Abstract
Poly(A) binding protein 2 (PABP2) of Arabidopsis thaliana was previously shown to interact with VPg-Pro of turnip mosaic virus (TuMV) and may consequently play an important role during infection. Subcellular fractionation experiments revealed that PABP2 was predominantly a cytoplasmic soluble protein in healthy plants. However, in TuMV-infected plants, a subpopulation of PABP2 was membrane associated or was localized in the nucleus. Confocal microscopy experiments indicated that PABP2 was partially retargeted to the nucleolus in the presence of TuMV VPg-Pro. In addition, the membrane association of PABP2 during TuMV infection resulted from the internalization of the host protein in 6K-VPg-Pro-induced vesicles, as shown by a combination of confocal microscopy and sucrose gradient fractionation experiments. This redistribution of an important translation initiation factor to the nucleolus and to membrane structure likely underlies two important processes of the TuMV replication cycle.
Cellular mRNAs are 5′ capped and 3′ polyadenylated. The cap structure (m7GpppN, where N represents any nucleotide) and the poly(A) tail are involved in translation initiation by interacting with cap-binding eukaryotic initiation factor 4E (eIF4E) and poly(A) binding protein (PABP), respectively. eIF4E is part of the eIF4F complex, which also contains eIF4G and eIF4A. eIF4G functions as a scaffold protein that binds many factors involved in recruiting the 40S ribosomal subunit to mRNAs, among which are eIF4E and PABP (56). The interaction between eIF4G, eIF4E, and PABP and with the extremities of the mRNA leads to the circularization of the latter, which purportedly increases the efficiency of translation (12).
Viruses target translation initiation factors in order to take over the protein synthesis machinery of the infected cells (4). Several studies have shown that eIF4E is often directly or indirectly inactivated during virus-induced host translational shutdown (6, 15, 23). PABP is another frequent target of RNA viruses. For example, nonstructural protein 3 of rotaviruses binds the 3′ nonpolyadenylated end of the viral RNA. It also competes with PABP for eIF4G binding. Thus, it has been proposed that during rotavirus infection, nonstructural protein 3 evicts PABP from eIF4G, impairing the translation of cellular mRNAs while at the same time enhancing the translation of rotaviral mRNAs (60). Another strategy used by RNA viruses to shut down host translation is to cleave PABP. Cleavage has been observed during picornavirus (27, 28), calicivirus (26), and retrovirus (1) infections.
PABP may be inactivated to prevent host mRNA translation from taking place, but it is often required for viral RNA synthesis and translation. The closed-loop model of translation initiation that mediates the cross talk between the 5′ and 3′ ends of mRNA appears to also be pertinent for explaining the replication and/or translation of several viruses. Poliovirus replication requires genome circularization through a protein-protein bridge requiring, among several proteins, PABP (17). The PABP interaction with eIF4G, which interacts with picornavirus internal ribosome binding sites, stimulates viral translation, likely through circularization (37, 58). Finally, the involvement of PABP in genome circularization and in translation in coronaviruses has been suggested (55).
Potyviruses have a positive-sense single-stranded RNA genome of approximately 10 kb that is linked at its 5′ end to the viral protein VPg and is polyadenylated at its 3′ end (45). The genome encodes a single polyprotein that is processed by three viral proteinases. Genome circularization may take place through protein-RNA and protein-protein interactions involving RNA-linked VPg, eIF4E, eIF4G, PAPB, and the poly(A) tail. VPg has been shown to interact with eIF4E or its isomer eIF(iso)4E, depending on the virus-host combination (24, 38, 39, 49, 64). Several lines of evidence suggest that eIF4E plays an important role in potyvirus replication. Knockout Arabidopsis thaliana plants for eIF(iso)4E are resistant to potyviruses (9, 29). Recessive resistant genes against potyviruses have been shown to code for eIF4E (22, 41, 46, 47). Complementing those experiments was the demonstration that the virulence determinant for these recessive resistances was VPg (40, 42) and that the failure of eIF4E alleles to bind VPg correlated with resistance in most cases (22). Precursor forms of VPg (i.e., 6K-VPg-Pro and VPg-Pro) have been detected in turnip mosaic virus (TuMV)-infected plants (31). 6K-VPg-Pro, through its 6K domain, induces the formation of vesicles from the endoplasmic reticulum (ER) membranes, where RNA synthesis takes place (51). Recent data have shown that these vesicles were the site of interactions of eIF(iso)4E with 6K-VPg-Pro. Interestingly, a population of VPg-Pro was present in the nucleolus, where it also interacted with the translation factor (3).
VPg and its precursor forms interact with several other proteins (14), notably, the viral RNA-dependent RNA polymerase (RdRp) (7) and PABP2 (31). Given the involvement of this host protein during the replication of several viruses, we investigated the implication of PABP in TuMV infection. We examined the subcellular localization of PABP2 in TuMV-infected cells. Using cellular fractionation and confocal microscopy, we observed that a subpopulation of PABP2 was internalized in virus-induced vesicles or was transported into the nucleolus. Using coagroinfiltration experiments, we provide evidence suggesting that this redistribution was the result of the interaction of the host protein with 6K-VPg-Pro and VPg-Pro, respectively.
MATERIALS AND METHODS
Recombinant protein expression in Escherichia coli and purification.
Recombinant eIF(iso)4E of A. thaliana was purified as described previously (64). VPg-Pro was purified as described previously (36). pETPABPhis encodes a His-tailed PABP2 of A. thaliana (GenBank accession no. NM_119572) and was produced as follows. PCR was performed on total cDNA of A. thaliana using forward (5′-TATATACATATGGCTAGCCCGAATTCGATGGCGCAGGTTCAACTT-3′ [NheI and EcoRI sites are underlined]) and reverse (5′-TATATACTCGAGAGAGAGGTTCAAGGAAGC-39 [the XhoI is underlined]) primers. The amplified fragment was digested with NheI and XhoI and ligated into similarly restricted pET21b (Novagen), and the recombinant plasmid was introduced into E. coli BL21(DE3) cells. A culture of E. coli BL21(DE3) cells grown overnight was diluted 1:100 in fresh medium and incubated at 37°C until the optical density at 600 nm reached 0.6. Protein production was then induced at room temperature with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h. Purification by metal chelation was performed as described previously (36).
ELISA-based binding assay.
VPg-Pro was absorbed to wells of an enzyme-linked immunosorbent assay (ELISA) plate (100 μl of protein at 10 ng μl−1) by overnight incubation at 4°C, and wells were blocked with 5% BLOTTO in phosphate-buffered saline (PBS). Appropriate proteins were diluted in PBS with 1% BLOTTO and 0.2% Tween 20 and incubated for 1.5 h at 4°C with the previously coated wells. Detection of retained protein was achieved as in the ELISA with anti-T7-tagged or anti-His-tagged sera followed by a peroxidase-labeled goat anti-mouse immunoglobulin G (KPL). Wells were washed three times with 0.05% Tween 20 between incubations.
Plasmid construction for expression in plants.
Plasmids encoding VPg-Pro-green fluorescent protein (GFP), VPg-Pro-DsRed, VPg-Pro-ctGFP, 6K-VPg-Pro-GFP, and ntGFP-eIF(iso)4E were described previously (3). PABP was obtained by PCR (JFPABP [5′-TCGGGATCCGAAGCTTATGGCGCGGTT {the HindIII site is underlined}] and FTPABP [5′-TGCTCACCATGGGGGATCCGAGAGAGAGGTTCAGGA { the NcoI site is underlined}]) from pETT7PABP and ligated into HindIII/NcoI-restricted pGreen/EGFP (3). The fusion of eIF(iso)4E from A. thaliana with DsRed was obtained as follows. DsRed was obtained by PCR (JFDsRed [TCTAGAGGATCCCCCATGGCCTCCTCCGAGAAC {the BamHI is underlined}] and FTDsRed EcoRI [TAATTAAAGGAATTCTTACAGGAACGGTGGTGGCGGCCCA {the EcoRI site is underlined, and the stop codon is in boldface type}]) from pER-DsRed (16, 65) and ligated into a pSK/35S cassette by BamHI/EcoRI restriction sites. eIF(iso)4E [JFeIF(iso)4E (TCAACTTCTAGAAATATGGCGACCGAT; the XbaI site is underlined) and FTeIF(iso)4E (CTCGAAGGATCCGACAGTGAACCGGCT; the BamHI site is underlined)] was amplified from PCR2.1/eIF(iso)4E and ligated into XbaI/BamHI-restricted pSK/35S-DsRed. 35S-labeled DsRed and 35S-labeled eIF(iso)4E-DsRed were then transferred into pCam bia1380 (http://www.bioforge.net/forge/entry.jspa?externalID=161&categoryID=8). pKS/35SDsRed and pSK/35S-eIF(iso)4E-DsRed were digested with EcoRV and EcoRI, while pCambia1380 was digested with XhoI and blunted with T4 DNA polymerase followed by EcoRI digestion, resulting in pCambia/DsRed and pCambia/eIF(iso)4E-DsRed. pCambia/mCherry was amplified (JFmCherry [5′-TATAATATTCTAGAGGATCCATGGTGAGCAAGGGCGAGGA {the XbaI and BamHI sites are underlined}] and FTmCherry [5′-ATCTACTAGTTAACGAATTCTTACTTGTACAGCTCGTCCA {the EcoRI and HpaI sites are underlined, and the stop codon is in boldface type}]) from pRSET-B (54) and XbaI/HpaI was cloned into pCambia/DsRed, resulting in pCambia/mCherry. To obtain pCambia/PABP2-mCherry, PABP2 was amplified (JFPABP [5′-TCGGGATCCGAAGCTTATGGCGCGGTT {the HindIII site is underlined}] and FTPABP [5′TAATTATATTCTAGAGAGGTTCAAGGAAGCGA {the XbaI site is underlined}]) from pET-T7PABP2 and ligated into the pCambia/mCherry vector, which was previously digested with HindIII/XbaI.
Protein expression in plants.
Vectors containing genes for fluorescent fusion proteins were introduced into A. tumefaciens AGL1 by electroporation. Transformed cells were selected on kanamycin-ampicillin plates. Bacterial cultures grown overnight were centrifuged, and cells were resuspended in water supplemented with 10 mM MgCl2 and 150 μM acetosyringone. The resulting preparation was used to agroinfiltrate leaves from 3-week-old Nicotiana benthamiana plants, along with Agrobacterium-containing plasmids encoding P19 or HcPro. Plants were kept for 4 days in a growth chamber before observation.
Confocal microscopy.
Sections from agroinfiltrated leaves were cut out and placed in immersion oil on a microscope coverglass. The coverglass was then inverted over a microscope slide, presenting a depression above which was placed the leaf section. Individual cells were observed with a 40× oil immersion objective on a Radiance 2000 confocal microscope from Bio-Rad. Fluorescent proteins were excited with an argon-krypton laser. The data for green and red channels were collected simultaneously. Images were collected with a charge-coupled-device camera and treated with Adobe Photoshop or Image J (http://rsb.info.nih.gov/ij/) software.
Nucleus isolation.
Nucleus isolation was carried out according to a method described previously (13). Brassica perviridis plants (three-leaf stage) were infected with TuMV or mock inoculated with PBS. At 12 days postinoculation, leaves that developed next to the inoculated leaves were harvested. Leaf tissue (5 g) was cut into pieces with a blade and minced in 3 volumes (15 ml) of nucleus isolation buffer (NIB) (50 mM Tris-HCl [pH 7.2]-5 mM KCl-5 mM MgCl2-1 M sucrose-10 mM β-mercaptoethanol-0.2 mM phenylmethylsulfonyl fluoride). The homogenate was filtered through nylon membranes (160 μm and 41 μm) and subjected to centrifugation at 14,000 × g for 15 min at 4°C. The pellet was resuspended in 1 volume (5 ml) of NIB and homogenized. Seven milliliters of nuclei in 5% Percoll was layered onto a step gradient of Percoll with 5-ml layers of 15%, 30%, 45%, and 60% and subjected to centrifugation at 530 × g for 10 min and at 8,500 × g for 20 min at 4°C. Nuclei accumulated between the 60% and 45% Percoll layers and the 30% and 45% Percoll layers. Nuclei were diluted in 5 volumes of NIB, mixed gently by inversion, and subjected to centrifugation at 1,500 × g for 10 min. Nuclei were gently resuspended in 25 volumes of NIB and centrifuged again. Nuclei were resuspended in 100 μl of storage buffer (50 mM Tris-HCl [pH 8]-0.3 mM sucrose-5 mM MgCl2-1.5 mM NaCl-0.1 mM CaCl2-5 μM β-mercaptoethanol). Proteins from S14 and nucleus preparations were diluted 1:5 in loading sodium dodecyl sulfate (SDS) buffer, separated by SDS-polyacrylamide gel electrophoresis (PAGE), and detected by immunoblot analysis with the ECL-based secondary antibody system (Amersham).
Subcellular fractionation and membrane flotation assay.
B. perviridis plants (three-leaf stage) were infected with TuMV or mock inoculated with PBS. At 12 days postinoculation, leaves that developed next to the inoculated leaves were harvested. Plant tissues were extracted and fractionated into postnuclear (S3), soluble (S30), and membrane-enriched (P30) fractions as described previously (16, 51). Briefly, 1 g of tissue was ground in 4 ml of homogenization buffer (51). Nuclei, chloroplasts, cell wall, and debris were removed by centrifugation at 3,000 × g at 4°C for 10 min twice. The supernatant (S3) was centrifuged at 30,000 × g at 4°C for 30 min, resulting in soluble (S30) and crude membrane (P30) fractions. The pellets (P30) were resuspended in protein loading buffer (29) in a volume equal to that of the corresponding supernatant. Twenty microliters of S30 and P30 was analyzed by SDS-PAGE and detected by immunoblot analysis with the ECL-based secondary antibody system (Amersham).
Membrane flotation assays were conducted essentially as previously described (65). Briefly, P30 fraction was resuspended in 400 μl of NTE buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM EDTA), and 300 μl was mixed with 1.6 ml of 85% (wt/vol) sucrose in NTE buffer and overlaid with 7 ml of 65% sucrose in NTE buffer and then overlaid with 3.1 ml of 10% sucrose in NTE buffer. After centrifugation at 100,000 × g for 18 h in a Beckman SW41Ti rotor, 12 1-ml fractions were collected from the bottom of the tube, and 20 μl was analyzed by SDS-PAGE and detected by immunoblot analysis with the ECL-based secondary antibody system (Amersham).
Copurification of VPg-Pro.
The P30 fraction was resuspended in 4 ml of homogenization buffer containing 300 mM KCl and 40 mM octyl-β-glucopyranoside. The P30 fraction was washed at 30,000 × g for 20 min (4°C), and purification of VPg-Pro-containing complexes was performed by metal chelation as described previously (36). Immunoreactions were detected using the ECL-based secondary antibody system (Amersham).
Sucrose gradient.
The P30 fraction was resuspended in a volume of homogenization buffer equivalent to that used for the S30 fraction, layered onto a 9-ml 20 to 45% sucrose gradient containing the respective homogenization buffer, and subjected to centrifugation at 143,000 × g in a Beckman SW41 Ti rotor for 4 h at 4°C. Fractions (0.75 ml) were collected, diluted 1:5 in protein dissociation buffer, and subjected to immunoblot analysis following 12.5% SDS-PAGE. Immunoreactions were detected using the ECL-based secondary antibody system (Amersham).
RESULTS
Redistribution of PABP2 in TuMV-infected cells.
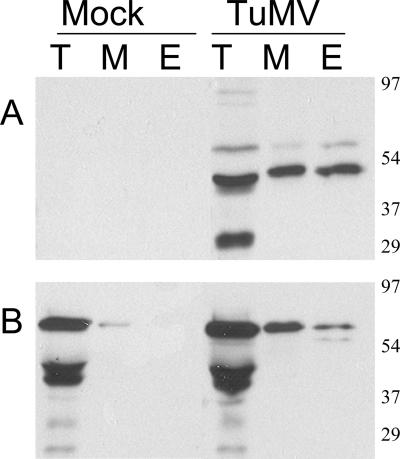
PABP2 has previously been shown to interact with VPg-Pro (31) and may consequently play an important role during the TuMV cell cycle. Its cellular distribution during infection was thus investigated by subcellular fractionation of extracts derived from mock-inoculated and TuMV-infected B. perviridis plants. B. perviridis was chosen because it is a natural host of TuMV and because it provides ample infected material for cellular fractionation experiments. Leaves that developed symptoms next to the inoculated ones were harvested 12 days postinfection, and soluble (S30) as well as membrane-enriched (P30) fractions were separated by centrifugation. To allow a quantitative evaluation of the distribution of the proteins among the two fractions, the P30 pellet was resuspended in the same volume as that of the S30 supernatant. Total, soluble, and membrane-associated proteins were separated by SDS-PAGE and subjected to immunoblot analysis. Fractions were analyzed with a rabbit serum raised against a recombinant form of PABP2 from A. thaliana. PABP2 was found mainly in the soluble fraction of mock-inoculated plants, although a small quantity was also observed in the membrane-enriched fraction (Fig. 1A). Generally, a somewhat higher level of PABP2 was noted in TuMV-infected cells. Most of the PABP2 was observed in the soluble fraction, but a significant quantity was also found to be associated with membranes.
FIG. 1.
Immunoblot analysis of soluble, membrane-associated, and nuclear proteins from healthy or TuMV-infected plants. B. perviridis plants were mock inoculated or infected with TuMV. (A) Twelve days after infection, total proteins (T) were extracted and soluble proteins (S) were separated from membrane-associated proteins (M) by centrifugation at 30,000 × g. Proteins were separated by SDS-PAGE and analyzed by Western blotting using a rabbit serum against PABP2. (B) Twelve days after infection, leaves were homogenized and centrifuged at 14,000 × g to separate the “soluble” fraction (S) from crude nuclei (N), which were further purified by Percoll gradient centrifugation. Proteins were separated by SDS-PAGE and analyzed by Western blotting using a rabbit serum against PABP2. The text on the left shows the electrophoretic migration positions of the indicated proteins.
The nuclear distribution of PABP2 was also investigated. The S14 fraction and Percoll gradient-purified nuclei were analyzed by immunoblot assay using antibodies raised against PABP2 (Fig. 1B). For the S14 fraction, 1/1,000 of the preparation was loaded onto the acrylamide gel, whereas one-fifth was loaded for the nuclear fraction. The PABP signal was detected in the S14 fraction of both mock- and TuMV-inoculated leaves. However, PABP2 was also detected in the nuclear-enriched fraction of TuMV-infected leaves but not in healthy leaves. These data then indicate that there is a cellular redistribution of PABP2 to the nucleus and membranes during TuMV infection.
Nuclear localization of PABP2 in the presence of VPg-Pro.
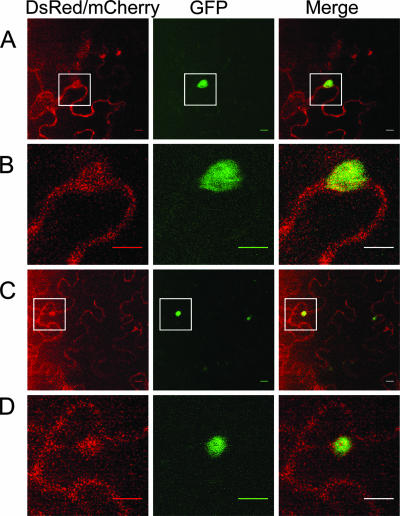
Since infected nuclei contain VPg-Pro (3, 50) and an in vitro interaction between this viral protein and PABP was reported previously (31), the localization of PABP2 in the nucleus might be driven by the presence of the viral protein in this cellular organelle. PABP2 was fused at its C-terminal end with GFP and expressed in N. benthamiana by agroinfiltration. Expression of the fusion protein was assessed by immunoblot analysis using a rabbit serum raised against PABP2, and a signal corresponding to the expected molecular mass for the fusion protein was observed (data not shown). Fluorescence was visualized 2 to 5 days postinfiltration by confocal microscopy. No notable differences in cellular localization were observed during this time period. Fluorescence was generally observed in 30 to 50% of the cells in the infiltrated area. A previously described ER-targeted fluorescent marker (ER-DsRed) (65), which showed the expected reticulate pattern (3), was coexpressed along with the PAPB2-GFP fusion to facilitate the identification of membrane structures and the nucleus. Fluorescence associated with PABP2-GFP was distributed exclusively in the cytoplasm (Fig. 2A). When PABP2-GFP was coexpressed with VPg-Pro-DsRed, green fluorescence was seen throughout the cytoplasm, but it was also observed in a subnuclear structure exactly where red fluorescence was emitted by VPg-Pro (Fig. 2B and C). Control experiments indicated that there was no green fluorescence emitted by VPg-Pro-DsRed under the experimental setup used (data not shown). A similar pattern was observed when PABP2 was fused to mCherry and VPg-Pro was fused to GFP (data not shown). To get a better indication of the nature of the subnuclear structure, VPg-Pro-DsRed was coexpressed with a GFP fusion of A. thaliana fibrillarin 2, which localizes in the dense fibrillar component of the nucleolus (2). Figure 2D shows that VPg-Pro colocalized with fibrillarin 2, suggesting that PABP2 was relocated in this substructure of the nucleolus in the presence of VPg-Pro.
FIG. 2.
Subcellular localizations of PABP2, eIF(iso)4E, and VPg-Pro. N. benthamiana leaves were infiltrated with A. tumefaciens, and expression of fluorescent proteins was visualized by confocal microscopy 4 to 5 days later. A. tumefaciens suspensions contained binary Ti plasmids encoding PABP2-GFP and ER-DsRed2 (A), PABP2-GFP and VPg-Pro-DsRed2 (B and C), VPgPro-DsRed2 and Atfib2-GFP (D), eIF(iso)4E-DsRed2 and GFP-ER (E), eIF(iso)4E-DsRed2 and VPg-Pro-GFP (F and G), and PABP2-mCherry, ntGFP-eIF(iso)4E, and VPg-Pro-ctGFP (H and I). Left panels show fluorescence emitted by the red channel only, while middle panels show fluorescence emitted by the green channel only, and right panels show the merge between the red and green channels. C, G, and I are close-up views of the squares depicted in B, F, and H, respectively. Bar, 10 μm.
The fluorescence pattern of eIF(iso)4E was similar to that of PABP2. The fluorescence of eIF(iso)4E-DsRed merged with that of GFP-ER around the nuclear membrane and the ER network, while no eIF(iso)4E was detected in the nucleus (Fig. 2E). A similar distribution pattern was observed when eIF(iso)4E was fused to GFP (3). When coexpressed with VPg-Pro, eIF(iso)4E was now also observed in the nucleolus (Fig. 2F and G). The possibility of a tripartite complex between VPg-Pro, eIF(iso)4E, and PABP2 was consequently investigated using a combination of bimolecular fluorescence complementation (BiFC) and colocalization experiments. VPg-Pro was fused to the C terminus (VPg-Pro-ctGFP) and eIF(iso)4E was fused to the N terminus [ntGFP-eIF(iso)4E] of GFP, and they were coexpressed with PABP2-mCherry in N. benthamiana leaves. Leaves were observed by confocal microscopy 4 to 5 days postinfiltration. Fluorescence generated from BiFC concomitant with that from mCherry was generally observed in 30 to 50% of the cells in the infiltrated area. An incubation period that was slightly longer than normal was required to observe the fluorescence emitted by PABP-mCherry, but the general localization pattern was similar to that observed with PABP2-GFP after 2 days (Fig. 2, compare B and C). As expected, the green fluorescence emitted by the complementation between VPg-Pro-ctGFP and ntGFP-eIF(iso)4E was observed in the nucleolus. This structure was also characterized by the presence of red fluorescence emitted by PABP2-mCherry, although PABP2 was also observed in the cytoplasm (Fig. 2H and I). Control experiments with VPg-Pro-ctGFP and ntGFP or with ctGFP and ntGFP-eIF(iso)4E showed no fluorescence (3).
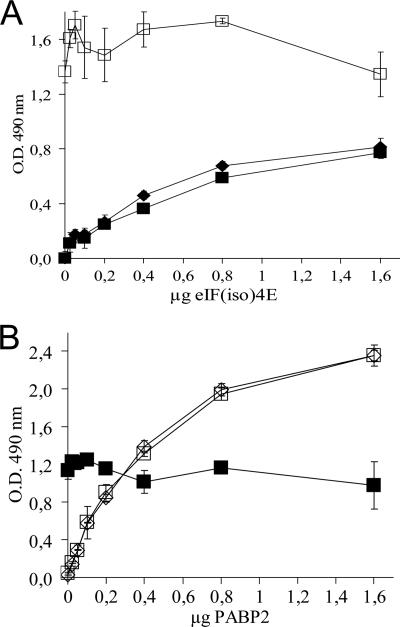
An ELISA-based binding assay has been used to show the in vitro interaction of PABP2 or eIF(iso)4E with VPg-Pro from TuMV (31). Here, this assay was used to determine if both initiation factors can bind simultaneously to VPg-Pro. Recombinant His-tagged PABP2 and T7-tagged eIF(iso)4E from A. thaliana were purified from E. coli by metal chelating or cap-binding chromatography, respectively. ELISA plate wells were then coated with VPg-Pro and were incubated with increasing concentrations of T7-tagged eIF(iso)4E in the absence or in the presence of a saturating binding concentration of His-tagged PABP2 or vice versa. Complex retention was detected using either an anti-T7-tagged or anti-His-tagged antibody. Figure 3A shows a saturation binding curve of VPg-Pro with eIF(iso)4E. A similar saturation curve was obtained when 1.6 μg of PABP2 was added to all concentrations of eIF(iso)4E. PABP2 was retained in the wells by VPg-Pro because a constant signal was detected with the anti-His-tagged antiserum. Control experiments using glutathione S-transferase, metal chelation, or cap-binding-purified E. coli lysate containing pET-21b showed that the interaction with VPg-Pro was specific for the initiation factors (data not shown) (31, 64). Similar results were obtained when ELISA plate wells were coated with VPg-Pro and were then incubated with increasing concentrations of His-tagged PABP2 in the absence or with a constant saturating concentration of T7-tagged eIF(iso)4E (Fig. 3B). eIF(iso)4E was also retained in the wells by VPg-Pro. This experiment then indicates that both PABP2 and eIF(iso)4E are not binding inhibitors of each other and that the VPg-Pro interaction domains are sufficiently far apart from each other so that both translation factors can bind the viral protein simultaneously. The above-described observations suggest that a trimolecular complex made up of PABP2, eIF(iso)4E, and VPg-Pro can take place in the nucleolus of TuMV-infected plants.
FIG. 3.
VPg-Pro interaction with PABP2 and eIF(iso)4E of A. thaliana demonstrated by ELISA-based binding assay. (A) Wells were coated with 1.0 μg purified VPg-Pro and then incubated with increasing concentrations of purified T7-tagged eIF(iso)4E with (▪) or without (⧫) His-tailed PAPB2 (1.6 μg). Retention of the complex was detected using anti-T7-tagged (filled symbols) or anti-His (open symbols) antibodies. (B) Wells were coated with 1.0 μg purified VPg-Pro and then incubated with increasing concentrations of purified His-tailed PAPB2 with (▪) or without (⧫) T7-tagged eIF(iso)4E (1.6 μg). Retention of the complex was detected using anti-T7-tagged (filled symbols) or anti-His (open symbols) antibodies. Values are means of four replicates from a typical experiment. Error bars represent standard deviations. O.D., optical density.
Membrane association of PABP2 in the presence of 6K-VPg-Pro.
6K-VPg-Pro was previously shown to induce the formation of cytoplasmic vesicles in which the interaction with eIF(iso)4E took place (3). To determine whether the membrane association of PABP2 during TuMV infection resulted from the formation of these vesicles, 6K-VPg-Pro-GFP was coexpressed with PABP2-mCherry in N. benthamiana leaves. Leaves were observed by confocal microscopy 4 to 5 days postinfiltration. The fluorescence produced by PABP2-mCherry was observed throughout the cytoplasm but showed a similar emission pattern where green fluorescence produced by 6K-VPg-Pro was observed (Fig. 4A and B). Similarly, fluorescence produced by eIF(iso)4E-DsRed was observed throughout the cytoplasm but showed a similar emission pattern where green fluorescence produced by 6K-VPg-Pro was observed (data not shown). The possibility of a tripartite complex between 6K-VPg-Pro, eIF(iso)4E, and PABP2 in these vesicles was investigated using a combination of BiFC and colocalization experiments. 6K-VPg-Pro was fused to the C terminus of GFP (6K-VPg-Pro-ctGFP), and eIF(iso)4E was fused to the N terminus of the GFP [ntGFP-eIF(iso)4E], and they were coexpressed with PABP2-mCherry in N. benthamiana leaves. Four to five days postinfiltration, leaves were observed by confocal microscopy. As expected, the green fluorescence emitted by the complementation between 6K-VPg-Pro-ctGFP and ntGFP-eIF(iso)4E was observed in vesicles. This structure was also characterized by the presence of red fluorescence emitted by PABP2-mCherry (Fig. 4C and D). Control experiments with 6K-VPg-Pro-ctGFP and ntGFP or with ctGFP and ntGFP-eIF(iso)4E showed no fluorescence (3). These experiments indicated that 6K-VPg-Pro might be responsible for the membrane association of PABP2 during TuMV infection and that a trimolecular complex made up of 6K-VPg-Pro, eIF(iso)4E, and PABP2 likely takes place in virus-induced vesicles.
FIG. 4.
Subcellular localization of eIF(iso)4E, PABP2, and 6K-VPg-Pro. N. benthamiana leaves were infiltrated with A. tumefaciens, and expression of fluorescent proteins was visualized by confocal microscopy 4 to 5 days later. A. tumefaciens suspensions contained binary Ti plasmids encoding PABP2-mCherry and 6K-VPg-Pro-GFP (A and B) or PABP2-mCherry, ntGFP-eIF(iso)4E, and 6K-VPg-Pro-ctGFP GFP (C and D). Left panels show fluorescence emitted by the red channel only, while middle panels show fluorescence emitted by the green channel only, and right panels show the merge between the red and green channels. B and D are close-up views of the squares depicted in A and C, respectively. Bar, 10 μm.
Characterization of the association of PABP with membranes in TuMV-infected cells.
The presence of PABP2 in P30 fractions of TuMV-infected leaves may result from a true membrane association or simply protein aggregation. To distinguish between these two possibilities, a membrane flotation assay was used (65). The membrane-enriched fraction (P30) was overlaid with a sucrose step gradient and subjected to centrifugation. Low-density membranes and proteins associated with these membranes float to the upper part of the gradient, while soluble proteins or aggregated proteins remain at the bottom. We used BiP (an endogenous ER luminal protein) (5) and 6K-VPg-Pro as membrane-associated protein controls. As shown in Fig. 5, BiP (A), 6K-VPg-Pro, and VPg-Pro (B) rose to the top of the gradient (fractions 8 and 9), indicating their membrane association. The detection of BiP in fractions 1 and 2 is likely the result of leakage from the ER lumen. Likewise, PABP2 was found in fractions 8 and 9 (Fig. 5C), confirming that it was membrane associated. PABP2 was also detected at high levels in fractions 1 and 2 and less so in fractions 3 to 7 and likewise may be the result of vesicle leakage during centrifugation (see below).
FIG. 5.
Membrane flotation assays. P30 fractions were used. Fractions were collected from the step sucrose gradient, and proteins present in each collected fraction were separated by SDS-PAGE (12%) and immunodetected with the anti-BiP (A), VPg-Pro (B), and PABP2 (C) antibodies.
To further characterize the type of membranes to which PABP2 was associated, TuMV-infected membrane-enriched fractions (P30) were analyzed on a 20 to 45% sucrose gradient. 6K-VPg-Pro and VPg-Pro were found at the bottom of the gradient (fractions 1 to 5) (Fig. 6A), where it was previously shown to contain ER membranes (30). The distribution of RdRp was found in the same fractions as those for 6K-VPg-Pro (Fig. 6B). This would indicate the position of the replication complex vesicles in the sucrose gradient. PABP2 was also detected in fractions 1 to 5 (Fig. 6C), indicating that the protein was associated with the ER membranes or ER-derived vesicles associated with replication complexes. A second peak of PABP2 was detected in fractions 11 to 15, corresponding to soluble protein fractions, and would indicate that PABP2 leakage from the vesicles took place during centrifugation.
FIG. 6.
Detection of proteins in membrane fractions following centrifugation in a sucrose gradient. The P30 fraction was prepared and centrifuged on 20 to 45% sucrose density gradients. The direction of sedimentation was from right to left, with fraction 15 representing the top of the gradient. Fractions were collected, proteins were separated by SDS-PAGE and electroblotted onto nitrocellulose, and the immunoblot was analyzed using anti-VPg-Pro (A), anti-RdRp (B), or anti-PABP2 (C) serum.
The cofractionation of PABP2 with 6K-VPg-Pro in the sucrose gradient suggests that the host protein might be associated with the vesicles induced by 6K-VPg-Pro through its association with the viral protein. VPg-Pro has an intrinsic capacity to bind to nickel-agarose resin, with the presence of a histidine tail being unnecessary and the binding to the resin being mediated by the VPg domain (36). Purification of 6K-VPg-Pro and VPg-Pro by metal chelation chromatography was thus attempted, and the copurification of PABP2 was evaluated. For TuMV-infected and mock-inoculated tissues, the solubilized membrane fraction was loaded on to a nickel-agarose column. After washing the resin, the bound proteins were eluted with 100 mM imidazole and analyzed by immunoblot assay following SDS-PAGE. Figure 7A shows that both 6K-VPg-Pro and VPg-Pro were effectively purified from infected tissues. Similarly, PABP2 was detected in the eluted protein fraction when the column was loaded with the membrane fractions from TuMV-infected leaves (Fig. 7B). The translation factor was not detected in the eluted protein fraction when membrane fractions from mock-inoculated plants were analyzed, even after prolonged film exposure (Fig. 7B and data not shown). PABP by itself is not purified by metal chelation chromatography (31). This experiment, combined with the confocal microscopy data, indicates that PABP2 is associated with 6K-VPg-Pro-induced vesicles.
FIG. 7.
Copurification of 6K-VPg-Pro/VPg-Pro and PABP2 by metal chelation chromatography. Membrane fractions were solubilized by the addition of 40 mM octyl-β-glucopyranoside loaded on to a column containing 0.4 ml nickel-agarose resin. Proteins were eluted with 100 mM imidazole. Total (T), membrane-associated (M), and eluted (E) proteins from mock-inoculated and TuMV-infected B. perviridis plants were separated by SDS-PAGE and electroblotted onto nitrocellulose. The membrane was probed using anti-VPg-Pro (A) and anti-PABP2 (B) sera.
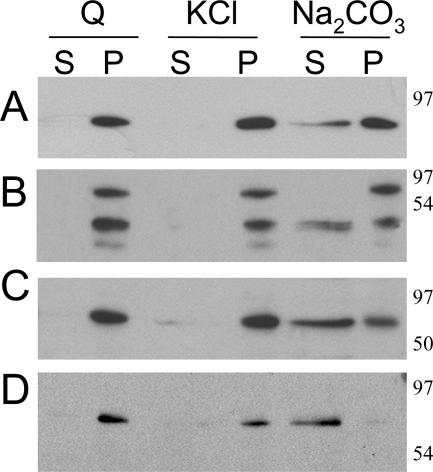
To determine the nature of the PABP2 association with the vesicles, P30 fractions were extracted with different agents known to dislodge peripheral or luminal membrane proteins. Treatment with 1.0 M KCl solubilizes peripheral proteins, while integral and luminal proteins remain associated with the membranes (48). Under high-pH conditions (0.1 M Na2CO3, pH 10.5), membrane vesicles are converted to open membrane sheets, allowing the release of peripheral and luminal proteins but not of integral membrane proteins (21). After treatment with various agents, the membranes were collected by centrifugation and analyzed by immunoblotting. The immunoblots were probed with antibodies against VPg-Pro, RdRp, PABP2, and BiP (a soluble ER luminal protein) (Fig. 8). As expected, 6K-VPg-Pro remained associated with the membrane pellet following extraction with 0.1 M Na2CO3 (pH 10.5) and 1 M KCl. In contrast, the luminal BiP protein, RdRp, PABP2, and a population of VPg-Pro were released into the supernatant after treatment with 0.1 M Na2CO3 (pH 10.5) but remained in the pellet after treatment with 1 M KCl. These results suggest that RdRp, PABP2, and VPg-Pro are luminal proteins of the 6K-VPg-Pro-induced vesicles.
FIG. 8.
Biochemical treatments of membrane-enriched fractions derived from TuMV-infected B. perviridis. Membrane-enriched (P30) fractions were treated with 1 M KCl or 0.1 M Na2CO3 (pH 10.5) for 30 min at 4°C. After separation of membrane-bound (P) and soluble (S) proteins, the presence of BiP (A), VPg-Pro (B), RdRp (C), and PABP2 (D) in these fractions was revealed by immunoblotting with the respective serum.
DISCUSSION
In this report, we have observed that the intracellular distribution of PABP2 was altered in TuMV-infected B. perviridis cells. In healthy plants, PABP2 was mostly soluble and was excluded from the nucleus. However, in TuMV-infected plants, PABP2 was also found in cytoplasmic virus-induced vesicles and in the nucleolus. Coexpression experiments suggested that the PABP2 redistribution in these compartments was the result of its interaction with 6K-VPg-Pro and VPg-Pro, respectively. This redistribution of an important translation initiation factor to the nucleolus and to membrane structures likely underlies two important processes in the TuMV replication cycle.
The first process may be the perturbation of cellular functions by the virus in order to take over cellular machineries necessary for replication. Although positive-sense RNA viruses replicate in the cytoplasm, there are several examples of proteins from such viruses that have been found in the nucleus, often in the nucleolus, or that transit through the nuclear pore complex to enter and exit the nucleus (for reviews, see references 18 and 19). For instance, Schaad et al. (50) previously observed the localization of tobacco etch virus VPg-Pro in the nucleolar region and showed that the inhibition of nuclear import of the viral protein prevented virus replication. The nucleolus is the site of transcription of rRNA, processing of pre-rRNAs, and biogenesis of preribosomal particles. Additionally, the nucleolus is involved in the stress response, cell cycle regulation, and gene silencing (34). The interaction of RNA viruses with the nucleolus could disrupt nucleolar function or recruit nucleolar proteins to aid in virus replication.
There are two classes of PABPs (33). One class is primarily nuclear and plays a role in the synthesis of poly(A) tails, but it also shuttles between the nucleus and the cytoplasm. The other class consists primarily of cytoplasmic PABPs. The latter class influences mRNA translation and decay, and it also shuttles between the nucleus and cytoplasm. A recent study has shown that mammalian PABPC1, a class 2 member, can bind nuclear pre-mRNA poly(A) tails (20). This protein may then have a role to play in pre-mRNA processing and stability and mRNA trafficking and nonsense-mediated mRNA decay. There are nine different gene families coding for PABPs in A. thaliana. PABP2 transcripts are the most abundant (44). In this study, it was found that PABP2 is mostly a cytoplasmic soluble protein, thus categorizing it as a class 2 PABP. However, in infected cells, or in the presence of VPg-Pro, a significant amount of PABP2 is redistributed to the nucleolus. A similar phenomenon was observed with eIF(iso)4E (3), and it appears that VPg-Pro could bind the two proteins simultaneously, as demonstrated by in vitro binding experiments and colocalization coupled with BiFC experiments. Similarly to PABP, eIF(iso)4E has been proposed to regulate mRNA export from the nucleus and may be required for the proofreading of transcripts before they are transported to the cytoplasm (57, 61). Consequently, the nuclear/nucleolar interaction of VPg-Pro with eIF(iso)4E and PABP2 may perturb the (transient) nuclear functions of these factors. This possibility may then explain the host gene shutoff that has been observed for potyvirus pea seedborne mosaic virus-infected cells (62). Additionally, VPg is involved in the long-distance transport of potyviruses (8, 52). Recently, it was shown for umbraviruses that the nucleolar localization of the viral transport protein was required for its function (25). Similar considerations could then be applicable to VPg.
The second process that may be regulated by the interaction of VPg-Pro with PABP and eIF(iso)4E is the coupling of viral translation with viral RNA synthesis. During replication in their host cells, positive-strand RNA viruses provoke membrane proliferation and modification that result in the formation of membrane-bound virus replication complexes (32). This reorganization is induced by a viral protein(s). For instance, brome mosaic virus helicase-like protein 1a induces the formation of spherular ER membrane invaginations (53). Similarly, tomato bushy stunt virus replication protein p33 causes the formation of peroxisomal ghosts (35). These novel organelles have been shown to contain virus replication proteins. These virus-induced vesicles would provide a protective environment against host factors that might degrade viral RNA (e.g., RNases) and optimal conditions for RNA synthesis. In the case of potyviruses, the 6K2 polypeptide is responsible for the formation of ER-derived vesicles (51), which can reach a size of 10 μm in diameter (3). It is through the fusion with this polypeptide that VPg-Pro (i.e., 6K-VPg-Pro) is membrane bound and can act as an anchoring point for other proteins such as viral RdRP as well as translation factors such as eIF(iso)4E and PABP2 (3; this study). The interaction of 6K-VPg-Pro with translation factors within virus-induced vesicles suggests that there might be a physical connection between viral translation and RNA synthesis.
A tight link between virus translation and replication exists. In the case of poliovirus, positive-strand RNA and newly synthesized viral proteins are initially dispersed throughout the ER during the earliest steps of the virus replication cycle. However, as translation progresses, individual poliovirus vesicle clusters emerge at the ER, coalesce, and mature into perinuclear fully active replication complexes (10). Egger et al. (11) showed previously that the formation of the replication complexes was formed in cis in a coupled process that involves viral translation, membrane modification, vesicle budding, and viral RNA synthesis. This is in agreement with previous reports that viral RNA must first be translated in order to replicate (43) and also with the suggestion that all components of the replication complex are delivered en bloc directly following translation (5, 59).
The fate of the protein synthesis machinery during the formation and maturation of the vesicles into replication complex organelles has not yet been addressed. One possibility is that ribosomes and translation factors remain associated with the ER. This implies that, at one point, viral translation terminates and that viral RNAs disengage from the ribosomes and become trapped within the virus-induced vesicles via specific interactions with an RNA-binding replication protein(s). The other possibility is that the protein synthesis machinery is internalized within the vesicles during their formation. As a consequence, translation factors are expected to be membrane associated. This is what has been observed for PABP2 during TuMV infection. This protein is mostly soluble in healthy plants. However, in infected cells, a significant proportion is membrane associated (as demonstrated by the membrane flotation and sucrose gradient experiments) and appears to be a luminal protein (as demonstrated by the Na2CO3 extraction experiment) of 6K-VPg-Pro-induced vesicles (as observed in the confocal microscopy experiments). Colocalization experiments coupled with BiFC have also shown that these same vesicles that contain PABP2 also include eIF(iso)4E. The presence of at least two translation initiation factors within 6K-VPg-Pro-induced vesicles thus points to the hypothesis that the protein synthesis machinery is trapped during the formation of these vesicles. Similarly, eIF3 and eEF1A have been shown to by copurified with active replication complexes of brome mosaic virus and tobacco mosaic virus. However, it is not known if viral RNA translation is proceeding in these vesicles. It is also plausible that PABP2 is needed to initiate the replication of the viral RNA with the vesicles. This possibility is supported by the interaction of PABP with the potyviral RdRP (63).
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada.
We are grateful to Marcel Desrosiers for help with the confocal microscope, A. Vitale for the anti-BiP serum, M. Echeverrıa for AtFibrillarin2-GFP, R. Tsien for mCherry, and H. Sanfaçon for the generous gift of ER-dsRed2. We finally thank H. Sanfaçon and K. A. White for critically reading the manuscript.
Footnotes
Published ahead of print on 1 August 2007.
REFERENCES
- 1.Alvarez, E., A. Castello, L. Menendez-Arias, and L. Carrasco. 2006. HIV protease cleaves poly(A)-binding protein. Biochem. J. 396:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barneche, F., F. Steinmetz, and M. Echeverria. 2000. Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 275:27212-27220. [DOI] [PubMed] [Google Scholar]
- 3.Beauchemin, C., N. Boutet, and J. F. Laliberte. 2007. Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of Turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J. Virol. 81:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushell, M., and P. Sarnow. 2002. Hijacking the translation apparatus by RNA viruses. J. Cell Biol. 158:395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collis, P. S., B. J. O'Donnell, D. J. Barton, J. A. Rogers, and J. B. Flanegan. 1992. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J. Virol. 66:6480-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connor, J. H., and D. S. Lyles. 2002. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J. Virol. 76:10177-10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darós, J.-A., M. C. Schaad, and J. C. Carrington. 1999. Functional analysis of the interaction between VPg-proteinase (NIa) and RNA polymerase (NIb) of tobacco etch potyvirus, using conditional and suppressor mutants. J. Virol. 73:8732-8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunoyer, P., C. Thomas, S. Harrison, F. Revers, and A. Maule. 2004. A cysteine-rich plant protein potentiates potyvirus movement through an interaction with the virus genome-linked protein VPg. J. Virol. 78:2301-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duprat, A., C. Caranta, F. Revers, B. Menand, K. S. Browning, and C. Robaglia. 2002. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 32:927-934. [DOI] [PubMed] [Google Scholar]
- 10.Egger, D., and K. Bienz. 2005. Intracellular location and translocation of silent and active poliovirus replication complexes. J. Gen. Virol. 86:707-718. [DOI] [PubMed] [Google Scholar]
- 11.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallie, D. R. 2002. Protein-protein interactions required during translation. Plant Mol. Biol. 50:949-970. [DOI] [PubMed] [Google Scholar]
- 13.Gaudino, R. J., and C. S. Pikaard. 1997. Cytokinin induction of RNA polymerase I transcription in Arabidopsis thaliana. J. Biol. Chem. 272:6799-6804. [DOI] [PubMed] [Google Scholar]
- 14.Guo, D., M. L. Rajamaki, M. Saarma, and J. P. Valkonen. 2001. Towards a protein interaction map of potyviruses: protein interaction matrixes of two potyviruses based on the yeast two-hybrid system. J. Gen. Virol. 82:935-939. [DOI] [PubMed] [Google Scholar]
- 15.Haghighat, A., Y. Svitkin, I. Novoa, E. Kuechler, T. Skern, and N. Sonenberg. 1996. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol. 70:8444-8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han, S., and H. Sanfacon. 2003. Tomato ringspot virus proteins containing the nucleoside triphosphate binding domain are transmembrane proteins that associate with the endoplasmic reticulum and cofractionate with replication complexes. J. Virol. 77:523-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiscox, J. A. 2003. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 95:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiscox, J. A. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5:119-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosoda, N., F. Lejeune, and L. E. Maquat. 2006. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol. Cell. Biol. 26:3085-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howell, K. E., and G. E. Palade. 1982. Hepatic Golgi fractions resolved into membrane and content subfractions. J. Cell Biol. 92:822-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang, B. C., I. Yeam, J. D. Frantz, J. F. Murphy, and M. M. Jahn. 2005. The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 42:392-405. [DOI] [PubMed] [Google Scholar]
- 23.Kentsis, A., E. C. Dwyer, J. M. Perez, M. Sharma, A. Chen, Z. Q. Pan, and K. L. Borden. 2001. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol. 312:609-623. [DOI] [PubMed] [Google Scholar]
- 24.Khan, M. A., H. Miyoshi, S. Ray, T. Natsuaki, N. Suehiro, and D. J. Goss. 2006. Interaction of genome-linked protein (VPg) of turnip mosaic virus with wheat germ translation initiation factors eIFiso4E and eIFiso4F. J. Biol. Chem. 281:28002-28010. [DOI] [PubMed] [Google Scholar]
- 25.Kim, S. H., E. V. Ryabov, N. O. Kalinina, D. V. Rakitina, T. Gillespie, S. Macfarlane, S. Haupt, J. W. Brown, and M. Taliansky. 2007. Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 26:2169-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuyumcu-Martinez, M., G. Belliot, S. V. Sosnovtsev, K. O. Chang, K. Y. Green, and R. E. Lloyd. 2004. Calicivirus 3C-like proteinase inhibits cellular translation by cleavage of poly(A)-binding protein. J. Virol. 78:8172-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuyumcu-Martinez, N. M., M. Joachims, and R. E. Lloyd. 2002. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J. Virol. 76:2062-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuyumcu-Martinez, N. M., M. E. Van Eden, P. Younan, and R. E. Lloyd. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol. Cell. Biol. 24:1779-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lellis, A. D., K. D. Kasschau, S. A. Whitham, and J. C. Carrington. 2002. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12:1046-1051. [DOI] [PubMed] [Google Scholar]
- 30.Leonard, S., J. Chisholm, J. F. Laliberte, and H. Sanfacon. 2002. Interaction in vitro between the proteinase of Tomato ringspot virus (genus Nepovirus) and the eukaryotic translation initiation factor iso4E from Arabidopsis thaliana. J. Gen. Virol. 83:2085-2089. [DOI] [PubMed] [Google Scholar]
- 31.Leonard, S., C. Viel, C. Beauchemin, N. Daigneault, M. G. Fortin, and J. F. Laliberte. 2004. Interaction of VPg-Pro of turnip mosaic virus with the translation initiation factor 4E and the poly(A)-binding protein in planta. J. Gen. Virol. 85:1055-1063. [DOI] [PubMed] [Google Scholar]
- 32.Mackenzie, J. 2005. Wrapping things up about virus RNA replication. Traffic 6:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mangus, D. A., M. C. Evans, and A. Jacobson. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayer, C., and I. Grummt. 2005. Cellular stress and nucleolar function. Cell Cycle 4:1036-1038. [DOI] [PubMed] [Google Scholar]
- 35.McCartney, A. W., J. S. Greenwood, M. R. Fabian, K. A. White, and R. T. Mullen. 2005. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17:3513-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menard, R., H. Chatel, R. Dupras, C. Plouffe, and J. F. Laliberte. 1995. Purification of turnip mosaic potyvirus viral protein genome-linked proteinase expressed in Escherichia coli and development of a quantitative assay for proteolytic activity. Eur. J. Biochem. 229:107-112. [DOI] [PubMed] [Google Scholar]
- 37.Michel, Y. M., A. M. Borman, S. Paulous, and K. M. Kean. 2001. Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation. Mol. Cell. Biol. 21:4097-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michon, T., Y. Estevez, J. Walter, S. German-Retana, and O. Gall. 2006. The potyviral virus genome-linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue. FEBS J. 273:1312-1322. [DOI] [PubMed] [Google Scholar]
- 39.Miyoshi, H., N. Suehiro, K. Tomoo, S. Muto, T. Takahashi, T. Tsukamoto, T. Ohmori, and T. Natsuaki. 2006. Binding analyses for the interaction between plant virus genome-linked protein (VPg) and plant translational initiation factors. Biochimie 88:329-340. [DOI] [PubMed] [Google Scholar]
- 40.Moury, B., C. Morel, E. Johansen, L. Guilbaud, S. Souche, V. Ayme, C. Caranta, A. Palloix, and M. Jacquemond. 2004. Mutations in potato virus Y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol. Plant-Microbe Interact. 17:322-329. [DOI] [PubMed] [Google Scholar]
- 41.Nicaise, V., S. German-Retana, R. Sanjuan, M.-P. Dubrana, M. Mazier, B. Maisonneuve, T. Candresse, C. Caranta, and O. LeGall. 2003. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus lettuce mosaic virus. Plant Physiol. 132:1272-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicolas, O., S. W. Dunnington, L. F. Gotow, T. P. Pirone, and G. M. Hellmann. 1997. Variations in the VPg protein allow a potyvirus to overcome va gene resistance in tobacco. Virology 237:452-459. [DOI] [PubMed] [Google Scholar]
- 43.Novak, J. E., and K. Kirkegaard. 1994. Coupling between genome translation and replication in an RNA virus. Genes Dev. 8:1726-1737. [DOI] [PubMed] [Google Scholar]
- 44.Palanivelu, R., D. A. Belostotsky, and R. B. Meagher. 2000. Arabidopsis thaliana poly(A) binding protein 2 (PAB2) functions in yeast translational and mRNA decay processes. Plant J. 22:187-198. [DOI] [PubMed] [Google Scholar]
- 45.Revers, F., O. Le Gall, T. Candresse, and A. J. Maule. 1999. New advances in understanding the molecular biology of plant/potyvirus interactions. Mol. Plant-Microbe Interact. 12:367-376. [Google Scholar]
- 46.Ruffel, S., M. H. Dussault, A. Palloix, B. Moury, A. Bendahmane, C. Robaglia, and C. Caranta. 2002. A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32:1067-1075. [DOI] [PubMed] [Google Scholar]
- 47.Ruffel, S., J. L. Gallois, B. Moury, C. Robaglia, A. Palloix, and C. Caranta. 2006. Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent pepper veinal mottle virus infection of pepper. J. Gen. Virol. 87:2089-2098. [DOI] [PubMed] [Google Scholar]
- 48.Sankaram, M. B., D. Marsh, L. M. Gierasch, and T. E. Thompson. 1994. Reorganization of lipid domain structure in membranes by a transmembrane peptide: an ESR spin label study on the effect of the Escherichia coli outer membrane protein A signal peptide on the fluid lipid domain connectivity in binary mixtures of dimyristoyl phosphatidylcholine and distearoyl phosphatidylcholine. Biophys. J. 66:1959-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaad, M. C., R. J. Anderberg, and J. C. Carrington. 2000. Strain-specific interaction of the tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two-hybrid system. Virology 273:300-306. [DOI] [PubMed] [Google Scholar]
- 50.Schaad, M. C., R. Haldeman-Cahill, S. Cronin, and J. C. Carrington. 1996. Analysis of the VPg-proteinase (NIa) encoded by tobacco etch potyvirus: effects of mutations on subcellular transport, proteolytic processing, and genome amplification. J. Virol. 70:7039-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaad, M. C., A. D. Lellis, and J. C. Carrington. 1997. VPg of tobacco etch potyvirus is a host genotype-specific determinant for long-distance movement. J. Virol. 71:8624-8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwartz, M., J. Chen, W. M. Lee, M. Janda, and P. Ahlquist. 2004. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc. Natl. Acad. Sci. USA 101:11263-11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 55.Shi, S. T., and M. M. Lai. 2005. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 287:95-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonenberg, N., and T. E. Dever. 2003. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 13:56-63. [DOI] [PubMed] [Google Scholar]
- 57.Strudwick, S., and K. L. Borden. 2002. The emerging roles of translation factor eIF4E in the nucleus. Differentiation 70:10-22. [DOI] [PubMed] [Google Scholar]
- 58.Svitkin, Y. V., H. Imataka, K. Khaleghpour, A. Kahvejian, H. D. Liebig, and N. Sonenberg. 2001. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA 7:1743-1752. [PMC free article] [PubMed] [Google Scholar]
- 59.Towner, J. S., M. M. Mazanet, and B. L. Semler. 1998. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J. Virol. 72:7191-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vende, P., M. Piron, N. Castagné, and D. Poncet. 2000. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J. Virol. 74:7064-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.von der Haar, T., J. D. Gross, G. Wagner, and J. E. McCarthy. 2004. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 11:503-511. [DOI] [PubMed] [Google Scholar]
- 62.Wang, D., and A. J. Maule. 1995. Inhibition of host gene expression associated with plant virus replication. Science 267:229-231. [DOI] [PubMed] [Google Scholar]
- 63.Wang, X., Z. Ullah, and R. Grumet. 2000. Interaction between zucchini yellow mosaic potyvirus RNA-dependent RNA polymerase and host poly-(A) binding protein. Virology 275:433-443. [DOI] [PubMed] [Google Scholar]
- 64.Wittmann, S., H. Chatel, M. G. Fortin, and J. F. Laliberte. 1997. Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology 234:84-92. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, S. C., G. Zhang, L. Yang, J. Chisholm, and H. Sanfacon. 2005. Evidence that insertion of tomato ringspot nepovirus NTB-VPg protein in endoplasmic reticulum membranes is directed by two domains: a C-terminal transmembrane helix and an N-terminal amphipathic helix. J. Virol. 79:11752-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]