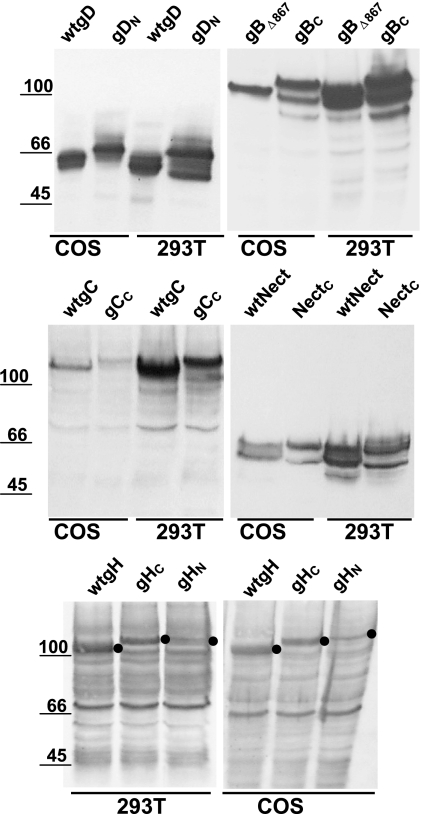
The mammalian expression plasmids for gH in the MTS vector, and gD and nectin1 in pcDNA3.1 (5, 31) were site-directed mutagenized 0 to 10 aa upstream of the stop codon, in order to generate restriction sites for the insertion of N or C amplimers. The sites were SphI for gH and BglII for gD or nectin1. Where necessary, the BglII site of pcDNA3.1 was preliminarily eliminated by digestion, filling in by T4-DNA polymerase and religation. N and C sequences were PCR amplified from pCMS-EGFP (Clontech) with the primer pairs (or variations thereof) 5′CCCAGATCTC CATGGTGAGC AAGGGCGAGG AGCTGT plus 5′GGGAAGCTTC TACTTGTCGG CCATGATATA GACGTTG or 5′CCCGCTAGCT CAGAAGAACG GCATCAAGGT GAACT plus 5′GGGAGATCTT ACTTGTACAG CTCGTCCATG CCGAGA, respectively. N amplimer was ligated with BglII-HindIII-digested gD plasmid or SphI-BglII-digested gH plasmid, generating gDN and gHN. The C-EGFP amplimer was ligated with digested nectin1 (BglII-XhoI) and gH (SphI-BglII) plasmids, generating NectC and gHC. The gC gene sequence was PCR amplified from DNA of HSV type 1 (F) with primers 5′AGATCTAGGC CTATGGCCCC GGGGCGGGTG GGCCTTGCCG TGGTCCTGTG GAGCCTG and 5′GAAGATGCGG CCGCTTAGCT AGCCGCCGAT GACGCTGCCG CGACTGTGAT GTGCG. The StuI-NheI-digested gC amplimer and the NheI-BglII-digested C amplimer were ligated with StuI-BglII-digested MTS vector. The gB-encoding plasmid in pcDNA3.1 was deleted of the endodomain sequences that carry endocytosis motifs, from aa 867 to the stop codon (gBΔ867), in order to maximize gB expression (1). The gBC chimera was generated by mixing gBΔ867 and C amplimers, generated with primer pairs 5′GGCTGGATCC TCCCCGTAGT CCCGCCATGC plus CCTTGATGCC GTTCTTCTGA GATCTCTTCT TCTTGGCCTT GTGTTC and 5′GAACACAAGG CCAAGAAGAA GAGATCTCAG AAGAACGGCA TCAAGG plus 5′GGGAAGCTTT TACTTGTACA GCTCGTCCAT GCCGAGA, followed by ligation with BamHI-HindIII-digested pcDNA3.1. Arrestin-transfected 293T or COS cells, mounted without fixation with Fluoromount, were observed with a Leica TCS-SL confocal microscope, set at 100% excitation at 488 nm with emission between 490 to 540 nm. Images were collected with a 63× 1.62 Leica oil immersion objective; confocal slices were 1.7 to 2.3 μm thick. For each experimental series—see Figure 2A to T and U to Z and Fig. 3A to F, G to I, J to L, and M to P—images were collected on the same day, under the same settings, applying 1,024- by 1,024-pixel resolution and an 8-bit intensity scale. Specifically, the first sample to be analyzed was the negative one, containing gCc; for subsequent observations of the samples belonging to the same series, the settings were then kept unmodified. Figure 1 shows the electrophoretic mobility of the N-EGFP or C-EGFP fusion proteins generated in this study, made in transfected COS or 293T cells, detected by Western blotting with monoclonal antibodies H170, H1817, H633, and CK6 to gD, gB, gC, and nectin1, respectively, and polyclonal antibodies to gH. As expected, all the fusion proteins exhibited a slower electrophoretic mobility relative to their respective wild-type versions.
FIG. 2.
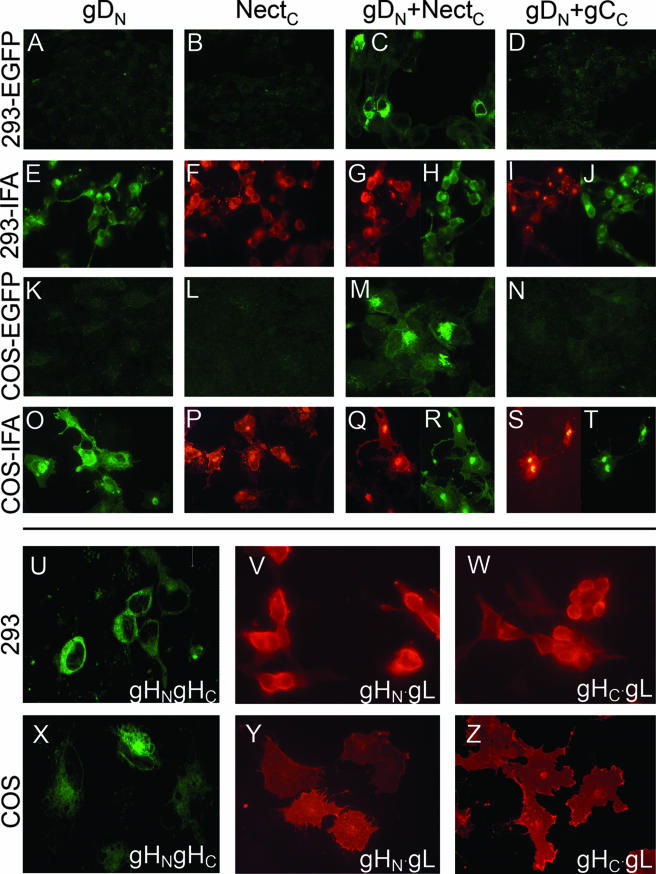
EGFP CA between gDN and NectC or between gHN+gHc, and lack of complementation between gDN and gCC. COS or 239T cells were transfected with the indicated plasmids, gDN (A, E, K, and O), NectC (B, F, L, and P), gDN+NectC (C, G, H, M, Q, and R), gDN+gCc (D, I, J, N, S, and T), and gHN+gHc (U to Z). (A to D, K to N, U, and X) CA. (E to J, O to T, V, W, Y, and Z) IFA. Green, gD; red, all others. Antibodies used were R8 to gD, R1.302 to nectin1, H633 to gC, and 53S to gH (reacts to a gL-dependent epitope).
FIG. 3.
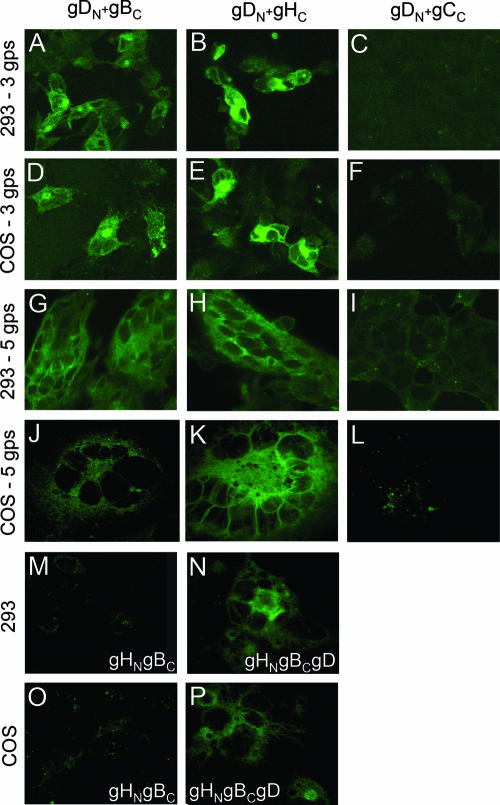
EGFP CA between HSV glycoproteins. COS or 239T cells were transfected with the indicated plasmids, gDN+gBC (A, D, G, and J), gDN+gHC (B, E, H, and K), gDN+gCC (C, F, I, and L), gHN+gBc (M and O), and gHN+gBc+gD (N and P). Cells transfected with three glycoproteins (gps) received, in addition, plasmids encoding wtgL or epidermal growth factor receptor, as appropriate. Cells transfected with five glycoproteins received gD, gBΔ867, gH, gL, and gC as the wild type or EGFP chimeras, as indicated.
FIG. 1.
Electrophoretic mobility of proteins fused to N- or C-EGFP portions, or their wild-type alleles, expressed in COS or 293T cells. Numbers on the left indicate the migration positions of molecular mass markers (in kilodaltons).
gD-nectin1 complex detection.
Inasmuch as EGFP-CA has been applied mainly to analysis of soluble mammalian or bacterial proteins (19), we first validated its application to membrane proteins—in particular, to HSV glycoproteins—by analysis of gD and its nectin1 receptor. Figure 2C and M documents complex formation between gDN and NectC as fluorescence emission from EGFP-CA in transfected 293T or COS cells. Cells were observed 36 h after arrestin-mediated transfection. Results with the two cell lines were essentially similar, although the level of expression and number of fluorescent cells was higher with 293T cells. In agreement with previous reports, the overall fluorescence emitted by complementation of split EFGP fragments was lower than that from the unsplit protein (19).
The specificity controls that validated the assay were as follows. First, neither gDN nor NectC emitted fluorescence when transfected singly (Fig, 2A, B, K, and L) or in combination with the wild-type alleles of nectin1 or gD (not shown), ruling out autofluorescence. Second, fluorescence was reconstituted only when the EGFP chimeric proteins were in a specific complex and not simply present in the same subcellular compartment. For this control, we selected gC, which is involved in virus attachment but not virus entry and is present in the same subcellular compartments as gD. The coexpression of gDN and gCC resulted in no or background fluorescence (Fig. 2D and N), ruling out the possibility that proteins that exhibit no specific interaction, but that are abundantly present in the same cellular compartment, give rise to EGFP complementation. We took advantage of the lack of EGFP complementation by gCC-containing samples and, in all experiments, used the gCC-containing sample to adjust the confocal microscope settings. The settings were then kept constant throughout the observation period of a same series of samples. Third, we ascertained by immunofluorescence assay (IFA) that all proteins were expressed, even those expressed singly (gDN, NectC) (Fig. 2E, F, O, and P) or in the gDN-gCC combination that did not yield EGFP fluorescence (Fig. 2I, J, S, and T). Importantly, the EGFP-glycoprotein chimeras were not hampered in plasma membrane localization (Fig. 2). We conclude that EGFP-CA fulfills the criteria for detection of specific interactions between membrane-bound proteins, particularly HSV-1 gD and its receptor.
Complexes between HSV glycoproteins.
The second series of experiments was performed with 293T and COS cells transfected with three membrane proteins in combinations that included gDN+gBC, gDN+gBΔ867, gDN+gHC+wtgL, gDN+wtgH+wtgL, and gDN+gCC. We used a form of gB deleted for endocytosis motifs, to maximize its expression and localization in exocytic and plasma membranes (1). Transfection mixtures were made equal in DNA amounts (900 ng/well, 300 ng/plasmid) by the addition of a plasmid encoding epidermal growth factor receptor 1 deleted of signaling sequences (26). This control ensured that exocytic membranes were loaded with comparable amounts of proteins. In both cell types observed 36 h after transfection, the gDN+gHC+wtgL combination resulted in a readily detectable fluorescence (Fig. 3B and E). The gDN+gBC combination gave rise to a somewhat weaker fluorescence (Fig. 3A and D) that nonetheless was much higher than the background fluorescence emitted by the gDN+gCC combination (Fig. 3C and F). Even though the subcellular localization cannot be clearly defined, EGFP appeared to localize to a perinuclear position, consistent with a Golgi compartment localization, to a cytoplasmic reticular compartment, consistent with endoplasmic reticulum, and to nuclear membranes. By IFA, all proteins resulted to be expressed, even those that did not yield EGFP fluorescence (not shown). We infer that gD can recruit gH to a complex. gD can also recruit gB to a complex. The gD-gH combination results in a stronger EGFP fluorescence than the gD-gB combination, possibly reflecting a stronger interaction, a more stable or longer half-life complex, a higher number of complexes at steady state, or peculiar behaviors of the fusion proteins.
Cells transfected with the quartet of gD, gB, gH, and gL form syncytia (30). A series of experiments was designed to verify whether the glycoprotein-EGFP chimeras were still functional in cell-cell fusion, and whether complexes were detectable under conditions that lead to cell-cell fusion. 293T or COS cells were cotransfected with combinations of five plasmids (1.25 μg/well, 250 ng/plasmid) encoding gD, gBΔ867, gH, gL, and gC or their EGFP chimeras. The transfected combinations included gDN+gBC+wtgH+wtgL+wtgC, gDN+gHC+wtgL+gBΔ867+wtgC, and gDN+gBΔ867+wtgH+wtgL+gCC. Cells were observed 24 h (293T) or 40 h (COS) after transfection. The results in Fig. 3G to L show that syncytia were formed for any combination, indicating that the EGPF-glycoprotein chimeras were not hampered in fusion activity. The strongest fluorescence was observed with the combination that included gDN+gHC (panels H and K). A somewhat weaker fluorescence was observed with the combination that included gDN+gBC (panels G and J), particularly in COS cells. No fluorescence above background level was observed with the combinations that included gDN+gCC (panels I and L). The stronger fluorescence in panels B relative to H and in panels E relative to K reflects (i) higher amounts of transfected DNA for each plasmid, (ii) a longer time interval after transfection (panels B versus H), (iii) the lack of dilution of complemented EGFP molecules consequent to fusion of transfected fluorescent cells with adjacent untransfected nonfluorescent cells, or (iv) possibly a longer half-life of the complexes when cell-cell fusion does not ensue.
We next tested whether gH and gB interact with each other and whether the interaction was dependent on the presence of gD. Cells were transfected with gHN+wtgL+gBC in the absence or presence of wtgD. Interaction between gHN-gBC was readily documented in the presence (Fig. 3N and P) but not in the absence (Fig. 3M and O) of gD.
While gB is known to be a trimer (17), the oligomeric state of gH is unknown. Here, we addressed the question of whether the split EGFP-CA was suitable to define the oligomeric state of gH. Cells were transfected with gHN+gHC+wtgL. As shown in Fig. 2U to Z, the CA readily documented that the gH·gL heterodimer (detected by its reactivity to MAb 53S) formed oligomeric structures in transfected cells.
Concluding remarks.
We validated the adaptation of the EGFP-CA to membrane proteins by first applying it to the gD-nectin1 interaction. The fluorescence emitted from the gDN-NectC combination was readily detectable, whereas that from the gDN-gCC combination was detected at only background levels, testifying to the assay specificity. For every series of observations, the gCC-containing sample was therefore used to adjust the confocal microscope settings. Samples exhibiting readily detectable fluorescence under these conditions were considered positive.
We detected a complex made of gD and gH, in agreement with coimmunoprecipitation data (23). The complex formed even in the absence of gB. In addition, we detected a complex made of gD and gB that formed even in the absence of gH·gL. We further documented the interaction between gH and gB; its gD dependence suggests that the interaction is triggered by gD. A notable property of the EGFP-CA as applied here was that complex formation between HSV glycoproteins was detected in intact cells, i.e., in the intracellular compartment and microenvironment and under the very conditions in which the interactions do occur. Importantly, the EGFP chimeric glycoproteins were not hampered in cell-cell fusion activity. Hence, the detected interactions were a faithful mirror of the interactions that take place under conditions that lead to cell-cell fusion. Inasmuch as EGFP reconstitution from split portions is an irreversible reaction, the assay does not allow us to infer whether the complexes between the HSV glycoproteins were stable or transient.
By taking advantage of the fact that CHO but not Vero cells lack the lipid ganglioside GM1, hemifusion, i.e., the mixing of the outer lipid leaflets of the virion envelope and cell membrane or cell-cell membranes, was differentiated from fusion, i.e., complete lipid mixing and content mixing (29). In that assay, gD and gH·gL are sufficient to induce hemifusion. Complete fusion additionally requires gB (29). Those findings support the view that HSV fusion occurs through steps, i.e., the juxtaposition of membranes and the triggering of fusion, hemifusion, and complete fusion. They do not shed light on the sequential order and mechanism of gB recruitment. Current data agree with those from the hemifusion study and, moreover, argue that hemifusion is carried out by the gD-gH·gL complex. Of note, the fact that three independent assays—coimmunoprecipitation, hemifusion and split EGFP CA—concordantly showed the interaction between gD and gH strongly substantiates the current approach.
Cumulatively, the current assay provides in situ evidence for the following. (i) gD recruits gH·gL and gB to complexes. (ii) gH and gB can be recruited to gD independently of one another. Thus, gD carries binding sites for both gH·gL and gB. The independent recruitment of these glycoproteins to gD is consistent with and substantiated by the observation that, at the outer nuclear membrane, virions deleted for gB but carrying gD+gH, or deleted for gH but carrying gD+gB are capable of fusion (6). (iii) Once gH·gL and gB are recruited to gD, they possibly interact with each other. (iv) gH·gL and gB are not necessarily recruited in a sequential order or one to the other. Current data support a model of HSV entry-fusion whereby gH and gB exert their activity through complex formation with gD, or following activation mediated by complex formation with gD.
Acknowledgments
We thank our colleagues G. Cohen and R. Eisenberg (University of Pennsylvania), T. Minson (Cambridge University), ande M. Lopez (INSERM, Marseille, France) for gifts of antibodies; C. Taddei and L. Dipietrangelo for invaluable assistance with confocal microscopy; the members of our laboratory for providing constructs; and Elisabetta Romagnoli for helpful assistance.
The work was supported by EU contract TargetHerpes-VI FP LSHG-CT-2006-037517, MIUR-PRIN-2005, University of Bologna RFO, and Fondi Roberto e Cornelia Pallotti from our Department.
Footnotes
Published ahead of print on 1 August 2007.
REFERENCES
- 1.Avitabile, E., G. Lombardi, T. Gianni, M. Capri, and G. Campadelli-Fiume. 2004. Coexpression of UL20p and gK inhibits cell-cell fusion mediated by herpes simplex virus glycoproteins gD, gH-gL, and wild-type gB or an endocytosis-defective gB mutant and downmodulates their cell surface expression. J. Virol. 78:8015-8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai, W. H., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campadelli-Fiume, G., M. Amasio, E. Avitabile, A. Cerretani, C. Forghieri, T. Gianni, and L. Menotti. 2007. The multipartite system that mediates entry of herpes simplex virus into the cell. Rev. Med. Virol. 17:313-326. [DOI] [PubMed] [Google Scholar]
- 4.Cocchi, F., L. Menotti, V. Di Ninni, M. Lopez, and G. Campadelli-Fiume. 2004. The herpes simplex virus JMP mutant enters receptor-negative J cells through a novel pathway independent of the known receptors nectin1, HveA, and nectin2. J. Virol. 78:4720-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin superfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farnsworth, A., T. W. Wisner, M. Webb, R. Roller, G. Cohen, R. Eisenberg, and D. C. Johnson. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. USA 104:10187-10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fusco, D., C. Forghieri, and G. Campadelli-Fiume. 2005. The pro-fusion domain of herpes simplex virus glycoprotein D (gD) interacts with the gD N terminus and is displaced by soluble forms of viral receptors. Proc. Natl. Acad. Sci. USA 102:9323-9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galdiero, S., A. Falanga, M. Vitiello, H. Browne, C. Pedone, and M. Galdiero. 2005. Fusogenic domains in herpes simplex virus type 1 glycoprotein H. J. Biol. Chem. 280:28632-28643. [DOI] [PubMed] [Google Scholar]
- 10.Galdiero, S., M. Vitiello, M. D'Isanto, A. Falanga, C. Collins, K. Raieta, C. Pedone, H. Browne, and M. Galdiero. 2006. Analysis of synthetic peptides from heptad-repeat domains of herpes simplex virus type 1 glycoproteins H and B. J. Gen. Virol. 87:1085-1097. [DOI] [PubMed] [Google Scholar]
- 11.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 12.Gianni, T., R. Fato, C. Bergamini, G. Lenaz, and G. Campadelli-Fiume. 2006. Hydrophobic α-helices 1 and 2 of herpes simplex virus gH interact with lipids, and their mimetic peptides enhance virus infection and fusion. J. Virol. 80:8190-8198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gianni, T., P. L. Martelli, R. Casadio, and G. Campadelli-Fiume. 2005. The ectodomain of herpes simplex virus glycoprotein H contains a membrane α-helix with attributes of an internal fusion peptide, positionally conserved in the Herpesviridae family. J. Virol. 79:2931-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gianni, T., L. Menotti, and G. Campadelli-Fiume. 2005. A heptad repeat in herpes simplex virus gH, located downstream of the α-helix with attributes of a fusion peptide, is critical for virus entry and fusion. J. Virol. 79:7042-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gianni, T., A. Piccoli, C. Bertucci, and G. Campadelli-Fiume. 2006. Heptad repeat 2 in herpes simplex virus-1 gH interacts with heptad repeat 1 and is critical for virus entry and fusion. J. Virol. 80:2216-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannah, B. P., E. E. Heldwein, F. C. Bender, G. H. Cohen, and R. J. Eisenberg. 2007. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J. Virol. 81:4858-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217-220. [DOI] [PubMed] [Google Scholar]
- 18.Hu, C. D., Y. Chinenov, and T. K. Kerppola. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789-798. [DOI] [PubMed] [Google Scholar]
- 19.Kerppola, T. K. 2006. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 7:449-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krummenacher, C., V. M. Supekar, J. C. Whitbeck, E. Lazear, S. A. Connolly, R. J. Eisenberg, G. H. Cohen, D. C. Wiley, and A. Carfi. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michnick, S. W. 2001. Exploring protein interactions by interaction-induced folding of proteins from complementary peptide fragments. Curr. Opin. Struct. Biol. 11:472-477. [DOI] [PubMed] [Google Scholar]
- 22.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Romero, P., A. Perez, A. Capul, R. Montgomery, and A. O. Fuller. 2005. Herpes simplex virus entry mediator associates in infected cells in a complex with viral proteins gD and at least gH. J. Virol. 79:4540-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rey, F. A. 2006. Molecular gymnastics at the herpesvirus surface. EMBO Rep. 7:1000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67:2285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rovero, S., A. Amici, E. D. Carlo, R. Bei, P. Nanni, E. Quaglino, P. Porcedda, K. Boggio, A. Smorlesi, P. L. Lollini, L. Landuzzi, M. P. Colombo, M. Giovarelli, P. Musiani, and G. Forni. 2000. DNA vaccination against rat her-2/Neu p185 more effectively inhibits carcinogenesis than transplantable carcinomas in transgenic BALB/c mice. J. Immunol. 165:5133-5142. [DOI] [PubMed] [Google Scholar]
- 27.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13-22. [DOI] [PubMed] [Google Scholar]
- 28.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian, R. P., and R. J. Geraghty. 2007. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. USA 104:2903-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou, G., E. Avitabile, G. Campadelli-Fiume, and B. Roizman. 2003. The domains of glycoprotein D required to block apoptosis induced by herpes simplex virus 1 are largely distinct from those involved in cell-cell fusion and binding to nectin1. J. Virol. 77:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]