Abstract
This study demonstrates that the envelope proteins of hepatitis B virus (HBV) could be incorporated into the lipid membrane of lentivirus pseudotype particles. The assembly procedure was initiated by the transfection of 293T cells with three plasmids: (i) a human immunodeficiency virus (HIV) packaging construct, (ii) a transfer plasmid expressing a reporter gene, and (iii) a plasmid expressing large (L), middle (M), and small (S) HBV envelope proteins. After 2 days, hepatitis B surface antigen and the antigenic forms of L, M, and S were detected at the cell surface by flow cytometry. Also, virus particles that were able to infect cultured primary human hepatocytes (PHH) were released. Under optimal conditions, 50% of PHH could be infected. In addition, the susceptibility of PHH and the resistance of other cell types to the pseudotype particles were similar to those observed for HBV and hepatitis delta virus (HDV), which shares the same L, M, and S. Furthermore, the infection of PHH by the pseudotype was sensitive to known inhibitors of HBV and HDV entry. These findings of specific and efficient infection of hepatocytes could be applicable to liver-specific gene therapy and may help clarify the attachment and entry mechanism used by HBV and HDV.
Thirty years ago, it was demonstrated that in an experimental situation, the envelope proteins of an unrelated animal virus could be incorporated into particles of vesicular stomatitis virus (VSV) to create pseudotypes (36). This concept was subsequently extended using retrovirus vectors that expressed no envelope protein but that could nonspecifically incorporate the envelope proteins of other viruses. Many examples of retrovirus pseudotyping have since been reported (9, 29, 34). Retrovirus vectors have the advantages of long-term expression of the transgene from the integrated provirus and the simplicity in modifying tropism by pseudotyping. Lentivirus vectors have an additional advantage among retroviruses in being able to infect both dividing and nondividing cells. As a consequence, these vectors are widely used in gene therapy and clinical trials to treat cancer, infectious diseases, vascular diseases, and monogenic diseases (9, 29).
In addition to gene therapy, pseudotyped retroviruses are commonly used to study virus entry mechanisms. This is because the expression of viral attachment proteins, separate from the viral replication machinery, allows the specific study of early events in the viral life cycle. For example, retrovirus vectors pseudotyped with hepatitis C virus (HCV) or severe acute respiratory syndrome-associated coronavirus envelope proteins closely resemble wild-type HCV or severe acute respiratory syndrome-associated coronavirus in their tropisms, entry mechanisms, and sensitivities to entry inhibitors (2, 14, 28). Furthermore, pseudotype viruses are safer than wild-type viruses and can be used in regular tissue culture facilities.
In contrast to studies with the envelope proteins of many animal viruses, little attention has been given to incorporating the envelope proteins of hepatitis B virus (HBV) into retrovirus particles. There are three HBV envelope proteins, known as large (L), middle (M), and small (S). They are co-C-terminal and share the entire S domain. Relative to S, M has an additional domain, pre-S2, at its N terminus. Similarly, relative to M, L has a pre-S1 domain.
Sung and Lai previously described the assembly of HBV envelope proteins onto a murine leukemia virus vector (30). The resulting pseudotype viruses were assayed on primary human hepatocytes. About 1 in 5,000 cells was infected, as indicated by the expression of a reporter gene encoded by the vector. A serious problem in these studies is that primary human hepatocytes are essentially nondividing cells, and murine leukemia virus-based vectors are unable to integrate their DNA into nondividing cells. Therefore, we made use of lentivirus vectors based on human immunodeficiency virus type 1 (HIV-1), a virus capable of infecting both dividing and nondividing cells (23, 33).
We describe here the production of HIV-based pseudotype viruses with HBV envelope proteins, including evidence for infection of primary human hepatocytes (PHH) with both efficiency and HBV envelope-directed specificity. Our findings have implications in three areas. First, they expand our understanding of the ways in which L, M, and S can be assembled into particles. Second, they will help clarify the as-yet-unknown mechanisms of HBV and hepatitis delta virus (HDV) attachment and entry (11). Third, they may facilitate the use of lentiviruses with HBV envelopes to deliver specific sequences to hepatocytes in vitro and maybe also in vivo as part of liver-specific gene therapy.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney 293T and human hepatoblastoma Huh7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Confluent monolayers of PHH on rat tail collagen in a 48-well configuration were obtained commercially (CellzDirect and Lonza Walkerville, Inc.) and maintained in Hepatostim medium supplemented with 10 ng/ml epidermal growth factor, receptor grade (both from BD Biosciences). Cryopreserved primary woodchuck hepatocytes (PWH) (a gift of William Mason) were cultured and infected as previously described (32). All cells were maintained at 37°C in 5% CO2. HDV and HBV were assembled and released from transfected cells as previously described (13). Woodchuck hepatitis virus was obtained from the serum of an infected animal (a gift from William Mason).
Flow cytometry.
293T cells were seeded at 6 × 106 cells per 10-cm-diameter plate. The next day, they were cotransfected, using calcium phosphate, with (i) pCMVΔR8.2, to provide HIV proteins including Gag and Pol (18); (ii) pHXCMVlacZWP, to provide the lacZ reporter gene (34); and (iii) pSVB45H (a gift from Don Ganem), to provide L, M, and S (4). The amounts of plasmid (see above) were 10, 20, and 30 μg, respectively. After 1 day, the cells were treated with 10 mM sodium butyrate to induce higher expression of the plasmids, and after one more day, the cells were detached from the monolayer with 0.5 mM EDTA in phosphate-buffered saline lacking calcium and magnesium and blocked with phosphate-buffered saline containing 5% fetal calf serum. Surface expression of HBV envelope proteins was detected using S-26, a monoclonal antibody specific for the HBV pre-S2 region (a gift from Vadim Bichko), followed by a dye-labeled secondary antibody. In order to detect intracellular envelope proteins, cells were permeabilized with a Cytofix/Cytoperm kit (BD Biosciences) prior to antibody staining. As a negative control, a nonspecific mouse monoclonal antibody of the same isotype was used as the primary antibody. For other studies, we substituted S-26 with an antibody specific for the S domain (Fitzgerald Industries). Cells were next incubated briefly with propidium iodide (PI), a dye that is taken up only by cells with a damaged plasma membrane and that stains the nuclear DNA. Cell suspensions were then washed and subjected to flow cytometry analysis using a FACScan apparatus (Becton-Dickinson). Data were analyzed using Flowjo 8.3.3 software. The percentages of PI-positive and -negative cells were determined for nonpermeabilized cells. Only PI-negative cells were analyzed for the expression of hepatitis B surface antigen (HBsAg) at the cell surface.
Lentivirus pseudotype production and characterization.
293T cells were seeded at 6 × 106 cells per 10-cm-diameter plate. Cells were cotransfected with three plasmids (see above) and induced with sodium butyrate, as described above. In some cases, pSVB45H was substituted with pSVBX24H to express only the HBV S protein or by two plasmids that separately express HBV S and L (13). Another substitution was done with a plasmid to express the VSV G protein (28). Two days after transfection, the medium was harvested, clarified, and used directly for infection or aliquoted and stored at −80°C. For the immunoblot assay depicted in Fig. 4, the cells were lysed in a solution containing 50 mM Tris (pH 8), 5 mM EDTA, 150 mM NaCl, and 1% Triton X-100. The lysates were centrifuged to remove nuclei prior to immunoblot analysis.
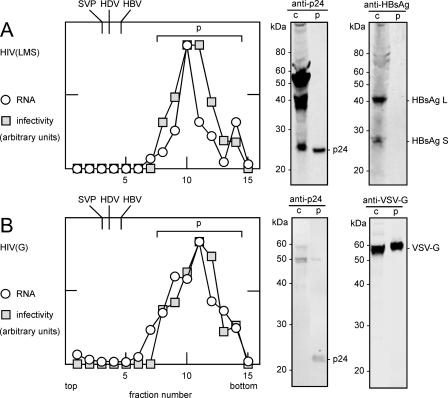
FIG. 4.
Rate-zonal sedimentation of particles released from transfected cells. A and B represent HIV(LMS) and HIV(G), respectively, assembled as described in the legend of Fig. 2. In each case, particles were collected from the medium and examined by rate-zonal sedimentation, as described in Materials and Methods. As controls, parallel gradients were performed to determine the peak sedimentation values for HBV SVP, HDV, and HBV, similarly to a method described previously (13), with results indicated at the top. For A and B, individual fractions were assayed (Table 1) by qPCR for LacZ genomic RNA and for infectivity on PHH. The results of both assays are expressed in arbitrary units. For each gradient, the peak region, indicated by p, was pooled and concentrated by ultracentrifugation. These pooled samples and aliquots of the cell lysates, indicated by c, were examined by immunoblotting to detect HIV p24, HBsAg, and VSV G, as indicated at the right. In parallel, a sample of MagicMark (Invitrogen) was electrophoresed to provide the molecular mass standards indicated at the left.
Immunoblot procedures.
Protein samples were heated at 70°C for 10 min in Laemmli buffer (16) prior to analysis on 12% precast gels (Duramide, Cambrex, or Nupage; Invitrogen). After electrophoresis and transfer onto nitrocellulose membranes, HBV envelope proteins, VSV G protein, and HIV p24 Gag protein were detected using rabbit anti-HBV S protein (Fitzgerald Industries), rabbit anti-VSV G (Bethyl), and monoclonal mouse anti-HIV p24 (Millipore), respectively, and infrared dye-labeled secondary antibodies (LI-COR). Detection and quantitation were achieved using a two-color infrared laser scanning apparatus and associated software (Odyssey; LI-COR).
Rate-zonal sedimentation.
The lentivirus particles as well as subviral particles (SVP), HDV, and HBV were examined by rate-zonal sedimentation using procedures similar to those described previously by Gudima et al. (13). We used gradients of 15 to 30% (wt/wt) sucrose in STE buffer (100 mM NaCl, 10 mM Tris [pH 8.0], 1 mM EDTA). After centrifugation in an SW41 rotor (Beckman) at 40 krpm for 80 min at 4°C, fractions of 0.75 ml were collected from the top. Aliquots of the gradient fractions were assayed for infectivity on PHH (as described below) as well as for HIV p24, VSV G, and HBsAg by immunoblotting or for the viral genomes by quantitative real-time reverse transcription-PCR (qPCR) assays. TaqMan primers and probes were used to assay for LacZ RNA, HDV genomic RNA, and HBV DNA (13).
Lentivirus quantitation.
RNA was extracted from pseudotype particles using Tri reagent (Molecular Research Center, Inc.) and treated with DNase (13), after which we quantitated the HIV RNA genome by qPCR using primers directed toward the 3′ end of the LacZ gene. To provide an absolute quantitation for this assay, a fragment of LacZ cDNA was amplified by PCR using a primer containing a T7 RNA polymerase promoter. From this, RNA was transcribed in vitro by T7 RNA polymerase using a Ribomax kit (Promega). The RNA product was extracted and gel purified, and known amounts were used to calibrate the qPCR and thus to determine the amount of lentivirus assembly in units of RNA genome equivalents (GE) per ml of medium.
Immunoprecipitation (IP) procedures.
Antibodies were first incubated with protein A-agarose beads (Invitrogen) in binding/washing buffer (150 mM NaCl, 50 mM Tris [pH 7.5], 1 mM MgCl2). Beads were washed three times to remove unbound antibodies before being incubated with HIV(LMS) or HIV (G) particles. Unbound virus was removed by washing the beads three times in binding/washing buffer. RNA was extracted from both immunoprecipitated particles and aliquots of the input material and assayed by qPCR to detect LacZ RNA.
Biological activity.
The infectivities of HIV pseudotype viruses were tested on PHH, PWH, 293T cells, Huh7 cells, and HepG2 cells. After 3 days, the cells were fixed with glutaraldehyde and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal; Fisher Scientific). Blue-stained cells were counted to determine the transducing units (TU). The cells were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) (Sigma) to detect all nuclei and thus deduce the percentage of infected cells. Digital images were collected using a light microscope with a 10× objective and processed using Photoshop 7.0 software (Adobe).
For the inhibition of infection, we used previously described immunoadhesins (IA): S1-IA and S1S2-IA (6). Briefly, these IA contain the Fc region of a rabbit immunoglobulin heavy chain to which either the pre-S1 domain of L or both the pre-S1 and pre-S2 domains have been fused at its N terminus. As another inhibitor, we used a chemically synthesized and myristoylated peptide derived from the N terminus of L, as designed and generously provided by Stephan Urban (10). All three inhibitors are known to block the infection of primary human hepatocytes by either HDV or HBV (6, 10). As a negative control, we used S1-IA/G2A G13A, a mutant IA that does not block virus infection (6). The inhibitors were added simultaneously with the HIV pseudotypes. For antibody neutralization studies, virus was incubated at room temperature for 30 min with antibody prior to infection. Typically, the infection of PHH was performed in triplicate. One set of cells was used to determine TU, as described above. Total RNA was extracted from the other two sets for assay of LacZ mRNA expression by qPCR, as described above. In Fig. 3, these data were normalized to the amount of input RNA and are presented as average values, with the range of values indicated by error bars.
FIG. 3.
Several proteins were tested for their ability to inhibit infection of PHH by either HIV(LMS) or HIV(G). Two of the inhibitors, indicated as S1-IA and S1S2-IA, were immunoadhesins containing the pre-S1 or the pre-S1 and pre-S2 domains of HBV L, as previously reported (6). The third inhibitor, indicated as peptide, was a chemically synthesized myristoylated pre-S1 peptide (10) (a gift from Stephan Urban). These inhibitors, at several concentrations as indicated, were added at the time of infection. Three days after infection, total cell RNA was extracted and assayed by qPCR to detect LacZ RNA. All infections were performed in duplicate, and the indicated levels of HIV infection, in arbitrary units, are average values and the ranges of these values.
RESULTS
Expression of HBV envelope proteins on the surface of transfected 293T cells.
The three envelope proteins of HBV, abbreviated here LMS, are cotranslationally inserted into the endoplasmic reticulum (ER) and, subsequently, within the ER or ER-Golgi intermediate compartment, facilitate HBV and HDV particle assembly and release (3, 15, 22, 31). However, others have reported experimental situations where at least some of the HBsAg can actually be detected at the cell surface (25, 30). Therefore, we undertook to confirm and extend such reports, with the ultimate aim of achieving the assembly of HIV vectors pseudotyped with LMS.
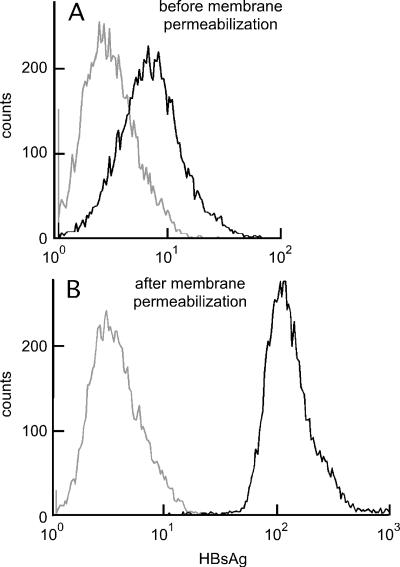
With this in mind, we carried out transient cotransfection of 293T cells with pSVB45H to express LMS along with one plasmid to express HIV Gag-Pol and another to express an HIV-based reporter RNA. At 2 days after transfection, the cells were collected and incubated with S-26, a pre-S2-specific mouse monoclonal antibody, followed by a dye-labeled secondary antibody. As a negative control, an aliquot of transfected cells was stained with a non-HBV monoclonal antibody of the same isotype. Cells were also stained with PI to detect cells that, through either damage or cell death, were permeable at the plasma membrane. The mean fluorescence intensity for such PI-positive cells was 50 times higher than that for the negative cells, and therefore, in the flow cytometry analysis, such cells, which represented 11% of the total, were easily gated out.
The results shown in Fig. 1A indicated that a significant fraction of the cells expressed HBsAg on the plasma membrane. Since the S-26 antibody recognizes an epitope within the pre-S2 region of L and M, we concluded that this region was not only in the plasma membrane but also exposed on the outside of the membrane. Figure 1B shows a separate analysis in which the cells were permeabilized prior to immunostaining. This further increased the mean fluorescence intensity by approximately 16-fold, consistent with the interpretation that the majority of the HBsAg were intracellular rather than on the cell surface. Similar results were obtained when 293T cells were transfected with the LMS plasmid only (data not shown).
FIG. 1.
Flow cytometry detected HBsAg on the surface of transfected 293T cells. At 2 days after cotransfection, as described in the text, cells were collected and stained for expression of HBsAg and also for incorporation of PI. In the analysis, cells that incorporated PI were excluded as being those with a disrupted plasma membrane. The black and gray lines refer to cells that were stained either with a pre-S2-specific mouse monoclonal antibody (S-26) or with a nonspecific mouse antibody of the same isotype, respectively. A and B refer to staining carried out before or after cell permeabilization, respectively.
The detection of at least some of the HBsAg on the cell surface is consistent with the ability to produce pseudotype particles with the pre-S domain exposed on the virion surface. We were also able to use a second antibody and detect surface expression of the S domain, which is present on L, M, and S (data not shown). Also, when cells were transfected with a construct that expressed only the S protein of HBV, we could detect expression at the cell surface (data not shown).
Production and quantitation of HIV pseudotype particles.
It is known that transfection of cells with retrovirus-based plasmids expressing Gag and Pol proteins and a reporter RNA genome can lead to the assembly and release of RNA-containing virus-like particles from the plasma membrane even when specific envelope proteins are not present (27). Given our results showing that HBsAg could be detected at the surface of transfected cells, we next asked whether LMS could be incorporated into HIV virus-like particles to produce pseudotype virus, designated here HIV(LMS).
293T cells were cotransfected as shown in Fig. 1. After 2 days, the culture medium was clarified, and RNA was extracted and assayed by qPCR to detect the HIV-based RNA genome that contained the LacZ gene as a reporter. This assay was calibrated relative to known amounts of an in vitro-transcribed LacZ RNA standard. Thus, we could deduce the titer in GE per ml. The titer of HIV(LMS) was compared with that of HIV(S), a pseudotype containing only HBV S, and that of HIV(G), a pseudotype with the envelope glycoprotein (G) of VSV. The results are summarized in Table 1. In this experiment, the amounts of the three pseudotyped particles exceeded 108 GE/ml. However, between separate experiments, the amounts of assembled particles ranged from 107 to 7 × 108 GE/ml.
TABLE 1.
Quantitation of HIV pseudotype assembly and infectivity
| Pseudotype | Assemblya (108 GE/ml) | Infectivityb (104 TU/ml)
|
|||
|---|---|---|---|---|---|
| PHH | PWH | Huh7 | 293T | ||
| HIV(G) | 1.4 | 4.0 | 2.8 | 48 | 440 |
| HIV(LMS) | 7.0 | 10 | <0.0005 | <0.0005 | <0.0005 |
| HIV(S) | 5.6 | <0.0005 | <0.0005 | <0.0005 | <0.0005 |
Assembly was quantitated by qPCR for the LacZ reporter sequence to determine the HIV GE/ml.
Infectivity was determined from assays at 3 days after infection of β-galactosidase-positive cells, where each positive cell is counted as 1 TU. The titer of the inoculum in TU/ml was deduced as the average result of infections performed in duplicate.
It is interesting that when comparable cotransfections were performed in Huh7 cells, LacZ-containing GE were not detected in the culture medium, that is, at least 500 times less (data not shown). This might indicate that HIV genome assembly (even independent of the incorporation of proteins into the envelope) is severely limited in this liver cell line.
Infectivity of pseudotype particles.
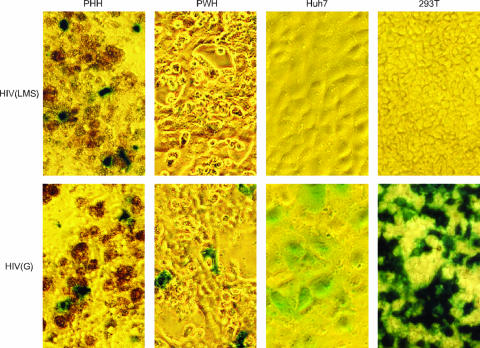
We tested the ability of these pseudotype particles to infect five different cell types: PHH, PWH, Huh7 cells, HepG2 cells, and 293T cells. Three days after infection, cells were fixed and stained with X-gal. Under these conditions, infected cells that expressed β-galactosidase produced blue staining, as visualized by light microscopy.
Representative results are shown in Fig. 2. HIV(LMS) infected PHH but not PWH, Huh7 cells, or 293T cells. It also failed to infect HepG2 cells, another liver cell line (data not shown). This tropism for HIV(LMS) is the same as that for HBV or HDV (11). In contrast, HIV(G) infected PHH, PWH, Huh7 cells, and 293T cells. This is consistent with the known broad tropism of HIV(G) (9). The higher infectivity in the cell lines may be due to an enhanced efficiency of integration and/or reporter expression in dividing cells. As a negative control, HIV(S) was unable to infect any of these cells (data not shown).
FIG. 2.
Light microscopy revealed infection of different cell types by HIV pseudotypes. HIV(LMS) or HIV(G) was used to infect PHH, PWH, Huh7 cells, or 293T cells. After 3 days, the cells were fixed and stained with X-gal. Infected cells were thus detected by blue staining. A brown signal was associated with some nonviable PHH.
In order to confirm that the PHH infected by HIV(LMS) were indeed hepatocytes, we made use of immunostaining to detect human albumin, a marker specific for hepatocytes, using procedures described previously for HDV infections (13). We observed that virtually all cells, including those that stained as positive for lacZ, were positive for albumin (data not shown).
Data shown Fig. 2 were quantitated to determine the number of TU per ml of virus-containing medium, as summarized in Table 1. We can deduce that for HIV(LMS) on PHH, there was only 1 TU per 7,000 GE. A similar result was reported previously for lentivirus vectors (26). A major contributor to the low infectivity per GE may be that the majority of GE-containing particles have either no or insufficient envelope proteins to be infectious.
For the experiment shown in Fig. 2, about 10% of PHH were infected by HIV(LMS). From a number of separate experiments, this percentage ranged from 1 to 50. This major variation was not due only to the source of virus, because we were able to use identical aliquots of virus that had been stored at −80°C to infect different sources of PHH and still got variable results. Such variation was also seen for the infection of PHH by HIV(G). We therefore consider that the major contribution to the variation was in the PHH. Certainly, there were differences in the livers from which these PHH were derived (e.g., age and health condition of donor and maintenance of liver after removal from the donor) and variations in the procedures used by different commercial suppliers to establish hepatocyte cultures. The importance of donor differences was previously observed for PHH with HIV pseudotype viruses bearing the HCV envelope proteins (2) and in mouse liver with HIV(G) (21).
Effect of inhibitors on the ability of pseudotype particles to infect PHH.
The above-described results showed that HIV(LMS), like HBV and HDV, infected PHH but not cell lines, supporting the interpretation that these viruses use the same attachment and entry mechanisms.
To provide an independent test of this interpretation, we made use of recent findings that certain molecules block HBV and HDV infections (6, 10) and asked whether the same applied to the pseudotype viruses.
As described recently, we have shown that certain IA containing the pre-S1 region of the HBV L can act as specific and potent inhibitors of HDV and HBV infection (6). Previous studies by others showed that chemically synthesized peptides can also inhibit infection (1, 10, 12). It is considered that these peptides are competitive inhibitors that interact with the host receptor(s) and thus block virus attachment and entry (11). We tested two immunoadhesins and one synthetic peptide. As summarized in Fig. 3, all three inhibitors, when present at 50 nM, blocked the infection of PHH by HIV(LMS). These dose requirements were similar to what we have observed for the inhibition of infection by HBV and HDV (6). As one negative control, we tested S1-IA/G2A G13A, a mutant HBV S1-IA that lacks an essential myristoylation site (6), and observed that it did not inhibit HIV(LMS) at 50 nM (data not shown). As an additional negative control, we observed that at 50 nM, one of the IA, S1S2-IA, was unable to inhibit the infection of PHH by HIV(G) (Fig. 3).
In summary, these inhibitor studies further support the interpretation that HIV(LMS) infects PHH via the same receptor(s) as that used by HBV and HDV.
Assembly and infectivity of HIV pseudotype particles containing different ratios of HBV L and S proteins.
In recent studies of HDV assembly, we demonstrated that the assembly of HDV RNA-containing particles and their subsequent ability to infect PHH could be controlled by the amounts of HBV L and S proteins available during assembly (13). Therefore, we tested whether a similar regulation was exerted over HIV pseudotype assembly and infectivity. The results of such a comparison are summarized in Table 2.
TABLE 2.
Effect of variations in the ratio of HBV L to S on particle assembly and infectivitya
| Ratio of L:S plasmids used for assembly | HIV
|
HDV
|
||
|---|---|---|---|---|
| Assembly (107 GE/ml) | Infectivity (TU/107 GE) | Assembly (107 GE/ml) | Specific infectivity | |
| 0:1 | 19.8 | <2.5 | 9.2 | <0.05 |
| 0.1:0.9 | 17.7 | 395 | 12.6 | 6.6 |
| 0.25:0.75 | 11.3 | 465 | 4.2 | 14.9 |
| 0.5:0.5 | 12.0 | 321 | 3.4 | 7.2 |
| 0.75:0.25 | 5.4 | 648 | 0.32 | 3.5 |
| 1:0 | 2.6 | <19 | <0.0018 | ND |
| 0:0 | 17.3 | <3 | <0.0018 | ND |
| 0:0 + VSV G | 14.0 | 210 | ND | ND |
HIV pseudotype assembly was initiated in 293T cells cotransfected with plasmids to express HBV L and S proteins in the series of ratios indicated plus two more plasmids to provide HIV proteins. As a positive control, HBsAg were replaced with VSV G. Particles were harvested from days 1 to 2. HDV assembly was done in Huh7 cells cotransfected with the same plasmids to express HBV L and S, together with pSVL(D3), to achieve HDV replication and assembly, as previously described (13). Particles were harvested from days 6 to 9. The RNA extracted from these viruses was quantitated by qPCR to determine the assembly titer in GE/ml. Also, equal amounts of GE were used to determine the infectivity on PHH. For the HIV pseudotypes, this infectivity was measured after 3 days by LacZ staining to deduce the titer in TU/107 GE. For HDV infectivity, the total cell RNA was extracted after 6 days, the yield of HDV GE/cell was determined by qPCR, and the specific infectivity was defined as the ratio of output GE/cell to the input multiplicity of GE/cell (13). The indicated values represent the averages of two measurements on duplicate samples. ND indicates values that were not determined.
Consider first the ability to achieve assembly. HIV pseudotype assembly was achieved even in the absence of the L and S proteins. This is expected for a retrovirus, since the envelope proteins are not needed for assembly, and yet it is quite different from HDV assembly, where envelope proteins are essential. However, when L and S were present, there were some similarities between pseudotype and HDV assembly. Specifically, when L alone was expressed, the assembly efficiency was reduced, probably due to its known cytotoxic effects (35). As the relative amount of S was increased, so was the assembly. These effects were more dramatic for HDV, where it is known that the increased expression of L reduces particle assembly (3, 13). In addition, the differences in the extent could in part be because pseudotype assembly was done in 293T cells, while that of HDV was done in Huh7 cells.
These various forms of HIV pseudotypes and HDV were then tested for infectivity on PHH. The results for the two viruses were similar. For example, in both cases, particles assembled with HBV S only were not infectious. The L protein had to be present for infectivity to be detected. Again, these results support the interpretation that the HIV pseudotypes infect PHH by a mechanism not distinguishable from that used by HDV.
Rate-zonal sedimentation of pseudotype particles.
Next, we used rate-zonal sedimentation to further characterize HIV(LMS) relative to HIV(G). From previous studies, we knew that rate-zonal sedimentation could be used to separate HBV SVP away from the larger HDV and HBV (13). We used a similar strategy to test both the size and protein composition of the pseudotype particles.
The time of sedimentation on gradients of 10 to 30% sucrose was reduced relative to those previously used for SVP, HDV, and HBV (13) since lentivirus particles were expected to be larger, with an expected sedimentation value of about 580S (27). After centrifugation, aliquots of the gradient fractions were assayed by qPCR for LacZ genomic RNA and for infectivity on PHH. The results for HIV(LMS) and HIV(G) are summarized in Fig. 4A and B, respectively. The distributions of genomic RNA and infectivity overlapped and were the same for both viruses. As expected, the distance sedimented by these pseudotypes was significantly greater than that for SVP, HDV, and HBV. Next, fractions containing HIV(LMS) and HIV(G) were pooled, as indicated by p, and examined by immunoblotting, in each case with comparisons to an aliquot of the cell lysate, as indicated by c. As shown in Fig. 4, we were able to detect p24 in both viruses and VSV G in HIV(G). However, for HIV(LMS), we could not detect HBsAg in the virus, even though they were detected in the cell lysate.
The inability to detect HBsAg in the HIV(LMS) particles was puzzling since our biological assays of infectivity provided strong but nevertheless indirect evidence for the presence of these proteins (Fig. 2 and 3 and Table 1). Therefore, we carried out a more sensitive assay in which HIV(LMS) and HIV(G) were immunoprecipitated by rabbit polyclonal antibodies directed against HBsAg and VSV G. The IP results (percentage calculated by the average number of antibody-selected genomes relative to the average number of input genomes, with the standard deviations indicated) showed that HIV(LMS) was immunoprecipitated 0.56% ± 0.25% and 0.33% ± 0.35% with anti-HBsAg and anti-VSV G, respectively, and that HIV(G) was immunoprecipitated 0.51% ± 0.07% and 3.25% ± 2.90% with anti-HBsAg and anti-VSV G, respectively (IP was performed in duplicate for each of the two viruses and for each of the two indicated rabbit polyclonal antibodies; RNA was extracted from both total input and IP samples and assayed by qPCR to detect the LacZ genome). The ability of either antibody to specifically immunoprecipitate the pseudotype with the corresponding envelope proteins was both low and not significantly above that obtained with the negative control antibody. Furthermore, HDV mixed with HIV(LMS) could specifically immunoprecipitate >50% of input HDV genomes but not a significant amount of the HIV genomes (data not shown). Thus, while this result validates the IP procedure, we are still left with the interpretation that most of the HIV pseudotypes do not contain sufficient envelope protein to allow IP. This is consistent with our previous result showing that for HIV(LMS), there was only 1 TU per 7,000 GE. Similarly, others previously reported only 1 TU in 10,000 GE for another lentivirus vector (26).
Therefore, as another approach to provide evidence for HBsAg on the surface of HIV(LMS), we tested whether HBsAg antibodies could neutralize the ability of particles to infect PHH. Incubation with the same HBsAg antibody described above reduced infection by 14-fold, while a control rabbit serum failed to neutralize infectivity. Also, M18/7, a pre-S1-specific antibody (a gift from Wolfram Gerlich) reduced infection by sevenfold, whereas a mouse isotype control antibody did not block infection. These examples of an ability to neutralize infectivity, when combined with the data described above, support the interpretation that while the HBsAg of HIV(LMS) may be insufficient for extensive IP, they are nevertheless still present on the surface of the particles and are essential for infectivity.
In summary, studies of the assembled HIV(LMS) by immunoblotting (Fig. 4) and IP (see above) were unable to detect HBsAg on the surface of the particles. Indeed, it might be that many of the assembled particles are deficient in or even devoid of HBsAg. Nevertheless, for the particles that do infect PHH, four lines of evidence support the interpretation that HBsAg are present and with pre-S sequences exposed on the surface of the particles. First, HIV(LMS) infected PHH and these cells only (Fig. 2 and Table 1), that is, the same tropism as HDV and HBV, which are known to depend upon HBsAg for attachment and entry (11). Second, HIV(S) was unable to infect PHH, again consistent with the need for pre-S sequences to be present on the surface. Third, several soluble forms of pre-S sequences were able to block the infection of PHH by HIV(LMS) (Fig. 3). Finally, two antibodies, specific for S and pre-S, respectively, were able to neutralize infectivity.
DISCUSSION
The studies presented here have significant implications in three different areas. The first area is in adding to our understanding of the complex process of HBV assembly that was suggested to take place in the ER (22) or ER-Golgi intermediate compartment (15). L, M, and S are inserted into the ER membrane cotranslationally, and for about half of the L protein, the pre-S region is then translocated into the ER lumen (19). This translocation event is independent of the HBV nucleocapsid and can be subsequently detected on secreted virions and SVP (5, 24). The dual topology of L supports its multiple roles in the viral life cycle, including attachment/entry and assembly (11).
Two previous studies detected HBsAg at the surface of transfected 293T cells by flow cytometry and immunocytochemistry analyses (25, 30). Our results by flow cytometry (Fig. 1) and infection (Fig. 2) support the interpretation that at least some of the pre-S has translocated and is exposed on the surface of both the cells and the released pseudotype particles. This surface expression of HBsAg might be a result of “leakage,” that is, incomplete ER retention, as has been suggested for the HCV envelope proteins (14).
It is well known that HIV capsid assembly and release are not dependent on envelope proteins (27) (Table 2). However, we observed a moderate reduction in HIV genome-containing particle production when increased amounts of L relative to S were used in pseudotyping (Table 2). This was probably due to a nonspecific cytopathic effect since previous studies showed that the overexpression of the HBV L protein leads to ER retention and cytotoxicity both in vitro and in vivo (8, 35). Furthermore, when L was used only in pseudotyping, the released genome-containing particles were not infectious (Table 2). We could not tell whether L only is not expressed on the cell surface or incorporated into HIV particles or whether infectivity requires S in addition to L. Flow cytometry and immunoblot analyses may help distinguish between these possibilities.
A second consequence of our studies is that they can help reveal the mechanisms by which HBV and HDV attach to and enter susceptible cells. Our data clearly show no difference in the specificities with which the HIV pseudotype infects cells relative to what we and others have previously observed for HBV and HDV infections (6, 11, 13). Specifically, (i) the HIV(LMS) virus infected PHH but not PWH, Huh7 cells, HepG2 cells, or 293T cells (Fig. 2 and data not shown); (ii) the HIV(S) virus, which has no pre-S domain, did not infect PHH (Tables 1 and 3); and (iii) certain immunoadhesins and chemically synthesized peptides containing pre-S sequences inhibited the infection of PHH by HIV(LMS) (Fig. 3). Thus, we expect that this pseudotype system provides a convenient approach to investigate the important and yet still unclear mechanisms of HBV and HDV attachment and entry.
A third consequence of our results is the potential use of lentivirus vectors with LMS as agents for efficient hepatocyte-specific gene delivery. Retroviruses have offered great promise in gene therapy, and lentiviruses have the additional advantage of infecting nondividing and quiescent cells (20, 29, 33). The HIV(LMS) virus was efficient in that it could infect up to 50% of cultured PHH. It was also specific, as mentioned above, in that it did not infect four other cell types that were tested (Fig. 2 and data not shown). Admittedly, more extensive in vivo studies are needed before we can justify the application of these lentivirus vectors in liver-specific gene delivery. With this caveat, our studies significantly advanced how lentivirus vectors might be used to target hepatocytes in the liver. While lentivirus vectors like HIV(G) have been used in experimental animals to target liver cells (23), such vectors can infect cells other than hepatocytes, even though some vectors have been modified to include liver-specific promoters (17). Compared to these vectors, HIV(LMS) provides additional liver specificity by infecting only PHH.
Obviously, lentivirus vectors with HBV envelopes offer significant advantages for battling liver-specific viruses, including HCV, HBV, and HDV, and for intervening in liver-specific deficiencies or diseases. For example, they can be used as part of combination therapies against chronic HBV/HDV by delivery to hepatocyte sequences that lead to the transcription of RNAs with antiviral activities, such as small hairpin RNAs (7), or to the translation of inhibitory proteins, such as HBV pre-S immunoadhesins (6).
Acknowledgments
J.T. was supported by grants AI-058269 and CA-06927 from the NIH and by an appropriation from the Commonwealth of Pennsylvania. This project was conceived by N.C., who was supported in part by an Elizabeth Knight Patterson fellowship.
Constructive comments on the manuscript were provided by Volker Bruss, Stephan Urban, William Mason, and Glenn Rall. We thank the Fox Chase Biochemistry and Biotechnology Facility and James Osterling of the Flow Cytometry Facility.
Footnotes
Published ahead of print on 1 August 2007.
REFERENCES
- 1.Barrera, A., B. Guerra, L. Notvall, and R. E. Lanford. 2005. Mapping of the hepatitis B virus pre-S1 domain involved in receptor recognition. J. Virol. 79:9786-9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruss, V. 2007. Hepatitis B virus morphogenesis. World J. Gastroenterol. 13:65-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. USA 88:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruss, V., X. Lu, R. Thomssen, and W. H. Gerlich. 1994. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 13:2273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chai, N., S. Gudima, J. Chang, and J. Taylor. 2007. Immunoadhesins containing pre-S domains of hepatitis B virus large envelope protein are secreted and inhibit virus infection. J. Virol. 81:4912-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, J., and J. M. Taylor. 2003. Susceptibility of human hepatitis delta virus RNAs to small interfering RNA action. J. Virol. 77:9728-9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chisari, F. V., P. Filippi, J. Buras, A. McLachlan, H. Popper, C. A. Pinkert, R. D. Palmiter, and R. L. Brinster. 1987. Structural and pathological effects of synthesis of hepatitis B virus large envelope polypeptide in transgenic mice. Proc. Natl. Acad. Sci. USA 84:6909-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cronin, J., X. Y. Zhang, and J. Reiser. 2005. Altering the tropism of lentiviral vectors through pseudotyping. Curr. Gene Ther. 5:387-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelke, M., K. Mills, S. Seitz, P. Simon, P. Gripon, M. Schnolzer, and S. Urban. 2006. Characterization of a hepatitis B and hepatitis delta virus receptor binding site. Hepatology 43:750-760. [DOI] [PubMed] [Google Scholar]
- 11.Glebe, D., and S. Urban. 2007. Viral and cellular determinants involved in hepadnaviral entry. World J. Gastroenterol. 13:22-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gripon, P., I. Cannie, and S. Urban. 2005. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J. Virol. 79:1613-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudima, S., Y. He, A. Meier, J. Chang, R. Chen, M. Jarnik, E. Nicolas, V. Bruss, and J. Taylor. 2007. Assembly of hepatitis delta virus: particle characterization including ability to infect primary human hepatocytes. J. Virol. 81:3608-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huovila, A. P., A. M. Eder, and S. D. Fuller. 1992. Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J. Cell Biol. 118:1305-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Levine, B. L., L. M. Humeau, J. Boyer, R. R. MacGregor, T. Rebello, X. Lu, G. K. Binder, V. Slepushkin, F. Lemiale, J. R. Mascola, F. D. Bushman, B. Dropulic, and C. H. June. 2006. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc. Natl. Acad. Sci. USA 103:17372-17377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 19.Ostapchuk, P., P. Hearing, and D. Ganem. 1994. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 13:1048-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park, F., K. Ohashi, W. Chiu, L. Naldini, and M. A. Kay. 2000. Efficient lentiviral transduction of liver requires cell cycling in vivo. Nat. Genet. 24:49-52. [DOI] [PubMed] [Google Scholar]
- 21.Park, F., K. Ohashi, and M. A. Kay. 2003. The effect of age on hepatic gene transfer with self-inactivating lentiviral vectors in vivo. Mol. Ther. 8:314-323. [DOI] [PubMed] [Google Scholar]
- 22.Patient, R., C. Hourioux, P.-Y. Sizaret, S. Trassard, C. Sureau, and P. Roingeard. 2007. Hepatitis B virus subviral envelope particle morphogenesis and intracellular trafficking. J. Virol. 81:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfeifer, A., T. Kessler, M. Yang, E. Baranov, N. Kootstra, D. A. Cheresh, R. M. Hoffman, and I. M. Verma. 2001. Transduction of liver cells by lentiviral vectors: analysis in living animals by fluorescence imaging. Mol. Ther. 3:319-322. [DOI] [PubMed] [Google Scholar]
- 24.Prange, R., and R. E. Streeck. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha, M. N., A. Tanaka, A. Jinno-Oue, N. Shimizu, K. Tamura, M. Shinagawa, J. Chiba, and H. Hoshino. 2005. Formation of vesicular stomatitis virus pseudotypes bearing surface proteins of hepatitis B virus. J. Virol. 79:12566-12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sastry, L., T. Johnson, M. J. Hobson, B. Smucker, and K. Cornetta. 2002. Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Ther. 9:1155-1162. [DOI] [PubMed] [Google Scholar]
- 27.Sharma, S., F. Murai, A. Miyanohara, and T. Friedmann. 1997. Noninfectious virus-like particles produced by Moloney murine leukemia virus-based retrovirus packaging cells deficient in viral envelope become infectious in the presence of lipofection reagents. Proc. Natl. Acad. Sci. USA 94:10803-10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmons, G., J. D. Reeves, A. J. Rennekamp, S. M. Amberg, A. J. Piefer, and P. Bates. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA 101:4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sinn, P. L., S. L. Sauter, and P. B. McCray, Jr. 2005. Gene therapy progress and prospects: development of improved lentiviral and retroviral vectors—design, biosafety, and production. Gene Ther. 12:1089-1098. [DOI] [PubMed] [Google Scholar]
- 30.Sung, V. M., and M. M. Lai. 2002. Murine retroviral pseudotype virus containing hepatitis B virus large and small surface antigens confers specific tropism for primary human hepatocytes: a potential liver-specific targeting system. J. Virol. 76:912-917. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Sureau, C. 2006. The role of the HBV envelope proteins in the HDV replication cycle. Curr. Top. Microbiol. Immunol. 307:113-131. [DOI] [PubMed] [Google Scholar]
- 32.Taylor, J., W. Mason, J. Summers, J. Goldberg, C. Aldrich, L. Coates, J. Gerin, and E. Gowans. 1987. Replication of human hepatitis delta virus in primary cultures of woodchuck hepatocytes. J. Virol. 61:2891-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verma, I. M. 1999. From reverse transcriptase to gene therapy: a marvelous journey. Harvey Lect. 95:43-66. [PubMed] [Google Scholar]
- 34.Watson, D. J., G. P. Kobinger, M. A. Passini, J. M. Wilson, and J. H. Wolfe. 2002. Targeted transduction patterns in the mouse brain by lentivirus vectors pseudotyped with VSV, Ebola, Mokola, LCMV, or MuLV envelope proteins. Mol. Ther. 5:528-537. [DOI] [PubMed] [Google Scholar]
- 35.Xu, Z., V. Bruss, and T. S. Yen. 1997. Formation of intracellular particles by hepatitis B virus large surface protein. J. Virol. 71:5487-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zavada, J., C. Dickson, and R. Weiss. 1977. Pseudotypes of vesicular stomatitis virus with envelope antigens provided by murine mammary tumor virus. Virology 82:221-231. [DOI] [PubMed] [Google Scholar]