Abstract
Histone deacetylase (HDAC) inhibitors such as valproic acid (VPA) induce the expression of quiescent proviral human immunodeficiency virus type 1 (HIV-1) and may deplete proviral infection in vivo. To uncover novel molecular mechanisms that maintain HIV latency, we sought cellular mRNAs whose expression was diminished in resting CD4+ T cells of HIV-1-infected patients exposed to VPA. c-Myc was prominent among genes markedly downregulated upon exposure to VPA. c-Myc expression repressed HIV-1 expression in chronically infected cell lines. Chromatin immunoprecipitation (ChIP) assays revealed that c-Myc and HDAC1 are coordinately resident at the HIV-1 long terminal repeat (LTR) promoter and absent from the promoter after VPA treatment in concert with histone acetylation, RNA polymerase II recruitment, and LTR expression. Sequential ChIP assays demonstrated that c-Myc, Sp1, and HDAC1 coexist in the same DNA-protein complex at the HIV promoter. Short hairpin RNA inhibition of c-Myc reduces both c-Myc and HDAC1 occupancy, blocks c-Myc repression of Tat activation, and increases LTR expression. These results expand the understanding of mechanisms that recruit HDAC and maintain the latency of HIV-1, suggesting novel therapeutic approaches against latent proviral HIV infection.
The molecular mechanisms that establish or maintain human immunodeficiency virus type 1 (HIV-1) latency are incompletely understood. Several mechanisms, which are not mutually exclusive, have been proposed to drive HIV-1 proviral quiescence or latency (14, 21, 37). As the HIV-1 provirus is often integrated into active regions of the host genome, the chromatin environment and the interaction of cellular and viral transcriptional regulators are likely to be critical for inducing and maintaining HIV-1 latency (5, 27, 37, 44). A surprisingly robust induction of HIV-1 transcription in response to deacetylase inhibitors has been observed (19, 22, 49).
Histone modifications remodel the chromatin of the viral promoter region but also regulate the functional properties of cellular and viral transcription factor binding to the HIV-1 long terminal repeat (LTR) (28, 40, 45, 50). By altering histones, recruiting other chromatin-remodeling factors, and altering the activity of transcription factors, histone deacetylases (HDACs) appear to be critical for the repression of HIV-1 transcription and the maintenance of HIV-1 latency (45, 50). In support of the clinical relevance of this model, we found that the administration of an HDAC inhibitor, valproic acid (VPA), in the context of intensified highly active antiretroviral therapy depleted latent HIV-1 infection in vivo (23), although this was not observed in a clinical cohort on standard antiretroviral therapy and incidentally coadministered VPA (41). Additional clinical studies to test this approach are ongoing.
Alternative therapeutic approaches to proviral HIV infection may be needed. As HDAC inhibition can modify the expression of a subset of human genes (45) and VPA modifies the activity of other host enzymes (26), we studied changes in host gene expression induced by VPA with resting CD4+ T cells of HIV-infected patients. We reasoned that perturbations in gene expression pathways might lead to the identification of novel mechanisms that maintained HIV proviral quiescence. We found that c-Myc expression was markedly downregulated when resting CD4+ lymphocytes from HIV-infected patients were exposed to VPA, suggesting that c-Myc may lead to the repression of HIV expression by a novel mechanism.
Myc is a member of the basic helix-loop-helix leucine zipper (bHLH-Zip) transcription factor family required for growth, proliferation, and differentiation. One member of the Myc family, the proto-oncogene c-Myc, is implicated in a variety of physiological processes, including the transcriptional regulation of many genes (6, 11, 12). When dimerized with the bHLH-Zip protein Max, c-Myc binds to the E-box sequence CACGTG and activates the transcription of target genes; however, Myc has also been shown to repress the transcription of several genes such as the C/EBPα, H-ferrin, IRP2, LFA-1 receptor, gadd45, platelet-derived growth factor, and p15Ink4b genes (1, 17, 18, 42, 47). One current model suggests that Myc represses transcription through functional interference with transcriptional activators (11). Alternatively, c-Myc may also block transcription by recruiting HDAC1 (39). c-Myc was recently shown to recruit the corepressor Dnmt3a to the p21Cip1 gene promoter and in association with DNA methyltransferase activity in vivo (3). Ectopic expression of c-Myc has been shown to decrease HIV-1 expression, but the molecular mechanism for this observation has not been determined (43).
To our surprise, we found that c-Myc is recruited to the HIV LTR by Sp1 and that c-Myc in turn recruits HDAC1 and blunts HIV promoter expression. This is the third mechanism reported to recruit HDAC1 to the HIV LTR and induce chromatin remodeling. This finding highlights a unique redundant use of this cellular enzyme by the HIV LTR to maintain quiescence in resting T cells and strongly suggests that mechanisms that recruit HDAC1 and maintain its activity at the LTR are important targets for antilatency therapies.
MATERIALS AND METHODS
Cell culture and VPA treatment.
HeLa-CAT-CD4 and J89 cells were maintained in Dulbecco's modified Eagle medium and RPMI 1640 medium, respectively, with 10% fetal bovine serum containing 100 μg/ml streptomycin and 100 units/ml of penicillin. HeLa-CAT-CD4 and primary resting CD4+ T cells (2 × 106 cells) were split into 100-mm culture dishes and grown overnight, VPA was added, and, after incubation for the appropriate time, the cells were collected. Proteins were extracted with Cellytic MT (Sigma, St. Louis, MO) for Western blotting or immunoprecipitation (IP), or the cells were cross-linked for chromatin IP (ChIP) assays.
Microarrays and analysis.
Microarrays were obtained from Agilent Technologies (Palo Alto, CA) and consisted of 23,500 60-mer oligonucleotides designed for broad coverage of the human genome. Lymphocytes were obtained by continuous-flow leukophoresis from stable, HIV-infected volunteers on antiretroviral therapy with plasma HIV-1 RNA levels of <50 copies/ml for more than 6 months and CD4 cell counts of >300 cells/ml. Committee-approved informed consent was obtained from all patients. The isolation of CD4 T cells was performed as described in detail previously (23, 37, 49). Total RNA from untreated, VPA-treated, or phytohemagglutinin (PHA)-treated resting CD4+ T cells obtained from HIV-infected, aviremic subjects was reverse transcribed, subjected to in vitro transcription, labeled with Cy3 and Cy5, and hybridized as described previously (15). For each treatment, RNA samples were processed and hybridized against a reference pool derived from RNA of the untreated cells from the same subject. Scanned intensities were normalized and converted to log ratio values, and significant (P < 0.01 with greater than a 1.5× to 2× difference) changes in gene expression were identified using the Rosetta Resolver system (Rosetta Biosoftware, Seattle, WA).
Transfection.
Cells (3 × 105) were seeded in 12-well plates and grown overnight. The expression plasmids pCMV-empty vector, pCMV-Tat, pCGN-c-Myc (a gift of W. Tansey, Cold Spring Harbor Laboratory) (38), pILIC-CAT, pNFA-CAT, and pSpC-CAT (34) were purified and cotransfected by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and then incubated for 48 to 96 h.
RNA preparation and real-time PCR.
Total RNA was isolated using TRIzol (Invitrogen), and the RNA was treated with DNase (RNase A-free; Invitrogen). Chloramphenicol acetyltransferase (CAT) expression was then assayed by SYBR-based real-time PCR (One-Step Real-Time RT-PCR kit; Applied Biosystems) with 0.1 μg total RNA and 50 nM specific primers for 40 cycles. The CAT primers were CAT-5F (5′-TTC GTC TCA GCC AAT CCC TGG GTG A-3′) and CAT-6R (5′-CCC ATC GTG AAA ACG GGG GCG AA-3′). The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) primers were GAPDH-3F (5′-CAC CAT GGA GAA GGC TGG GGC TCA-3′) and GAPDH-3R (5′-TGA CGA ACA TGG GGG CAT CAG CAG A-3′). The green fluorescent protein (GFP) primers were GFP-5Jor (5′-GGA GCG CAC CAT CTT CTT CA-3′) and GFP-3Jor (5′-AGG GTG TCG CCC TCG AA-3′). Monitoring mRNA expression of GFP is a more direct measure of transcription that is less likely to measure the secondary effects of c-Myc knockdown. The relative quantification was analyzed with Applied Biosystems relative quantification software.
ChIP and sequential ChIP assays.
The following antibodies were used: anti-acetyl-histone H4, anti-HP1α, anti-RNA polymerase CTD, anti-AceH4 (Upstate Biotechnology), anti-DNMT3a (IMGENEX, San Diego, CA), anti-HDAC1 (Abcam, Cambridge, MA), and anti-c-Myc (Santa Cruz Biotech, Santa Cruz, CA). ChIP assays were performed as previously described (14). PCR was carried out with the following primers: LTR7F (5′-TGG AGG TTT GAC AGC CGC CTA-3′) and LTR8R (5′-AGG CTT AAG CAG TGG GTT CCC TA-3′). The following primers were used for the LTR downstream PCR. For HeLa-CAT-CD4 cells, the primers were DS1 (5′-TTC GTC TCA GCC AAT CCC TGG GTG A-3′) and DS2 (5′-CCC ATC GTG AAA ACG GGG GCG AA-3′). For J89 cells, the downstream primers used were reported previously (45).
A quantitative real-time PCR assay was performed to analyze the enrichment of the proteins specific to the HIV-1 LTR region. The following primers were used for quantitation: LTR-7F (5′-AGC CCT CAG ATC CTG CAT ATA AGC A-3′) and LTR-8R (5′-TAG CCA GAG AGC TCC CAG GCT CAG A-3′). The relative changes in ChIP products are quantitated by real-time PCR of a fraction of the input and of immunoprecipitated promoter fragments. A ΔCT value was calculated for each sample by subtracting the threshold cycle (CT) value for the input from the CT value obtained for the ChIP sample. A ΔΔCT value was then calculated by subtracting the ΔCT value for the immunoprecipitated sample from the ΔCT value for the corresponding control IP with isotype immunoglobulin G (IgG). Differences (n-fold) (ChIP relative to control IgG ChIP or untreated ChIP relative to treated ChIP) were then determined by raising 2 to the ΔΔCT power.
For sequential ChIP assays, the initial ChIP was performed with the indicated antibodies, chromatin was eluted with dithiothreitol, and a second ChIP was then carried out. Amplified products were quantitated by real-time PCR. The relative binding of the indicated proteins to the HIV-1 LTR region was calculated from real-time PCR data as described above.
RNA interference.
Human Sp1 small interfering RNA expression plasmid pKD-Sp1-v1 and its mismatch vector pKD-NegCon-v1 were purchased from Upstate, while human c-Myc small interfering RNA expression plasmid pSUPER-c-Myc and its mismatch control vector pSUPER-GL2 were gifts from Ernest Martinez (10). HeLa-CAT-CD4 or J89 cells (3 × 105) were seed into six-well plates, and after incubation overnight, 2.5 μg of the plasmids was transfected into the cells with Lipofectamine 2000 or TransIT-Jurkat reagent (Mirus, Madison, WI). At 4 days, the cells were collected, and proteins were extracted with Cellytic MT (Sigma) for Western blotting, or the cells were cross-linked for ChIP assays.
RESULTS
Expression of c-Myc is reduced in resting CD4+ T cells of aviremic HIV-infected patients following exposure to VPA.
To discover additional pathways that maintain latency, we sought genes whose expression in primary cells was decreased by VPA. Under institutional review board-approved informed consent, resting CD4+ T cells were collected from three HIV-1-infected patients and on two occasions from a fourth patient. In all patients, viremia was stably suppressed by antiretroviral therapy. We exposed patients' cells for 6 h to 2 μg/ml PHA and 20 U/ml interleukin-2 (IL-2) or 1 mM VPA and IL-2, conditions in which HIV is routinely recovered from patients' resting CD4+ T cells in outgrowth assays (49, 50). As a negative control, cells were exposed to IL-2 (20 U/ml) alone, a condition in which the recovery of HIV is not observed. Total RNA was obtained and profiled by microarray.
We found that in primary resting CD4+ T cells from these HIV-infected patients, c-Myc gene expression was significantly downregulated by VPA. Overall, only 42 genes were consistently downregulated (P < 0.01) by an average of more than threefold, and 157 genes were consistently upregulated (P < 0.01) by an average of more than threefold. c-Myc was strikingly downregulated (Fig. 1A): only 24 of the 23,757 reporters studied were downregulated to a quantitatively greater extent.
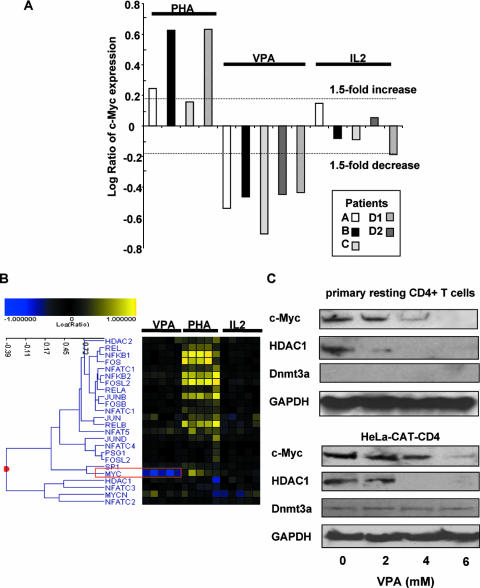
FIG. 1.
Expression of c-Myc in latently HIV-1-infected patient cells and HeLa-CAT-CD4 cells after VPA treatment. (A) c-Myc expression is significantly decreased upon exposure to VPA (P value for a change of >1.5-fold is <0.01). (B) In the resting CD4+ T cells of four HIV-infected patients, c-Myc RNA expression levels uniformly and substantially decreased following VPA exposure (red box). c-Myc RNA expression was unaffected by 20 U/ml IL-2 and upregulated modestly in some patients' cells following mitogen activation. (C) Decreased c-Myc protein expression 48 h after treatment with VPA. HeLa-CAT-CD4 cells or resting CD4+ T cells isolated from HIV-infected, highly active antiretroviral therapy-treated donors were treated with the amount of VPA indicated. The Western blot shows protein expression of c-Myc, HDAC1, the histone demethylase Dnmt3a, and GAPDH.
In contrast, the expression of the other transcription factors of known relevance to HIV promoter activation (e.g., Rel/NF-κB, Sp1, Fos, Jun, and NFAT) (7, 13, 27) was unchanged (Fig. 1B). As expected, mitogen activation induced the expression of a panoply of genes associated with HIV LTR activation, such as Sp1 and NF-κB (Fig. 1B). PHA also upregulated c-Myc expression (Fig. 1A), but presumably, this effect is overwhelmed by the multiple activation pathways induced by mitogen.
The downregulation of c-Myc expression after treatment with HDAC inhibitors has been reported for cancer cells (2, 20, 25) and HIV-infected cell lines (45). Furthermore, de la Fuente et al. (7) previously reported that mRNA expression of c-Myc was upregulated in latently HIV-1-infected ACH2 cells compared with uninfected CEM cells. The overexpression of c-Myc has been shown to decrease the expression of an HIV LTR reporter plasmid in Jurkat T cells (43). These findings suggest the possibility that c-Myc may downregulate HIV-1 transcription and promote HIV-1 latency. To confirm the changes observed in microarray analyses, protein expression of c-Myc was measured in HeLa-CAT-CD4 cells and in resting CD4+ T cells isolated from aviremic, HIV-infected volunteers after treatment with VPA. The protein level of c-Myc was reduced by VPA, in concert with decreased c-Myc mRNA expression. Due to an effect at the posttranscriptional level, HDAC1 protein expression was also diminished in the cell line and patient cells (Fig. 1C), although HDAC1 mRNA expression was not directly downregulated (Fig. 1B).
VPA alters chromatin structure by depleting several members of the structural maintenance of chromatin protein family, proteins associated with the structural maintenance of chromatin, DNA methyltransferases, and heterochromatin proteins (8, 30, 33, 48). The DNA methyltransferase Dnmt3a was recently found to be required for c-Myc inhibition of the p21 promoter (3). However, as shown in Fig. 1C, there was no significant change in the expression of the Dnmt3a protein in HeLa-CAT-CD4 cells.
c-Myc negatively regulates HIV-1 transcription.
To examine the role of c-Myc in HIV-1 LTR-directed transcriptional regulation, c-Myc was transiently coexpressed with HIV Tat in HeLa-CAT-CD4 cells, and 48 h after transfection, absolute CAT RNA expression was assayed using real-time PCR. c-Myc itself had no significant effect on the low level of basal HIV-1 LTR expression but induced significant and reproducible inhibition of Tat-activated HIV-1 LTR expression (Fig. 2A). Tat-activated expression of LTR-driven CAT mRNA was decreased by c-Myc in a dose-dependent fashion (Fig. 2B), while the transient expression of Tat itself was not affected by c-Myc.
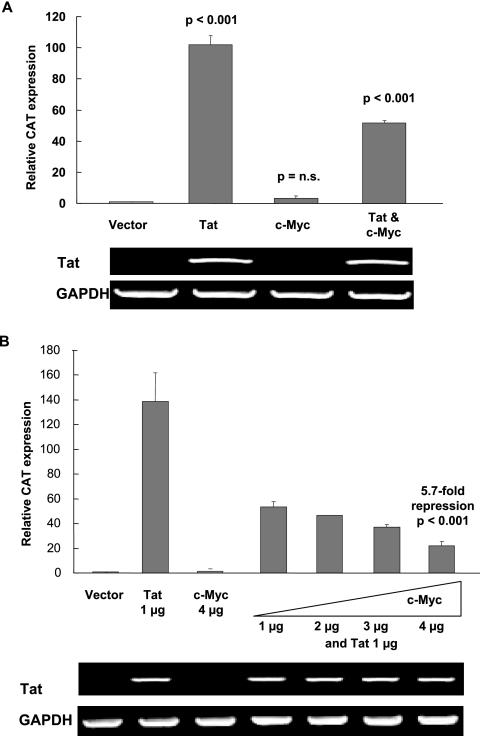
FIG. 2.
Overexpression of c-Myc negatively regulates HIV-1 LTR transcription. (A) c-Myc blunts Tat-activated LTR expression in HeLa-CAT-CD4 cells. pCMV vector, pCMV-Tat, pGCN-c-Myc, or pGCN-c-Myc plus pCMV-Tat were transfected into HeLa-CAT-CD4 cells. CAT reporter gene expression was assayed by real-time PCR after 48 h. RNA PCR demonstrates that GAPDH (loading control) and transfected Tat expression levels are equivalently unaffected by c-Myc. Relative CAT reporter gene expression was normalized to a value of 1.0 for pCMV-transfected cells. Bars indicate standard errors of the means for three independent experiments. P values are given for comparisons with vector control. n.s., not significant. (B) Dose-dependent inhibition of Tat activation by c-Myc. LTR expression was monitored in the presence of increased ratios of pGCN-c-Myc to pCMV-Tat (1:1, 1:2, 1:3, and 1:4). At the highest dose of c-Myc vector, Tat activation was repressed 5.7-fold (P < 0.001).
Both c-Myc and HDAC1 are tethered to the HIV-1 LTR promoter region.
Through the recruitment of corepressors, DNMT3a or HDACs, c-Myc can inhibit the expression of its target p21, cad, or NDRG2 gene (3, 34). To strengthen the hypothesis that c-Myc plays a direct molecular role in the repression of HIV LTR expression, ChIP assays were done to seek evidence of c-Myc occupancy at the HIV-1 promoter. HeLa-CAT-CD4 cells were treated with 4 mM VPA or vehicle for 4 h, at which time cells were cross-linked and sonicated, DNA-protein complexes were collected by centrifugation, and ChIP was performed. c-Myc was present at the nucleosome 1 (Nuc-1) region of the HIV LTR in the basal state, and occupancy was decreased in cells exposed to the HDAC inhibitor VPA (Fig. 3A). As previously demonstrated (14), HDAC1 was also present at Nuc-1 in the basal state, and occupancy diminished after VPA exposure. In concert with reduced c-Myc and HDAC1 occupancy, the binding of acetylated histone H4 increased (Fig. 3A). Although the methyltransferase Dnmt3a interacts with c-Myc at other promoters, it was not detected at the HIV LTR and therefore served as a negative control. RNA polymerase II occupancy at the Nuc-1 region and a downstream site (45) also increased after VPA exposure. Notably, the specificity of c-Myc and HDAC1 ChIP was illustrated by the lack of occupancy at the downstream site.
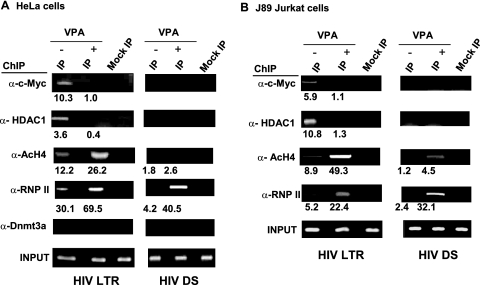
FIG. 3.
Recruitment of c-Myc to the HIV-1 LTR region. (A) Binding of c-Myc, HDAC1, acetylated H4, and RNA polymerase II to the Nuc-1 region of the HIV-1 LTR in HeLa-CAT-CD4 cells. VPA exposure resulted in significantly decreased occupancy of HDAC1 and c-Myc, increased occupancy of RNA polymerase II and acetylated histone H4 at the Nuc-1 region, and an increase in only RNA polymerase II detection downstream (DS) of the initiation region. Amplified products were quantitated by real-time PCR. The relative binding of indicated proteins to the HIV-1 LTR region was measured after normalization with that of input DNA and IgG. (B) Binding of c-Myc and HDAC1 to HIV-1 LTR in latently HIV-1-infected J89 lymphocytic cells. VPA exposure again resulted in significantly decreased occupancy of HDAC1, c-Myc, acetylated histone H4, and RNA polymerase II near Nuc-1 and little change at the downstream site. Amplified products were quantitated by real-time PCR. The relative binding of indicated proteins to the HIV-1 LTR region was measured after normalization with that of input DNA and IgG. Assays were performed in triplicate (or more), and the significance of changes was tested by a two-sided t test. P values are <0.05 for all changes of at least twofold and <0.01 for all changes of at least threefold.
Extending these findings in the context of a lymphocytic cell line, we observed similar binding of c-Myc and HDAC1 to the HIV-1 LTR region in latently HIV-1-infected J89 Jurkat T cells (Fig. 3B). As in the HeLa-CAT-CD4 cell line, c-Myc and HDAC1 recruitment was disrupted after VPA exposure. Again, no significant binding of Dmnt3a to the HIV-1 LTR was observed, and c-Myc and HDAC1 were not detected downstream of the Nuc-1 region. These results, together with the findings described above, suggest that c-Myc is a negative regulator of HIV-1 gene transcription via the recruitment of HDAC1 to the HIV-1 LTR promoter region.
c-Myc is associated with HDAC1.
HDAC inhibitors displace HDAC1 from the HIV-1 LTR Nuc-1 region and increase the acetylation of histone H4 at Nuc-1 (14). Both c-Myc mRNA and protein levels diminished when the cells were treated with VPA (Fig. 1). Therefore, c-Myc may recruit HDAC1 to the LTR, and HDAC inhibitors may act both by blocking HDAC enzyme action, thus inhibiting factor interactions upregulated by acetylation, and by decreasing the cellular levels of key repressor factors, including c-Myc. c-Myc may act by direct interactions with HDAC1 or may exist in a complex that includes HDAC1.
We validated the previously reported ability of c-Myc and HDAC1 to directly interact (3, 34), as in HeLa-CAT-CD4 and J89 cells. We found c-Myc present in complexes immunoprecipitated by anti-HDAC1, and conversely, HDAC1 was present in complexes immunoprecipitated by anti-c-Myc (not shown).
To demonstrate that c-Myc and HDAC1 coexist within the same protein complex resident in the Nuc-1 region of the HIV LTR, sequential ChIP assays were performed. In such assays, an initial ChIP was performed with an antibody that recognizes one protein. The precipitated chromatin-DNA complex was washed and eluted, and a second IP was performed with the same antibody, a second antibody, or control isotype IgG.
When ChIP was first performed with anti-c-Myc (Fig. 4), sequential ChIP showed the occupancy of HDAC1 in the same protein-DNA complex. As the cross-linking distance of formaldehyde is 4 Å, these results do not prove a direct interaction of these proteins but provide evidence of close occupancy at the LTR initiation region. The specificity of these results is illustrated by the lack of significant IP using Dnmt3a, a cellular factor that can interact with c-Myc but for which there is no evidence of occupancy at the Nuc-1 region of the LTR. The finding that the IP product is decreased when a different antibody is used during the second ChIP (e.g., less product for ChIP with c-Myc/c-Myc than with c-Myc/HDAC1) may illustrate that not all c-Myc-containing complexes include HDAC1 or might simply be an artifact of differing antibody affinities for epitopes that are accessible within the DNA-protein complex under study. Specificity was again demonstrated, as these complexes were not detected downstream of Nuc-1.
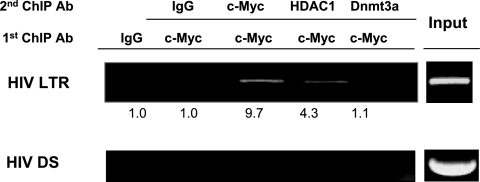
FIG. 4.
c-Myc and HDAC1 are found in the same complex at the HIV LTR initiator region. Sequential ChIP assays were performed after an initial IP with anti-c-Myc. Protein-DNA complexes were detected near the Nuc-1 region after a second IP with anti-c-Myc or anti-HDAC1 but not in the downstream (DS) region and not with isotype IgG or antibody (Ab) against the methyltransferase Dnmt3a. Real-time PCR quantitation of DNA IP, as detailed in Materials and Methods, is indicated.
Binding of c-Myc to the HIV-1 LTR promoter is mediated by Sp1.
ChIP assays demonstrated that c-Myc bound the HIV-1 LTR, but the mechanism of its recruitment was unclear. Given that LSF/YY1 and NF-κB p50 can recruit HDAC1 (46, 50), a link for c-Myc to HDAC1 recruitment was unproven. Although a putative E box in the initiator region (Inr) has been proposed to be a site for c-Myc binding, there is no evidence that c-Myc binding occurs at that site (18). Furthermore, while we have demonstrated an association between c-Myc and the HIV-1 LTR promoter, c-Myc can be recruited to promoters by other transcription factors or corepressors without direct promoter binding (32).
Given the host transcription factors known to play central roles at the LTR Nuc-1 region, a prime candidate among the network of c-Myc binding partners with potential to recruit c-Myc to the HIV LTR was Sp1. Sp1 has been shown to directly interact with c-Myc (12). To determine if Sp1 recruits c-Myc to the HIV-1, LTR reporter assays were performed (24).
Wild-type HIV-1 LTR CAT plasmids (pILIC-CAT) or an LTR reporter lacking Sp1 binding sites (pSpA-CAT) was cotransfected into HeLa cells with and without Tat and c-Myc expression vectors (Fig. 5A). While the activation potential of the HIV LTR is diminished when Sp1 sites are mutated, such constructs can be activated or respond to Tat, and viral clones encoding Sp1 mutations can replicate (24, 31, 35).
FIG. 5.
Sp1 mediates c-Myc regulation of HIV-1 LTR expression. Sp1 sites are required for c-Myc-mediated repression, and Sp1 and c-Myc coexist within a complex at the LTR. (A) Expression plasmids pCMV-empty vector, pCMV-Tat, and pcDNA-c-Myc and the wild-type (wt) pILIC-CAT, pSpA-CAT, and pNFA-CAT reporter plasmids were cotransfected into HeLa cells using Lipofectamine 2000 reagent. CAT expression was analyzed by real-time RT-PCR. Tat-activated LTR RNA expression is displayed, with normalization to the cells transfected with pCMV-Tat for each of the reporter assays. PCR products for the GAPDH loading control and transfected Tat are shown. Error bars indicate standard errors of the means for three independent experiments. (B) Sequential ChIP assays were performed after an initial IP with anti-Sp1. A protein-DNA complex near Nuc-1 was recovered after a second IP with anti-Sp1, anti-c-Myc, or anti-HDAC1 (P < 0.01) but not at the downstream region or with isotype IgG or antibody (Ab) against the methyltransferase Dnmt3a. Real-time PCR quantitation of DNA IP, as detailed in Materials and Methods, is indicated.
c-Myc expression inhibited wild-type HIV-1 LTR expression, as measured by real-time PCR quantitation of LTR-driven CAT mRNA, but had no significant effect on the Tat-driven expression of the mutant HIV-1 LTR reporter lacking Sp1 binding sites. In cells transfected with a mutant HIV-1 LTR reporter lacking NF-κB binding sites (pNFA-CAT), c-Myc was still able to repress HIV-1 expression, although as previously demonstrated (31), the activation of HIV-1 with Tat was relatively lower than that of wild-type and Sp1-deleted mutant HIV-1 LTR. This suggests that Sp1 recruits c-Myc to the HIV-1 LTR with subsequent recruitment of HDAC1 and LTR repression.
To demonstrate that c-Myc and Sp1 coexist within the same protein complex resident in the Nuc-1 region of the HIV LTR, sequential ChIP assays were again performed. When ChIP was first performed with anti-Sp1 (Fig. 5B), sequential ChIP showed the occupancy of c-Myc and HDAC1 in the same protein-DNA complex. Again, these results do not prove a direct interaction of these proteins but provide evidence of close co-occupancy at the LTR initiation region. The specificity of these results was illustrated by the lack of significant IP using Dnmt3a or at the downstream site.
shRNA inhibition of Sp1 expression reduces c-Myc and HDAC1 occupancy and repression of LTR expression.
To directly demonstrate the role of Sp1 in c-Myc recruitment to the HIV-1 LTR region in vivo, we used a short hairpin RNA (shRNA) that specifically depleted Sp1 protein expression and tested if the knockdown of Sp1 would disrupt c-Myc binding to the HIV-1 promoter. Transfection of pKD-Sp1-v1 specifically depleted Sp1 in HeLa-CAT-CD4 cells, whereas transfection of a vector encoding a scrambled sequence did not (Fig. 6A). However, although c-Myc expression was unchanged (Fig. 6A) after Sp1 knockdown, when measured by ChIP in the same extract, the occupancy of Sp1, c-Myc, and HDAC1 at the LTR was significantly decreased (Fig. 6B).
FIG. 6.
c-Myc occupancy at the HIV-1 LTR and its repression of Tat activation require Sp1. (A) Specific shRNA-mediated knockdown of Sp1 protein expression. HeLa-CAT-CD4 cells were transfected with pKD-sp-v1 plasmids directed against Sp1 or pKD-Neg-con-v1 (control) plasmids. Sp1, c-Myc, and GAPDH protein levels were assessed by immunoblotting. (B) Reduced occupancy of Sp1, c-Myc, and HDAC1 at the LTR initiator region upon Sp1 depletion. ChIP assays were performed after Sp1 knockdown, and changes in binding were quantitated by real-time PCR. (C) Blockade of c-Myc inhibition of Tat-mediated HIV-1 transcription following Sp1 knockdown. As expected, Tat activation is less efficient after Sp1 knockdown, but a 20-fold increase of RNA expression over the basal level was still measured. However, c-Myc expression had no effect of Tat activation after Sp1 knockdown. PCR products for the GAPDH loading control and transfected Tat are shown. Error bars indicate standard errors of the means for three independent experiments. n.s, not significant. (D) Specific shRNA-mediated knockdown of c-Myc protein expression. (E) Similarly, occupancy of c-Myc and HDAC1 is reduced at the LTR initiator region upon c-Myc depletion. (F) shRNA in the J89 Jurkat T-cell line directed against c-Myc depletes c-Myc protein levels and results in the significant upregulation of HIV-1 LTR expression. (G) Knockdown of c-Myc in J89 cells results in the significant upregulation of HIV LTR expression.
To measure the effect of Sp1 knockdown on the c-Myc inhibition of HIV-1 activation, a cotransfection of c-Myc and Tat expression vectors was performed along with the active and control Sp1 shRNA vectors described above. Control Sp1 shRNA did not affect Tat activation or c-Myc inhibition of Tat activation. As expected, Tat activation was blunted by Sp1 shRNA, although RNA expression was still activated 20-fold above baseline by Tat. However, after Sp1 knockdown, c-Myc did not inhibit Tat activation (Fig. 6C). Tat expression was unchanged by RNA interference targeted at Sp1. These findings demonstrate that Sp1 is critical for both c-Myc binding to the HIV-1 LTR and the repression of Tat-dependent HIV-1 activation by HDAC1 recruitment mediated by c-Myc.
We then used an shRNA that specifically depleted c-Myc protein expression and tested if the knockdown of c-Myc would diminish HDAC1 recruitment to the HIV-1 promoter. Transfection of pSUPER-Myc827 specifically depleted c-Myc (10), whereas transfection of a vector encoding a scrambled sequence did not (Fig. 6D). c-Myc knockdown resulted in a significant decrease in the occupancy of c-Myc and HDAC1 at the LTR (Fig. 6E).
Consistent with these findings, the knockdown of c-Myc in the J89 Jurkat cell line (Fig. 6F), encoding an intact provirus expressing Tat, results in the upregulation of LTR-driven GFP expression (Fig. 6G). The extent of LTR induction is similar to that previously seen when HDAC1 recruitment by LSF/YY1 was inhibited (5).
DISCUSSION
While many factors may be involved in HIV-1 latency, the chromatin environment plays an important role in maintaining proviral quiescence (29). As HIV-1 integrates most frequently into genes that are transcribed in resting CD4+ T cells (13), chromatin at the proviral promoter may play an important role in maintaining quiescence in the minority of proviral genomes that enter latency. Although we initially sought new mechanisms that drive proviral quiescence, here, we discovered a novel additional mechanism that may contribute to the maintenance of proviral latency. In VPA-treated cells from latently HIV-1-infected patients, we found that the mRNA expression of c-Myc was significantly downregulated. Reduced protein expression was confirmed in primary resting CD4+ cells obtained from HIV-infected patients treated with VPA. Gene reporter analysis further confirmed c-Myc repression of HIV-1 expression.
c-Myc is associated with HDAC1 in HeLa-CAT-CD4 cells, and in concert with LTR upregulation induced by the HDAC inhibitor VPA, c-Myc binding to the HIV-1 promoter was disrupted. Both decreased binding of c-Myc to the HIV-1 promoter and decreased protein levels of c-Myc following VPA treatment may contribute to the downregulation of c-Myc occupancy at the HIV LTR. However, reduced protein levels of c-Myc were seen only after 48 h of VPA exposure, whereas occupancy was significantly decreased within 4 h. c-Myc occupancy might be diminished by increased histone acetyltransferase activity after VPA exposure or other direct or secondary consequences of histone deacetylase inhibition.
Several models are suggested for c-Myc repression of target genes. Among them, c-Myc appears to be recruited to the HIV-1 promoter by its interaction with Sp1. The core promoter region of the HIV-1 LTR contains three tandem Sp1 binding sites and two NF-κB elements located upstream of the TATA box of HIV-1. The insensitivity of the Sp-1 binding site mutant HIV-1 LTR to suppression by ectopically expressed c-Myc indicated that Sp-1 might mediate c-Myc binding to the HIV-1 LTR. This was supported by the specific shRNA-mediated inhibition of Sp-1 expression.
As YY1 can interact with c-Myc (43), it is possible that these factors interact in a supercomplex to recruit HDAC to the LTR. However, while this might be so, each factor appears to be limiting for HDAC recruitment, as the knockdown of c-Myc or p50 or small-molecule inhibition of LSF binding (and thereby YY1 recruitment) is capable of disrupting the binding of HDAC1 to the HIV-1 LTR and activating HIV-1 expression (5, 46).
Of course, the complexities of HIV LTR regulation require further study. For example, while c-Myc downregulation accompanies VPA-mediated LTR depression, c-Myc upregulation accompanies mitogen-mediated LTR activation. This paradox is not fully understood but is likely related to the markedly different global gene expression patterns induced by VPA and mitogen activation (Fig. 1B).
Similarly, c-Myc downregulation is suggested to be partially required for the lung Krüppel-like factor (LKLF)-mediated programming of quiescence in T-cell function, at least in a Jurkat cell line system. Again, however, several other gene pathways in addition to c-Myc mediate the effects of LKLF on Jurkat cells (4).
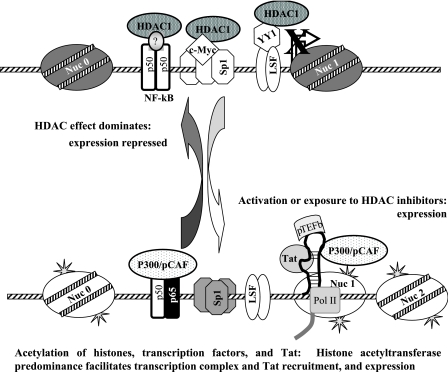
When HDAC inhibitors are administered to latently HIV-1-infected cells, transcriptional repression mediated by HDACs along with other corepressors is abrogated, leading to the expression HIV from an integrated genome (5, 14, 36, 45). Several mechanisms (Fig. 7) have been reported to recruit HDAC1 to the HIV LTR (5, 14, 16, 46). The relative importance of each of these mechanisms in vivo is unclear so far, although a specific molecular inhibition of HDAC recruitment via either LSF/YY1 was sufficient to disrupt latent HIV infection in the resting CD4+ T cells of HIV-infected patients (19).
FIG. 7.
Model of convergent recruitment of HDAC1 to maintain HIV-1 latency. Proviral latency is maintained, in part, by the action of several transcription factors that recruit HDAC1 to the HIV-1 LTR, preserving the deacetylation of histones H3 and H4. This state is disrupted by deacetylase inhibition, which simultaneously decreases the occupancy of repressors, acetylates histones, and facilitates the action of activator factors.
The apparent evolution within the HIV promoter of multiple mechanisms through which HDAC1 is recruited is striking and of high potential therapeutic significance. HDAC1 is recruited to the highly conserved initiator region of the HIV promoter by three distinct transcription factor complexes. These complexes are comprised of canonical factors that are ubiquitous in cell types infected by HIV-1 and required for both basal and activated promoter expression. It would seem less likely, therefore, that a subpopulation of latently infected cells would be unresponsive to this redundant mechanism of regulation or that the HIV LTR could function in the absence of these core factors that are capable of recruiting HDAC1. This suggests that potent inhibitors specific for HDAC1 in the HIV LTR might be broadly effective therapeutics to disrupt latent HIV infection and might avoid toxicities that could accompany the global inhibition of other members of the HDAC family. Therapeutics that do not directly inhibit HDAC but that prevent its occupancy or action at the HIV LTR may be considered as an alternate or an additional approach. For example, small-molecule reagents that inhibit c-Myc have entered early clinical testing in oncology (9).
Ultimately, just as the replicative stage of the HIV life cycle is attacked at multiple steps, the DNA stage of the HIV life cycle might be attacked by the administration of antilatency therapeutics in combination or in series to disrupt the maintenance of latency and induce the death of productively infected cells.
Acknowledgments
We are grateful to N. Archin and S. Choudhary for helpful discussions; M. Cheema, A. Hartmann-Duff, and D. Parker for technical assistance; L. Ngo for clinical coordination; and HIV-seropositive volunteers for sample donations.
This study was supported by NIH grants AI-45297 and AI-64074 to D.M.M., by UNC CFAR P30 AI50410, and by GCRC grant R00046.
Footnotes
Published ahead of print on 1 August 2007.
REFERENCES
- 1.Amundson S. A., Q. Zhan, L. Z. Penn, and A. J. Fornace, Jr. 1998. Myc suppresses induction of the growth arrest genes gadd34, gadd45, and gadd153 by DNA-damaging agents. Oncogene 17:2149-2154. [DOI] [PubMed] [Google Scholar]
- 2.Bali, P., P. George, P. Cohen, J. Tao, F. Guo, C. Sigua, A. Vishvanath, S. S. Annavarapu, W. Fiskus, L. Moscinski, P. Atadja, and K. Bhalla. 2004. Superior activity of the combination of histone deacetylase inhibitor LAQ824 and the FLT-3 kinase inhibitor PKC412 against human acute myelogenous leukemia cells with mutant FLT-3. Clin. Cancer Res. 10:4991-4997. [DOI] [PubMed] [Google Scholar]
- 3.Brenner, C., R. Deplus, C. Didelot, A. Loriot, E. Vire, C. De Smet, A. Gutierrez, D. Danovi, D. Bernard, T. Boon, P. G. Pelicci, B. Amati, T. Kouzarides, Y. de Launoit, L. Di Croce, and F. Fuks. 2005. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 24:336-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckley, A. F., C. T. Kuo, and J. M. Leiden. 2001. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc-dependent pathway. Nat. Immunol. 2:698-704. [DOI] [PubMed] [Google Scholar]
- 5.Coull, J. J., G. He, C. Melander, V. C. Rucker, P. B. Dervan, and D. M. Margolis. 2002. Targeted derepression of the human immunodeficiency virus type 1 long terminal repeat by pyrrole-imidazole polyamides. J. Virol. 76:12349-12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang, C. V., L. M. Resar, E. Emison, S. Kim, Q. Li, J. E. Prescott, D. Wonsey, and K. Zeller. 1999. Function of the c-Myc oncogenic transcription factor. Exp. Cell Res. 253:63-77. [DOI] [PubMed] [Google Scholar]
- 7.de la Fuente, C., F. Santiago, L. Deng, C. Eadie, I. Zilberman, K. Kehn, A. Maddukuri, S. Baylor, K. Wu, C. G. Lee, A. Pumfery, and F. Kashanchi. 2002. Gene expression profile of HIV-1 Tat expressing cells: a close interplay between proliferative and differentiation signals. BMC Biochem. 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Detich, N., V. Bovenzi, and M. Szyf. 2003. Valproate induces replication-independent active DNA demethylation. J. Biol. Chem. 278:27586-27592. [DOI] [PubMed] [Google Scholar]
- 9.Devi, G. R., T. M. Beer, C. L. Corless, V. Arora, D. L. Weller, and P. L. Iversen. 2005. In vivo bioavailability and pharmacokinetics of a c-MYC antisense phosphorodiamidate morpholino oligomer, AVI-4126, in solid tumors. Clin. Cancer Res. 11:3930-3938. [DOI] [PubMed] [Google Scholar]
- 10.Faiola, F., X. Liu, S. Lo, S. Pan, K. Zhang, E. Lymar, A. Farina, and E. Martinez. 2005. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol. Cell. Biol. 25:10220-10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gartel, A. L. 2006. A new mode of transcriptional repression by c-myc: methylation. Oncogene 25:1989-1990. [DOI] [PubMed] [Google Scholar]
- 12.Gartel, A. L., X. Ye, E. Goufman, P. Shianov, N. Hay, F. Najmabadi, and A. L. Tyner. 2001. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. USA 98:4510-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han, Y., K. Lassen, D. Monie, A. R. Sedaghat, S. Shimoji, X. Liu, T. C. Pierson, J. B. Margolick, R. F. Siliciano, and J. D. Siliciano. 2004. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78:6122-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, G., and D. M. Margolis. 2002. Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol. Cell. Biol. 22:2965-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Shoemaker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19:342-347. [DOI] [PubMed] [Google Scholar]
- 16.Imai, K., and T. Okamoto. 2006. Transcriptional repression of human immunodeficiency virus type 1 by AP-4. J. Biol. Chem. 281:12495-12505. [DOI] [PubMed] [Google Scholar]
- 17.Inghirami, G., F. Grignani, L. Sternas, L. Lombardi, D. M. Knowles, and R. Dalla-Favera. 1990. Down-regulation of LFA-1 adhesion receptors by C-myc oncogene in human B lymphoblastoid cells. Science 250:682-686. [DOI] [PubMed] [Google Scholar]
- 18.Izumi, H., C. Molander, L. Z. Penn, A. Ishisaki, K. Kohno, and K. Funa. 2001. Mechanism for the transcriptional repression by c-Myc on PDGF beta-receptor. J. Cell Sci. 114:1533-1544. [DOI] [PubMed] [Google Scholar]
- 19.Kashanchi, F., J. C. Melpolder, J. S. Epstein, and M. R. Sadaie. 1997. Rapid and sensitive detection of cell-associated HIV-1 in latently infected cell lines and in patient cells using sodium-n-butyrate induction and RT-PCR. J. Med. Virol. 52:179-189. [PubMed] [Google Scholar]
- 20.Kuefer, R., M. D. Hofer, V. Altug, C. Zorn, F. Genze, K. Kunzi-Rapp, R. E. Hautmann, and J. E. Gschwend. 2004. Sodium butyrate and tributyrin induce in vivo growth inhibition and apoptosis in human prostate cancer. Br. J. Cancer 90:535-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lassen, K., Y. Han, Y. Zhou, J. Siliciano, and R. F. Siliciano. 2004. The multifactorial nature of HIV-1 latency. Trends Mol. Med. 10:525-531. [DOI] [PubMed] [Google Scholar]
- 22.Laughlin, M. A., G. Y. Chang, J. W. Oakes, F. Gonzalez-Scarano, and R. J. Pomerantz. 1995. Sodium butyrate stimulation of HIV-1 gene expression: a novel mechanism of induction independent of NF-kappa B. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 9:332-339. [PubMed] [Google Scholar]
- 23.Lehrman, G., I. B. Hogue, S. Palmer, C. Jennings, C. A. Spina, A. Wiegand, A. L. Landay, R. W. Coombs, D. D. Richman, J. W. Mellors, J. M. Coffin, R. J. Bosch, and D. M. Margolis. 2005. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet 366:549-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonard, J., C. Parrott, A. J. Buckler-White, W. Turner, E. K. Ross, M. A. Martin, and A. B. Rabson. 1989. The NF-κB binding sites in the human immunodeficiency virus type 1 long terminal repeat are not required for virus infectivity. J. Virol. 63:4919-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, X. N., Q. Shu, J. M. Su, L. Perlaky, S. M. Blaney, and C. C. Lau. 2005. Valproic acid induces growth arrest, apoptosis, and senescence in medulloblastomas by increasing histone hyperacetylation and regulating expression of p21Cip1, CDK4, and CMYC. Mol. Cancer Ther. 4:1912-1922. [DOI] [PubMed] [Google Scholar]
- 26.Loscher, W. 2002. Basic pharmacology of valproate: a review after 35 years of clinical use for the treatment of epilepsy. CNS Drugs 16:669-694. [DOI] [PubMed] [Google Scholar]
- 27.Lusic, M., A. Marcello, A. Cereseto, and M. Giacca. 2003. Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J. 22:6550-6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marban, C., L. Redel, S. Suzanne, C. Van Lint, D. Lecestre, S. Chasserot-Golaz, M. Leid, D. Aunis, E. Schaeffer, and O. Rohr. 2005. COUP-TF interacting protein 2 represses the initial phase of HIV-1 gene transcription in human microglial cells. Nucleic Acids Res. 33:2318-2331. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Marcello, A. 2006. Latency: the hidden HIV-1 challenge. Retrovirology 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchion, D. C., E. Bicaku, A. I. Daud, D. M. Sullivan, and P. N. Munster. 2005. Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Cancer Res. 65:3815-3822. [DOI] [PubMed] [Google Scholar]
- 31.Margolis, D. M., A. B. Rabson, S. E. Straus, and J. M. Ostrove. 1992. Transactivation of the HIV-1 LTR by HSV-1 immediate-early genes. Virology 186:788-791. [DOI] [PubMed] [Google Scholar]
- 32.Marhin, W. W., S. Chen, L. M. Facchini, A. J. Fornace, Jr., and L. Z. Penn. 1997. Myc represses the growth arrest gene gadd45. Oncogene 14:2825-2834. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell, C. P., Y. Chen, M. Kundakovic, E. Costa, and D. R. Grayson. 2005. Histone deacetylase inhibitors decrease reelin promoter methylation in vitro. J. Neurochem. 93:483-492. [DOI] [PubMed] [Google Scholar]
- 34.Pal, S., R. Yun, A. Datta, L. Lacomis, H. Erdjument-Bromage, J. Kumar, P. Tempst, and S. Sif. 2003. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 23:7475-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parrott, C., T. Seidner, E. Duh, J. Leonard, T. S. Theodore, A. Buckler-White, M. A. Martin, and A. B. Rabson. 1991. Variable role of the long terminal repeat Sp1-binding sites in human immunodeficiency virus replication in T lymphocytes. J. Virol. 65:1414-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quivy, V., E. Adam, Y. Collette, D. Demonte, A. Chariot, C. Vanhulle, B. Berkhout, R. Castellano, Y. de Launoit, A. Burny, J. Piette, V. Bours, and C. Van Lint. 2002. Synergistic activation of human immunodeficiency virus type 1 promoter activity by NF-κB and inhibitors of deacetylases: potential perspectives for the development of therapeutic strategies. J. Virol. 76:11091-11103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quivy, V., and C. Van Lint. 2002. Diversity of acetylation targets and roles in transcriptional regulation: the human immunodeficiency virus type 1 promoter as a model system. Biochem. Pharmacol. 64:925-934. [DOI] [PubMed] [Google Scholar]
- 38.Salghetti, S. E., S. Y. Kim, and W. P. Tansey. 1999. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 18:717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satou, A., T. Taira, S. M. Iguchi-Ariga, and H. Ariga. 2001. A novel transrepression pathway of c-Myc. Recruitment of a transcriptional corepressor complex to c-Myc by MM-1, a c-Myc-binding protein. J. Biol. Chem. 276:46562-46567. [DOI] [PubMed] [Google Scholar]
- 40.Sheridan, P. L., T. P. Mayall, E. Verdin, and K. A. Jones. 1997. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev. 11:3327-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siliciano, J. D., J. Lai, M. Callender, E. Pitt, H. Zhang, J. B. Margolick, J. E. Gallant, J. Cofrancesco, Jr., R. D. Moore, S. J. Gange, and R. F. Siliciano. 2007. Stability of the latent reservoir for HIV-1 in patients receiving valproic acid. J. Infect. Dis. 195:833-836. [DOI] [PubMed] [Google Scholar]
- 42.Staller, P., K. Peukert, A. Kiermaier, J. Seoane, J. Lukas, H. Karsunky, T. Moroy, J. Bartek, J. Massague, F. Hanel, and M. Eilers. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 3:392-399. [DOI] [PubMed] [Google Scholar]
- 43.Stojanova, A., C. Caro, R. J. Jarjour, S. K. Oster, L. Z. Penn, and R. J. Germinario. 2004. Repression of the human immunodeficiency virus type-1 long terminal repeat by the c-Myc oncoprotein. J. Cell. Biochem. 92:400-413. [DOI] [PubMed] [Google Scholar]
- 44.Van Lint, C. 2000. Role of chromatin in HIV-1 transcriptional regulation. Adv. Pharmacol. 48:121-160. [DOI] [PubMed] [Google Scholar]
- 45.Van Lint, C., S. Emiliani, and E. Verdin. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 5:245-253. [PMC free article] [PubMed] [Google Scholar]
- 46.Williams, S. A., L. F. Chen, H. Kwon, C. M. Ruiz-Jarabo, E. Verdin, and W. C. Greene. 2006. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 25:139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, K. J., A. Polack, and R. Dalla-Favera. 1999. Coordinated regulation of iron-controlling genes, H-ferritin and IRP2, by c-MYC. Science 283:676-679. [DOI] [PubMed] [Google Scholar]
- 48.Xu, Y., S. Voelter-Mahlknecht, and U. Mahlknecht. 2005. The histone deacetylase inhibitor suberoylanilide hydroxamic acid down-regulates expression levels of Bcr-abl, c-Myc and HDAC3 in chronic myeloid leukemia cell lines. Int. J. Mol. Med. 15:169-172. [PubMed] [Google Scholar]
- 49.Ylisastigui, L., N. M. Archin, G. Lehrman, R. J. Bosch, and D. M. Margolis. 2004. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 18:101-108. [DOI] [PubMed] [Google Scholar]
- 50.Ylisastigui, L., J. J. Coull, V. C. Rucker, C. Melander, R. J. Bosch, S. J. Brodie, L. Corey, D. L. Sodora, P. B. Dervan, and D. M. Margolis. 2004. Polyamides reveal a role for repression in latency within resting T cells of HIV-infected donors. J. Infect. Dis. 190:1429-1437. [DOI] [PubMed] [Google Scholar]