Abstract
The biogenesis of multivesicular bodies (MVBs) is topologically equivalent to virion budding. Hence, a number of viruses exploit the MVB pathway to build their envelope and exit from the cell. By expression of dominant negative forms of Vps4 and Vps24, two components of the MVB pathway, we observed an impairment in infectious herpes simplex virus (HSV) assembly/egress, in agreement with a recent report showing the involvement in HSV envelopment of Vps4, the MVB-specific ATPase (C. M. Crump, C. Yates, and T. Minson, J. Virol. 81:7380-7387). Furthermore, HSV infection resulted in morphological changes to MVBs. Glycoprotein B (gB), one of the most highly conserved glycoproteins across the Herpesviridae family, was sorted to MVB membranes. In cells expressing the dominant negative form of Vps4, the site of intracellular gB accumulation was altered; part of gB accumulated as an endoglycosidase H-sensitive immature form at a calreticulin-positive compartment, indicating that gB traffic was dependent on a functional MVB pathway. gB was ubiquitinated in both infected and transfected cells. Ubiquitination was in part dependent on ubiquitin lysine 63, a signal for cargo sorting to MVBs. Partial deletion of the gB cytoplasmic tail resulted in a dramatic reduction of ubiquitination, as well as of progeny virus assembly and release to the extracellular compartment. Thus, HSV envelopment/egress and gB intracellular trafficking are dependent on functional MVB biogenesis. Our data support the view that the sorting of gB to MVB membranes may represent a critical step in HSV envelopment and egress and that modified MVB membranes constitute a platform for HSV cytoplasmic envelopment or that MVB components are recruited to the site(s) of envelopment.
Multivesicular bodies (MVBs) constitute a central station in the endocytic-lysosomal pathway. They are responsible for the biosynthetic delivery of hydrolases to lysosomes as well as for the sorting of a number of cell surface receptors, destined to degradation in the lysosome (69). At the ultrastructural level, early endosomes appear predominantly as tubulovesicular structures, whereas late endosomes, which are capable of fusing with lysosomes, exhibit a multivesicular aspect and, for this reason, are named MVBs. The transition between these two endosomal compartments occurs by involution of the limiting membrane to form intraluminal vesicles. When the MVBs fuse with lysosomes, the intraluminal vesicles and their contents are degraded. Both the lipid and protein compositions of the endosome change along the pathway to lysosomes (75). A major signal for sorting of cargoes along the MVB pathway is ubiquitination (41). In Saccharomyces cerevisiae, MVB biogenesis requires a total of 17 yeast class E Vps proteins (47). Vps23 and two other class E Vps proteins form a cytosolic complex termed ESCRT-I (endosomal sorting complex required for transport I), which recognizes ubiquitinated endosomal cargo (41). ESCRT-I activates another soluble class E Vps complex called ESCRT-II, which in turn is required to initiate the assembly of the ESCRT-III complex on endosomal membranes (3). ESCRT-III, the core of the apparatus that drives membrane curvature and MVB vesicle formation, is formed by four structurally related class E Vps proteins that exhibit homology to human chromatin modifying proteins (CHMP) (3). Finally, to enable the recycling of MVB machinery, ESCRT-III recruits the AAA-type ATPase Vps4, a class E Vps protein that disassembles and thereby recycles the ESCRT machinery (3). Overexpression of ATPase-defective Vps4 proteins induces the formation of enlarged endosomes and dysfunctional MVBs that are defective in the sorting and recycling of endocytosed substrates (“class E” phenotypes) (3). The Vps4 mutants also prevent normal ESCRT protein trafficking, because these proteins are trapped on the surfaces of the aberrant MVBs (3). On the other hand, dominant negative forms of the ESCRT-III component Vps24/CHMP3, essential for vesicle invagination (47), like Vps24 fused to a bulky tag such as red fluorescent protein, induce class E-like phenotypes (81).
Numerous enveloped RNA viruses, including retroviruses (81, 29), rhabdoviruses (36, 45), filoviruses (35, 37, 44), arenaviruses (83), and possibly ortho- and paramyxoviruses (82), hijack the endocytic pathway in particular membranes of MVBs as platforms for the assembly of the virion envelope and for virus egress. Essentially, two mechanisms for the recruiting of virion proteins to the MVB pathway were highlighted. One is based on the interaction of ESCRT components with structural proteins carrying specific amino acidic motifs named late domains (L-domains), which serve as binding sites for MVB components, escort the virion proteins to the MVB membranes, and enable envelope formation and virus budding (18, 58). The other mechanism involves the ubiquitination of virion components. Different riboviruses hijack different MVB proteins and thus enter the MVB assembly pathway at different steps (18, 35, 58). The best known example is that seen with retroviruses. Gag proteins typically carry L-domains with one of the tetrapeptide sequences PTAP, PPxY, and YPDL (58). In human immunodeficiency virus, the L-domain located in the p6 protein is able to recruit TSG101, the mammalian homolog of the yeast ESCRT-I component Vps23 (29, 49). The simple recruitment of the ESCRT machinery by p6 is sufficient to induce membrane curvature and fission and to drive the formation and release of virus-like particles, even in the absence of viral components other than Gag (29). Viral L-domains are critical also for exploiting the MVB-linked ubiquitination machinery. Evidence is particularly compelling in the case of retroviruses, which incorporate ubiquitin molecules, some of which are individually linked to Gag (50, 60-62, 72, 80). The recruitment of ubiquitin ligase activity by L-domains correlates with virus release (7, 38, 48, 79, 85). In addition, TSG101 is an ubiquitin-binding protein (70, 71). A proteasome inhibitor that decreases the levels of free ubiquitin in the cytoplasm blocks the release of certain retroviruses (61-64, 74, 79).
Relative to riboviruses, much less is known about how DNA-enveloped viruses, including herpesviruses, acquire their envelope. Herpesviridae are large DNA-containing enveloped viruses that share a number of properties, including basic mechanisms of virus entry, progeny virus assembly, and exit from the cell. Herpesviruses enter the cell by direct fusion with the plasma membrane or endocytic membranes (13, 14). Progeny nucleocapsids are assembled in the nucleus and exit this compartment by budding at the inner nuclear membrane (51-54). The subsequent steps in virus assembly and release remain controversial (12, 14, 54). The currently most credited view envisions that virions are de-enveloped at the outer nuclear membrane and that the de-enveloped capsids released into the cytoplasm undergo a second envelopment (51-54). The site of cytoplasmic envelopment has not been completely characterized. The Golgi apparatus, trans-Golgi network (TGN) membranes, and occasionally endosomes are usually indicated as platforms for envelopment (1, 6, 14, 34, 46, 54).
Key unanswered questions on herpesvirus envelopment/egress center on (i) how the curvature of the membrane is destined to become the envelope attained, (ii) which membranes serve as platforms for secondary envelopment, and (iii) which molecular interactions between viral and cellular proteins drive virion envelopment/egress. The aims of this study were (i) to investigate whether components of the MVB biogenesis pathway play a role in herpes simplex virus (HSV) envelopment/egress and (ii) to determine the molecular mechanisms that the virus puts in place in order to exploit the MVB membranes as platforms for building its envelope and to achieve virion egress. While this paper was in preparation, it was reported that a functional Vps4, the ATPase that releases ESCRT components from assembled MVBs, is required for HSV envelopment (17). We report that (i) Vps24/CHMP3, a component of the ESCRT-III complex essential for vesicle invagination (69), and Vps4 are critical for HSV envelopment/egress; (ii) the MVB compartment is modified following HSV infection; (iii) glycoprotein B (gB) accumulates at MVB membranes, and a functional MVB biogenesis is required for glycoprotein trafficking and maturation; (iv) gB is ubiquitinated particularly by the lysine 63 residue of ubiquitin; and (v) partial deletion of the gB cytoplasmic tail results in a dramatic reduction of ubiquitination and progeny virus envelopment and egress to the extracellular compartment. The results indicate that MVB membranes serve as platforms for HSV envelopment/egress and that sorting of gB to MVB membranes may represent a critical step in the envelopment/egress process.
MATERIALS AND METHODS
Mammalian expression plasmids.
The pBJ5-Vps4E228Q plasmid encodes the human Vps4-A protein with a C-terminal FLAG epitope containing an E228Q mutation. The pBJ5-HA-Ub plasmid expresses a hemagglutinin (HA)-tagged version of wild-type (wt) ubiquitin, while the pBJ5-HA-UbK48R and pBJ5-HA-UbK63R constructs encode HA-tagged ubiquitin characterized by the replacement of lysine 48 and lysine 63, respectively, with an arginine. The pDsRed-Vps24 construct contains the CHMP3/Vps24-encoding sequence inserted into the pDsRed2-N1 vector (Clontech). All the above-named constructs have previously been described (80, 81). The pgBwt-MTS plasmid encodes gB of HSV type 1 (HSV-1) strain F (2). The gBΔ867-MTS plasmid expresses a truncated form of gB in which the 37 C-terminal amino acids of the C tail are placed downstream of a stop codon (2). The gH-MTS and gL-MTS plasmids encode gH and gL of HSV-1 strain F, respectively (2).
Cell lines and viral strains.
African green monkey kidney cells (Vero cells, ATCC number CCL-81; COS-7 cells, ATCC number CRL-1651) and human kidney cells (293T cells, ATCC number CRL-11268) were grown in Dulbecco's modified Eagle's medium with the addition of 10% heat-inactivated fetal calf serum (complete medium).
The HSV type 1 (HSV-1) strain F was kindly provided by B. Roizman (University of Chicago, Chicago, IL). Recombinant HSV-1 R8102 was constructed by N. Markowitz and B. Roizman (University of Chicago) by inserting a cassette containing a lacZ gene under the control of the ICP-27 promoter (325-bp BamHI-HinfI fragment from the left end of the HSV-1 BamHI B fragment) at the UL3-UL4 boundary (5). Viruses were grown and subjected to titer determination by plaque assay of Vero cells, as previously described (11). 293T cells or COS-7 cells were infected with HSV-1 at the appropriate multiplicities of infection (MOI). The viral inoculum was removed after 1.5 h of adsorption, virus absorbed to cells was inactivated with pH 3 citrate buffer wash, and the cells were overlaid with Dulbecco's modified Eagle's medium containing 2% serum. At different time points, medium and cells were harvested separately and the virus was titrated in Vero cells (11). The KΔt ΔgB HSV was previously described (9).
Entry assay.
For infectivity determinations, transfected 293T cells (1 × 105) were plated in 24-well tissue culture dishes and infected with HSV-1 R8102 at the MOI of 3 PFU/cell. After 6 h of incubation, infected cells were visualized using the β-galactosidase (β-Gal) substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; GIBCO Laboratories). Briefly, washed cells were fixed (phosphate-buffered saline [PBS] containing 2% formaldehyde and 0.2% glutaraldehyde), permeabilized (2 mM MgCl2 containing 0.01% deoxycholate and 0.02% Nonidet P-40 [NP-40]), and incubated with buffered X-Gal (0.5 mg/ml). Alternatively, infected cells were solubilized in PBS containing 0.5% NP-40 and the β-Gal substrate ortho-nitrophenyl-β-d-galactopyranoside (3 mg/ml) and the reaction was quantified by spectrometry (2).
HSV-1 glycoprotein immunoprecipitation and Western blot analysis.
The cells were lysed in PBS* (PBS containing 1% NP-40 and 1% deoxycholate) containing protease inhibitors (0.1 mM Nα-p-tosyl-l-lysine chloromethyl ketone-0.1 mM tosylsulfonyl phenylalanyl chloromethyl ketone). Immunoprecipitations were carried out with the appropriate antibodies from lysates of infected or transfected cells, as previously described (32). The immunocomplexes were harvested with protein A-Sepharose (Sigma). Proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were transferred to a Hybond ECL nitrocellulose membrane (GE Healthcare) and detected with appropriate antibodies and enhanced-chemiluminescence reagents (GE Healthcare). Anti-gB antibody H1817 (Rumbaugh-Goodwin Institute for Cancer Research, Plantation, FL) or anti-gH 53S (31) was used for the immunoprecipitation, while anti-HA antibody HA.11 (Covance), anti-gH antibody H12 (66), antiubiquitin antibody (Sigma), anti-HSV-1 polyclonal antiserum (DAKO), enhanced-chemiluminescence anti-mouse immunoglobulin G-horseradish peroxidase (GE Healthcare), and enhanced-chemiluminescence anti-rabbit immunoglobulin G-horseradish peroxidase (GE Healthcare) were used for Western blotting analysis. The membrane used for ubiquitin immunoblot analysis was autoclaved in deionized water for 30 min as described previously to expose latent antigenic sites on ubiquitin prior to the incubation with the antibody (55, 56).
Nucleic acid extraction, real-time PCR, and reverse transcription-PCR (RT-PCR) assays.
DNA extraction was performed as follows. The cells were incubated in 250 μl lysis buffer (0.1% Triton X-100, 0.1% SDS, 20 mg/ml proteinase K, 10 mM Tris-HCl-1 mM EDTA) at 56°C for 1 h and boiled for 10 min. Quantitative PCR testing was performed on 5-μl aliquots. Briefly, the extracted DNA was assayed with a sequence detector system (ABI PRISM 7700) in 25 μl of PCR mixture containing 12.5 μl TaqMan universal master mix, 15 pmol of each primer, and 10 pmol of the probe. The primers amplified a fragment of the HSV UL30 gene (forward primer, 5′-ACATCATCAACTTCGACTGG-3′; reverse primer, 5′-CTCAGGTCCTTCTTCTTGTCC-3′). The fluorogenic probe sequence was 5′-FAM-ATGGTGAACATCGACATGTACGG-TAMRA-3′, where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine. Thermal cycling conditions were one cycle of 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. A standard curve was made by six serial dilutions (from 5 × 106 to 50 copies) of a control plasmid containing the region amplified by the primers. The detection threshold was 10 genomic copies per reaction. The HSV genomic copy number of the samples was calculated automatically with 7700 ABI PRISM SDS software and then expressed as numbers of viral DNA copies per cell. The number of cells present in the PCR mix was determined by real-time quantitative-PCR amplification of a β-globin sequence gene (Applied Biosystems), as recommended by the manufacturer.
[35S]methionine labeling.
293T cells transfected with the appropriate construct were infected with HSV-1(F) at the MOI of 10 PFU/cell. At 8 h postinfection, the cells were washed and incubated in methionine-free medium for 1 h, followed by [35S]methionine labeling for 15 h. Cells were harvested and lysed in radioimmunoprecipitation assay buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% NP-40, 0.5% sodium deoxycholate, 0.05% SDS). Protein extracts were analyzed by SDS-PAGE and autoradiography.
Immunofluorescence analysis.
293T cells (1.5 × 105) were grown on glass coverslips. When required, the cells were either infected with HSV-1 or transfected with the appropriate constructs by using the Lipofectamine 2000 reagent (Invitrogen). The cells were then fixed at different time points with either 4% paraformaldehyde for 10 min at room temperature and then with 0.1% Triton X-100 in PBS or acetone-methanol (1:1, vol/vol) for 5 min at −20°C. Samples were incubated with appropriate primary and secondary antibodies diluted with bovine serum albumin (1%, wt/vol) in PBS. The primary antibodies used were monoclonal antibody (MAb) to gB (Virusys), MAb 53S to gH (31), rabbit polyclonal antibody (PAb) to lysosome-associated membrane protein 1 (LAMP-1; H-228, sc-5570; Santa Cruz Biotechnologies Inc.), and rabbit PAb to calreticulin (ab4; Abcam). Fluorescein isothiocyanate-conjugated anti-mouse or anti-rabbit immunoglobulin G antibody (Sigma) or Alexa Fluor 568 goat anti-mouse or anti-rabbit immunoglobulin G antibody (Invitrogen) was used as a secondary antibody. The cells were observed with a confocal microscope (Leica).
endo H treatment.
For the endoglycosidase H (endo H) treatment, 293T cells cotransfected with the pgBwt-MTS and pBJ5-Vps4E228Q plasmids or the pBJ5 construct were lysed in PBS* and the cell lysates were subjected to gB immunoprecipitation, as described above. The immunoprecipitated proteins that bound to the Sepharose beads were resuspended in 30 μl of 1× denaturing buffer (0.5% SDS and 0.04 M dithiothreitol; New England Biolabs), and the reaction mixture was heated at 100°C for 10 min. Next, 4 μl of 10× G5 reaction buffer (0.5 M sodium citrate [pH 5.5]; New England Biolabs), 3 μl of endo H (500,000 U/ml; New England Biolabs), and water were added up to 40 μl. The reaction mixture was incubated at 37°C for 1 h. Twenty-five microliters of each sample was analyzed by SDS-PAGE, followed by Western blotting analysis.
Infectivity complementation assay.
The infectivity complementation assay was performed as previously described (16). Briefly, COS-7 cells were transfected with plasmids encoding wt gB or gBΔ867 and epidermal growth factor receptor (EGFR) as a negative control. Four hours later, cells were infected with HSV-KΔt (3 PFU/cell). The viral inoculum was removed after 1.5 h of adsorption, virus adsorbed to cells was inactivated with pH 3 citrate buffer wash, and cells were overlaid with Dulbecco's modified Eagle's medium containing 1% serum. Twenty-four hours later, medium and cells were harvested separately and progeny virus was titrated in complementing gB-expressing D6 cells (9).
RESULTS
A functional MVB pathway is required at a late step of HSV-1 replication.
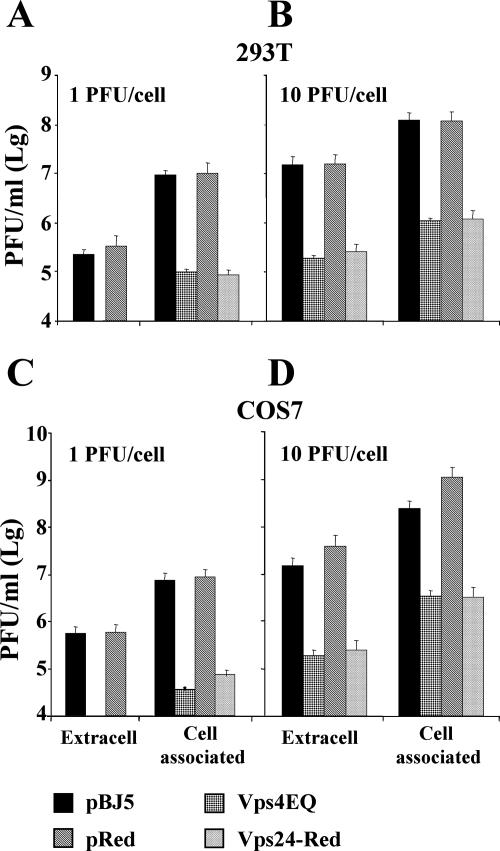
The most specific way to interfere with MVB pathway biogenesis is to use dominant negative versions of essential components of the pathway. Therefore, in order to determine whether the MVB compartment plays a role in HSV-1 replication, 293T and COS-7 cells were transfected with a construct expressing a dominant negative form of Vps4 (Vps4E228Q) or a construct encoding the dominant negative version of Vps24/CHMP3 (Vps24-Red). Both dominant negative mutants have been extensively characterized and are known to block the biogenesis of MVBs (81). Twelve hours after transfection, cells were infected with HSV-1(F), at two MOI (1 or 10 PFU/cell). The yield of cell-associated and extracellular virus at 36 h after infection was determined by plaque assay in Vero cells. The results showed a significant reduction in the production of cell-associated as well as released virus in the presence of either one of the two dominant negative proteins, irrespectively of the MOI used (Fig. 1A and B). The decrease was observed in both cell lines, irrespective of the fact that virus production was higher in COS-7 than in 293T cells (Fig. 1, compare panels A, B to C, and D).
FIG. 1.
Effect of Vps4E228Q and Vps24-Red on the yield of extracellular and cell-associated HSV-1. 293T cells (A, B) and COS-7 cells (C, D) were transfected either with an empty vector (pBJ5 or pDsRed2-N1) or with constructs expressing either Vps4E228Q or Vps24-Red. Twelve hours after transfection, the cells were infected with HSV-1 at 1 PFU/cell (A, C) or 10 PFU/cell (B, D). At 36 h postinfection, the extracellular (Extracell) and cell-associated virus were titrated by plaque assay in Vero cells.
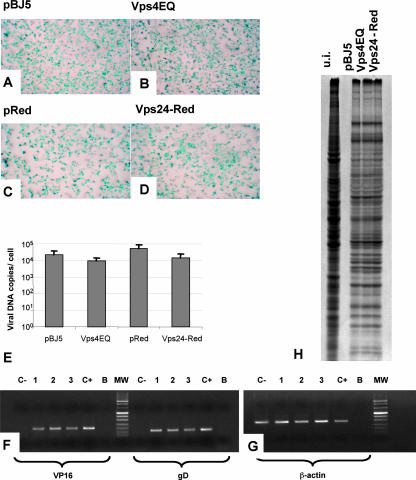
Inasmuch as expression of the dominant negative versions of Vps4 or Vps24 might affect receptor distribution at the cell surface and thus decrease the amount of virus taken into cells, we next determined whether the expression of Vps4E228Q or Vps24-Red reduced virus entry. 293T cells expressing either Vps24-Red or Vps4E228Q were infected with the HSV-1 recombinant R8102 (5), which carries the β-Gal gene under the immediate early ICP27 promoter (3 PFU/cell). Numerous studies have shown that the extent of β-Gal expression is a direct measure of the extent of virus entry into the cells (16). As reported in Fig. 2A to D, no significant difference in numbers of infected cells was observed between cells expressing the dominant negative version of the two proteins and mock-transfected cells.
FIG. 2.
Vps4E228Q (Vps4EQ) or Vps24-Red does not affect the early steps of HSV-1 replication. (A to D) 293T cells were transfected with plasmids expressing the dominant negative forms of Vps4 (B) or Vps24 (D) cellular protein or their respective empty vector, pBJ5 (A) or pDsRed2-N1 (pReD) (C). Twelve hours after transfection, the cells were infected with the R8102 recombinant HSV-1, carrying the lacZ gene under the control of the ICP-27 immediate promoter (3 PFU/cell) (5). Six hours later, the infected cells were stained with X-Gal. The panels illustrate light microscope images (magnification, ×1.5). (E) Quantification of HSV-1 genomic copies by real-time PCR assay. 293T cells were transfected with the constructs expressing either Vps4E228Q or Vps24-Red or the corresponding empty vectors. Twelve hours posttransfection, the cells were infected with HSV-1 (10 PFU/cell). Twenty-four hours later, the amount of viral DNA was evaluated by real-time PCR assay. Results are expressed as numbers of viral DNA copies per cell. (F and G) Total RNA was extracted from 293T cells transfected with the indicated plasmids. The RT-PCR assay was performed with primers specific for VP16 and gD (F) or the β-actin genes (G). Lanes: 1, 293T cells transfected with pBJ5 and infected with HSV-1; 2, 293T cells transfected with pBJ5-Vps4E228Q and infected with HSV-1; 3, 293T cells transfected with pDsRed-Vps24 and infected with HSV-1; C+, Vero cells infected with HSV-1; C−, uninfected 293T cells; B, no-template control; MW, molecular weight markers. (H) 293T cells transfected with the indicated plasmids were infected with HSV-1 strain F, labeled with [35S]methionine, and harvested 24 h after infection. Radiolabeled proteins were detected by autoradiography following SDS-PAGE. u.i., uninfected 293T cells.
To better define the step in HSV replication affected by the expression of the dominant negative versions of Vps4 and Vps24, we next determined whether the expression of the dominant negative mutants affected viral DNA replication or late gene transcription. To this end, the amount of viral DNA present in transfected infected cells 24 h after HSV-1(F) infection (10 PFU/cell) was measured by real-time PCR, using primers that anneal to the HSV-1 DNA polymerase gene. The viral DNA copies differed very slightly between cells expressing either Vps4E228Q or Vps24-Red and the mock-transfected controls (Fig. 2E). Transcription of the late gene was determined by RT-PCR, by employing oligonucleotides annealing to either the VP16- or the glycoprotein D-encoding sequence. The results show no significant reduction in late gene expression (Fig. 2F to G) in cells expressing the dominant negative versions of Vps4 and Vps24. According to this finding, no impairment in viral protein synthesis, measured as [35S]methionine incorporation, was observed (Fig. 2H). This series of results indicates that the decrease in the amount of progeny HSV assembled and released into the extracellular compartment in the presence of dominant negative versions of Vps4 or Vps24 was not due to a defect in virus entry consequent, for example, upon the unavailability or mislocalization of viral receptors, or to a major defect in viral DNA replication and transcription or in viral mRNA translation. The results further show that the decrease in virus release was MOI and cell type independent. Overall, the results confirm and extend previous findings on the role of the Vps4 ATPase in virus envelopment and release (17) and imply that not only the activity of Vps4 but more generally a functional MVB pathway is required for a step in virus replication that takes place after late gene expression and prior to the release of progeny virus into the extracellular medium. This step has been defined by electron microscopy to be at the stage of cytoplasmic envelopment (17).
The morphology of the MVB compartment is modified in HSV-1-infected cells.
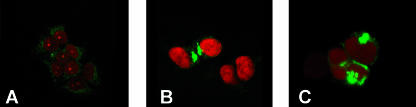
In HSV-1 infected cells, various compartments are modified in order to render the cell a suitable microenvironment for virus replication, envelopment, and release (1, 14, 34). We searched for modifications of the late endosomes or MVB compartment by immunofluorescence with an antibody directed to LAMP-1, a marker of this organelle (27). Confocal microscopy showed that, in uninfected cells, LAMP-1 was dispersed throughout the cytoplasm, as expected. By contrast, in infected cells (12 h and 24 h postinfection), LAMP-1 staining was concentrated in a perinuclear region and appeared to be overall increased, possibly reflecting an augmentation of the compartment itself (Fig. 3B and C). The morphological changes are consistent with an involvement of the MVB pathway in HSV replication.
FIG. 3.
Effect of HSV infection on the morphology of the MVB compartment. 293T cells were infected with HSV-1 and analyzed at 12 or 24 h after infection by immunofluorescence with PAb to LAMP-1. The cell nuclei were stained with propidium iodide. Cells were observed with a Leica confocal microscope at a ×63 magnification objective. (A) Uninfected 293T cells. (B) 293T cells 12 h after HSV-1 infection. (C) 293T cells 24 h after HSV-1 infection.
gB colocalizes with the MVB membranes, and its maturation and intracellular trafficking are dependent on a functional MVB biogenesis process.
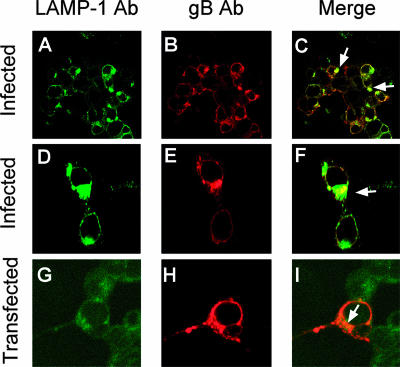
Of the several glycoproteins encoded by HSV, gB, one of the four essential glycoproteins, is known to possess endocytosis motifs and to undergo recycling from the plasma membrane to an endocytic compartment that has not been clearly identified (2). This property is conserved in most herpesviruses (20, 24, 40, 73). We reasoned that gB may be sorted to the MVB compartment. To test this hypothesis, we analyzed by immunofluorescence whether gB colocalizes with the LAMP-1 MVB marker. In infected cells, gB colocalized in part with the MVB membranes, which therefore represent a site of gB accumulation (Fig. 4A to F). Similar results were obtained with 293T cells transfected with a construct expressing wt gB (pgBwt-MTS) (Fig. 4G to I). Thus, the colocalization of gB with MVB membranes is an intrinsic property of gB.
FIG. 4.
gB accumulates in part at MVB membranes in HSV-infected and -transfected cells. (A to F) 293T cells were infected with HSV-1 (10 PFU/cell). Twenty-four hours later, the cells were stained with MAb to gB (Virusys) and PAb to LAMP-1 and analyzed by confocal microscopy at a ×63 magnification objective. (G to I) 293T cells were transfected with a construct expressing gB (pgBwt-MTS). Fourty-eight hours later, the cells were stained with MAb to gB (Virusys) and a PAb to LAMP-1 and analyzed by confocal microscopy. The arrows point to colocalization spots. (A to C) Low magnification; (D to F) high magnification.
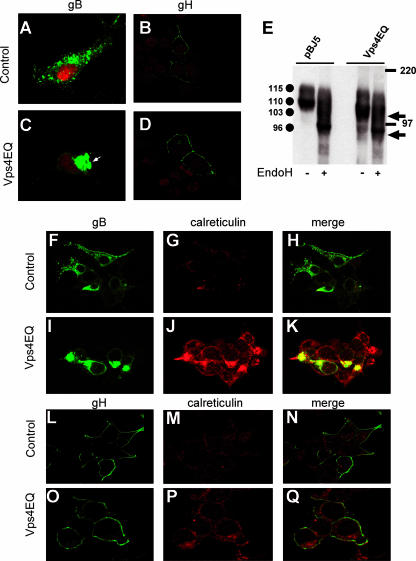
As a further evidence, we asked whether gB localization was altered in cells in which MVB biogenesis was affected by the expression of the dominant negative versions of Vps4. 293T cells were transfected with pgBwt-MTS and pBJ5-Vps4E228Q or the corresponding empty vector pBJ5. Confocal microscopy showed that in the pBJ5-Vps4E228Q-transfected cells, gB localized to a single perinuclear region (Fig. 5B). This localization contrasted with gB's distribution at discrete vesicles observed in pBJ5-transfected cells, which exemplifies the typical distribution pattern of this glycoprotein in both infected and transfected cells (Fig. 5A). The localization at MVBs was a typical property of gB not observed with gH, another essential glycoprotein for HSV and a partner of gB in virus entry (13). In infected cells, gH accumulates preferentially at the plasma membrane (32). As seen in Fig. 5C and D, gH distribution was not affected by the expression of a dominant negative version of Vps4. To better define that the site of gB accumulation depends on correct MVB biogenesis, we analyzed the electrophoretic mobility of the gB accumulated in cells expressing Vps4E228Q. Figure 5E shows that in pBJ5-transfected cells, gB was present as two bands with apparent molecular masses of 115 and 110 kDa, in agreement with previous data (2). In Vps4E228Q-transfected cells, the predominant form was not the 110 Mr form but a 103 Mr form, absent from pBJ5-transfected cells. This form of gB represented an immature form of the glycoprotein, as assessed by sensitivity to endo H digestion (Fig. 5E). In agreement with the accumulation of an immature form of gB, immunofluorescence analysis highlighted that in 293T cells expressing Vps4E228Q, gB localized at a site that coincides in part with the distribution of calreticulin (Fig. 5H). Cumulatively, the results indicate a mislocalization of gB and a specific defect in glycoprotein maturation in cells in which MVB biogenesis was blocked. Indeed, the modifications to glycoprotein maturation, trafficking, and distributions were observed specifically with gB and not with gH (Fig. 5D and L to Q).
FIG. 5.
Effect of blocking MVB biogenesis on gB and gH localization and maturation. 293T cells were cotransfected with a construct expressing Vps4E228Q (Vps4EQ) or the empty vector pBJ5, along with the pgBwt-MTS plasmid, expressing wt gB (A and C), or gH-MTS-gL-MTS plasmids encoding gH/gL (B and D). At 48 h after transfection, the cells were stained with MAb H1817 to gB (A and C) or MAb 53S to gH (B, D) and analyzed by confocal microscopy. Nuclei were stained with propidium iodide. (E) 293T cells were cotransfected with a construct expressing Vps4E228Q or the empty vector pBJ5, along with the pgBwt-MTS plasmid. Forty-eight hours after transfection, gB was immunoprecipitated from the cell lysates and treated with endo H (+) or left undigested (−). Samples were analyzed by SDS-PAGE and Western blotting with PAb to the major HSV-1 glycoproteins. Circles identify the gB forms exhibiting the indicated apparent Mrs. Arrows identify the electrophoretic mobilities of endo H-digested immature forms of gB. The electrophoretic mobilities of the molecular mass markers are reported. (F to Q) 293T cells cotransfected with pBJ5-Vps4E228Q along with either pgBwt-MTS (I to K) or gH-MTS-gL-MTS (O to Q) were analyzed by confocal microscopy, with MAb to gB (Virusys) or MAb 53S to gH and a PAb to calreticulin, as indicated. 293T cells cotransfected with the empty vector pBJ5 together with either the pgBwt-MTS plasmid (F to H) or the gH-MTS-gL-MTS plasmids (L to N) were used as a control.
gB is ubiquitinated in infected and in transfected cells, and its ubiquitination involves ubiquitin residues implicated in endocytosis.
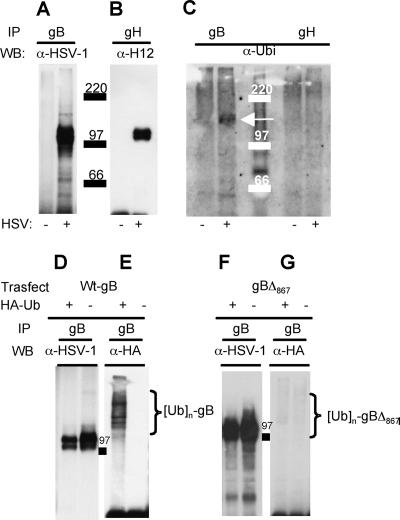
It is well established that ubiquitination provides a key signal for endosomal sorting of membrane proteins to membranes destined to give rise to MVBs (41). To shed light on the mechanism by which gB is sorted to the MVB membranes, we asked whether gB is ubiquitinated. gB and gH, as a control, were immunoprecipitated from lysates of HSV-1(F)-infected cells with MAb H1817 and MAb 53S, respectively. Immunoprecipitated proteins were analyzed by Western blotting with a PAb to the major HSV-1 envelope proteins, MAb H1817 to gH or an antibody to ubiquitin. Figure 6A to C show that gB reacted with the ubiquitin-specific antibody but that no signal was present in the case of gH. To confirm whether gB was ubiquitinated and to investigate whether gB ubiquitination took place in the absence of additional viral proteins, gB was ectopically expressed together with an HA-tagged version of ubiquitin and immunoprecipitated with the MAb H1817 to gB. Figure 6D to E show that gB exhibited multiple ubiquitinated forms also in transfected cells.
FIG. 6.
HSV-1 gB is ubiquitinated in infected and in transfected cells. (A to C) Electrophoretic and Western blotting analyses of gB and gH immunoprecipitated (IP) from lysates of HSV-1-infected (+) or uninfected (−) 293T cells by means of MAb H1817 to gB or MAb 53S to gH. The immunoprecipitated proteins were separated by SDS-PAGE, followed by Western blotting (WB) with the indicated antibodies (α-HSV-1, PAb to the major HSV-1 glycoproteins; α-H12, MAb to gH antibody; α-Ubi, PAb to ubiquitin). (C) The arrow points to the band of ubiquitinated gB. Numbers identify the electrophoretic mobilities of the molecular mass markers. (D to G) 293T cells were cotransfected with a construct expressing wt gB (pgBwt-MTS) (D, E) or a truncated form of gB (gBΔ867) (F, G). Cells were also transfected with a construct encoding HA-tagged wt ubiquitin (HA-Ub) or the corresponding pBJ5 empty vector, indicated as HA-Ub + or HA-Ub −, respectively. Forty-eight hours posttransfection, gB was immunoprecipitated with MAb H1817. The separated proteins were analyzed by Western blotting with the antibodies listed above. Braces identify polyubiquitinated forms of gB. 97 indicates the electrophoretic mobility of the molecular mass marker.
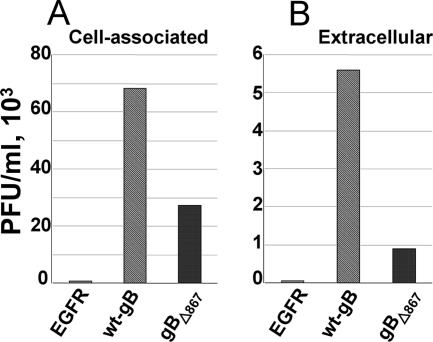
The cytoplasmic tail of gB is known to carry signals for gB trafficking (6, 20, 23, 39, 59). We asked whether the gB cytoplasmic tail represents the site for ubiquitin addition and whether it plays any role in progeny virus release to the extracellular medium. We made use of a construct expressing a truncated form of gB in which the 37 most-C-terminal amino acids of the C tail are placed downstream of a stop codon (gBΔ867-MTS). gBΔ867 localizes to a large extent to the plasma membrane and fails to localize at the cytoplasmic vesicles typical of wt-gB accumulation (2). gBΔ867-MTS was expressed in 293T cells together with the HA-tagged version of ubiquitin. Forty-eight hours later, gB was immunoprecipitated from the cell lysates with MAb H1817 to gB. The results reported in Fig. 6F and G show that gBΔ867 was significantly less ubiquitinated than wt gB, indicating that signals for ubiquitin addition reside mainly in the gB cytoplasmic tail. The role of the gB cytoplasmic tail in HSV egress was measured by complementing gB deletion virions with gBΔ867 or wt gB as a control. As a further control, the deletion virus was complemented with an unrelated glycoprotein, EGFR. Figure 7 shows dramatic decreases in the amounts of cell-associated and extracellular virus when virions were complemented with gBΔ867 instead of wt gB.
FIG. 7.
Yield of cell-associated and extracellular virions complemented with gBΔ867, wt gB, or EGFR. The ΔgB HSV KΔt (9) was grown in 293T cells transiently expressing wt gB, gBΔ867, or EGFR as a negative control. At 24 h after infection, the cell-associated progeny virus (A) or virus released in the extracellular medium (B) was titrated in gB-complementing D6 cells.
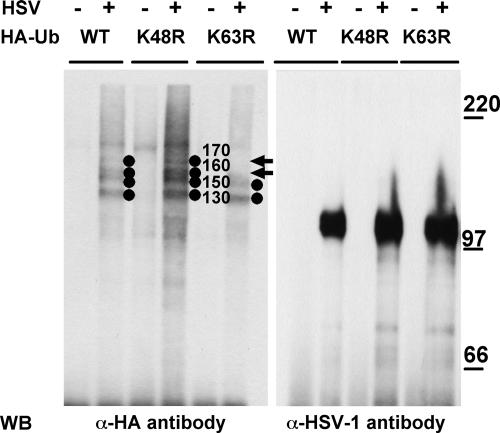
Ubiquitin is a modifying group that functions in diverse cellular processes (43). Polyubiquitin chains, linked to the target protein through lysine 48, mark proteins for proteasomal degradation (15, 42, 68). In contrast, ubiquitin chains linked through the ubiquitin residue lysine 63 impart roles that do not involve the proteasome (19, 25, 77) and include the ubiquitin-dependent endocytosis of certain plasma membrane proteins (28, 78). In order to determine which ubiquitin residues were engaged in gB ubiquitination, we transfected 293T cells with constructs expressing ubiquitin mutants in which either lysine 48 or lysine 63 were individually replaced with arginine. Twenty-four hours later, the cells were infected with HSV-1 and the cell lysates were immunoprecipitated with MAb to gB. The results in Fig. 8 show that in cells transfected with the K48R ubiquitin mutant, gB exhibited four bands with apparent molecular masses of 170, 160, 150, and 130 kDa. In cells transfected with the K63R mutant, the 170- and possibly the 160-kDa bands were absent (Fig. 8). This result provides evidence that gB ubiquitination involves in part the Lys63 ubiquitin residue implicated in endocytosis.
FIG. 8.
Effect of ubiquitin mutants on gB ubiquitination. To detect ubiquitin conjugates, 293T cells were transfected with expression vectors encoding the following HA-tagged forms of ubiquitin (HA-Ub): the wt, K48R mutant, or K63R mutant. Twenty-four hours later, the cells were infected with HSV-1 (10 PFU/cell) (+) or mock infected (−). Twenty-four hours postinfection, gB was immunoprecipitated from cell lysates. Western blotting (WB) was performed with the indicated antibodies (α-HA, MAb to HA; α-HSV-1, PAb to major HSV-1 glycoproteins). Circles identify the ubiquitinated forms of gB; values to the right of the circles indicate their apparent Mrs. Arrows point to the 170- and 160-kDa forms of ubiquitinated gB absent from the K63R sample. The values to the right identify the migration positions of molecular mass markers (in kilodaltons).
DISCUSSION
The aim of this work was to determine whether the machinery involved in MVB biogenesis plays a role in HSV-1 envelopment/egress and to identify possible molecular mechanisms responsible for the recruitment of viral proteins to the MVB pathway. MVB biogenesis was specifically blocked through the use of a dominant negative version of Vps24/CHMP3 (Vps24-Red), an essential component of the ESCRT-III complex responsible for vesicle invagination during MVB biogenesis, and of Vps4 (Vps4E228Q), the ATPase that releases the ESCRT components after vesicle budding has taken place. Thus, the two mutants block MVB biogenesis at two different steps in the assembly pathway (69).
We observed the following. (i) A block in MVB biogenesis resulted in a dramatic reduction in the yield of cell-associated and extracellular virus; the effects were MOI and cell line independent. The reductions in virus yield and egress were not consequent to a reduction in virus entry due, for example, to mislocalization or reduced availability of virus receptors and took place after viral DNA replication and late gene transcription and translation. The results indicate that both Vps24 and Vps4 are critical for efficient HSV-1 assembly/release, very likely for HSV envelopment. Inasmuch as both the cell-associated and the extracellular virus yields were reduced, the block is likely exerted at the envelopment step. The results confirm the involvement of Vps4 reported recently (17) and extend it by highlighting a critical role of yet another component of the MVB machinery that functions at a step of MVB biogenesis earlier than that regulated by Vps4. The results presented here fully agree with the detailed electron microscopy analysis showing that the step in HSV-1 replication affected by expression of the dominant negative version of Vps4 is virion envelopment (17).
(ii) The MVB compartment was morphologically altered in HSV-infected cells, and appeared to be enlarged compared to that of uninfected cells. Thus, the MVB compartment should be added to the list of organelles that are modified in HSV-infected cells, a list that includes the Golgi apparatus, the cytoskeleton, the nucleoskeleton, the nucleoplasm, and some other organelles (1, 14, 34). It is generally assumed that such modifications contribute to rendering the cell a suitable microenvironment for HSV-1 replication and release.
(iii) To shed light on possible molecular mechanism(s) underlying the involvement of the MVB biogenesis machinery in HSV envelopment/egress, we asked whether HSV encodes structural proteins that are sorted to the MVB compartment. We reasoned that a likely candidate could be gB, a glycoprotein highly conserved among members of the Herpesviridae family (67) and an essential glycoprotein for HSV entry into the cell, production of infectious viral particles, in vivo pathogenesis, and neuroinvasiveness (4, 8-10, 30, 57, 76, 84). The gB cytoplasmic tail carries endocytosis motifs and syncytial mutations (6, 20, 23, 39, 59). The protein recirculates from the plasma membrane to vesicles that tend to enlarge and are positive for some endosomal markers (2, 6, 39, 59). The nature of these vesicles has not been fully characterized. Point or deletion mutations that impair gB endocytosis are associated with altered intracellular transport and with reduced infectious progeny (6, 13, 24, 40). We observed that gB accumulated at membranes positive for the MVB marker LAMP-1. The gB-LAMP-1 colocalization was observed in both HSV-infected and gB-transfected cells, highlighting that this is an intrinsic property of the glycoprotein. A role for the MVB pathway in determining the site of gB accumulation was further supported by the finding that a block in MVB biogenesis through expression of the dominant negative Vps4 mutant resulted in the accumulation of an endo H-sensitive immature form of gB at a morphologically distinct site, partly positive for calreticulin. Overall, these results indicate that gB accumulates at MVB membranes and that the intracellular trafficking of gB requires a correct MVB biogenesis process. These properties were specific to gB and were not observed with gH.
(iv) A well-known mechanism for recruitment of membrane proteins, e.g., receptors, to MVBs is ubiquitination, which represents a signal for endosomal sorting of membrane proteins to the MVBs and for the targeting of membrane proteins to lysosomal degradation (68, 75). Particularly critical to the latter process is Lys63-dependent ubiquitination (28). Our results show that gB was modified by ubiquitination. This modification was peculiar to gB, as gH did not undergo such modification. gB ubiquitination was observed both in infected and in transfected cells, again highlighting that this is an intrinsic property of the glycoprotein. A ubiquitin mutant carrying the K63R substitution showed that gB ubiquitination involved in part K63, a residue implicated in endocytosis and not in proteasome-dependent degradation (28).
(v) A wealth of studies indicate that the domain critical for gB trafficking is the cytoplasmic tail, known to carry endocytosis motifs and syncytial mutations (2, 6, 14, 20, 26). A gB mutant lacking the 37 most C-terminal amino acids of the cytoplasmic tail (gBΔ867) and impaired in its ability to accumulate at cytoplasmic vesicles (2), was significantly less ubiquitinated than the wt protein. Remarkably, virions carrying in their envelope gBΔ867 were impaired most likely in cytoplasmic envelopments and egress (Fig. 7). Thus, the gB cytoplasmic tail carries most of the signals for gB ubiquitination and, at the same time, plays a critical role in virus envelopment and release. This finding is in agreement with a recent detailed quantitative study showing that virions lacking gB exhibit a reduced ability to egress from the cell (22), as well as with complementation studies with gB mutants (6, 20). Taken together, recent and current data underscore the idea that components normally involved in MVB biogenesis play a critical role in the envelopment of herpes simplex virions at cytoplasmic membranes and point to gB as a candidate protein that links HSV envelopment-egress to the MVB pathway. In the case of human immunodeficiency virus type 1, the effects of the interaction of Gag with TSG101, an ESCRT-I component, vary in different cell lines. Thus, in monocytic cells, the interaction leads to the virus budding directly into the MVB compartment (65). In contrast, in lymphocytic cells, the interaction leads to the recruitment of MVB components to the plasma membrane, which in this way becomes the site of budding (65). The site of gB accumulation as well as its ubiquitination makes it likely that the membranes of the MVBs serve as platforms for HSV envelopment, at least in some cells. Alternatively, the site of envelopment may be membranes other than those of the MVBs, to which components involved in MVB biogenesis are recruited through the intervention of gB and possibly additional viral proteins; candidates are the tegument proteins, predicted to contain L-domain motifs (17). A detailed electron microscopy analysis showed that the HSV budding compartment lacks the characteristic morphology of MVBs (17), consistent with a previous study of pseudorabies virus (33). This finding may be explained by the modifications to the compartment induced by the infection observed in our study, or it may imply that membranes other than those of the MVBs are the actual site of envelopment, once MVB components are recruited to them. Deletion of gB does not totally prevent HSV envelopment but reduces the amount of virus assembly and release to the extracellular medium (22). Given the proteomic complexity of HSV, and of herpesviruses in general, it is possible that the mechanisms that govern HSV envelopment and egress are redundant, a view supported by studies of the effect of multiple glycoprotein deletions (21, 22). It is also possible that more than one type of cell membrane serves as a platform for the recruitment of cell and viral components to achieve the ultimate goal of virus envelopment and release. Finally, given that gB is one of the most highly conserved proteins among members of the Herpesviridae family, the involvement of MVB components for virion envelopment/egress is likely to be a feature common to members of the family.
Acknowledgments
We thank our colleagues G. Cohen and R. Eisenberg (University of Pennsylvania) and T. Minson (Cambridge University) for gifts of antibodies, Valeria Bergonzini for assistance with the confocal microscopy images, Elena Sartori for help and technical support, Bettina Strack and Jens von Einem for helpful discussion, Jacob Cohen for critical reading of the manuscript, and Dario Masiero and Massimo Fioretti for artwork.
Alessandra Comin is a Ph.D student in the Virology and Microbial Biotechnologies program at the University of Padua.
This work was supported by grants from Cassa di Risparmio di Padova e Rovigo and the University of Padua; grant MIUR-PRIN-2005 to A. Calistri, G.P., and C.P.; and NIH grant AI29873 to H.G. The studies at the University of Bologna were supported by EU contract TargetHerpes-VI FP LSHG-CT-2006-037517, grant MIUR-PRIN-2005, and the Fondi Roberto e Cornelia Pallotti. C.S. was supported by a postdoctoral fellowship from the Azienda Ospedaliera of Padova, Padova Hospital.
Footnotes
Published ahead of print on 8 August 2007.
REFERENCES
- 1.Avitabile, E., S. Di Gaeta, M. R. Torrisi, P. L. Ward, B. Roizman, and G. Campadelli-Fiume. 1995. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol. 69:7472-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avitabile, E., G. Lombardi, T. Gianni, M. Capri, and G. Campadelli-Fiume. 2004. Coexpression of UL20p and gK inhibits cell-cell fusion mediated by herpes simplex virus glycoproteins gD, gH-gL, and wild-type gB or an endocytosis-defective gB mutant and downmodulates their cell surface expression. J. Virol. 78:8015-8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babst, M. 2005. A protein's final ESCRT. Traffic 6:2-9. [DOI] [PubMed] [Google Scholar]
- 4.Baghian, A., L. Huang, S. Newman, S. Jayachandra, and K. G. Kousoulas. 1993. Truncation of the carboxy-terminal 28 amino acids of glycoprotein B specified by herpes simplex virus type 1 mutant amb1511-7 causes extensive cell fusion. J. Virol. 67:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beitia Ortiz de Zarate, I., K. Kaelin, and F. Rozenberg. 2004. Effects of mutations in the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B on intracellular transport and infectivity. J. Virol. 78:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, S. M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bzik, D. J., B. A. Fox, N. A. DeLuca, and S. Person. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185-190. [DOI] [PubMed] [Google Scholar]
- 9.Cai, W. Z., S. Person, S. C. Warner, J. H. Zhou, and N. A. DeLuca. 1987. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J. Virol. 61:714-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai, W. H., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calistri, A., C. Parolin, and G. Palù. 2003. Herpes simplex virus type 1 can either suppress or enhance human immunodeficiency virus type 1 replication in CD4-positive T lymphocytes. J. Med. Virol. 70:163-170. [DOI] [PubMed] [Google Scholar]
- 12.Campadelli-Fiume, G., and B. Roizman. 2006. The egress of herpesviruses from cells: the unanswered questions. J. Virol. 80:6716-6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campadelli-Fiume, G., M. Amasio, E. Avitabile, A. Cerretani, C. Forghieri, T. Gianni, and L. Menotti. 2007. The multipartite system that mediates entry of herpes simplex virus into the cell. Rev. Med. Virol. [Epub ahead of print.] [DOI] [PubMed]
- 14.Campadelli-Fiume, G., and L. Menotti. 2007. Entry of alphaherpesviruses into the cell, p. 93-112. In A. Arivin, E. Mocarski, P. S. Moore, B. Roizman, R. Whitley, and K. Yamanishi (ed.), Human herpesviruses. Cambridge University Press, Cambridge, United Kingdom. [PubMed]
- 15.Chau, V., J. W. Tobias, A. Bachmair, D. Marriott, D. J. Ecker, D. K. Gonda, and A. Varshavsky. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243:1576-1583. [DOI] [PubMed] [Google Scholar]
- 16.Cocchi, F., L. Menotti, V. Di Ninni, M. Lopez, and G. Campadelli-Fiume. 2004. The herpes simplex virus JMP mutant enters receptor-negative J cells through a novel pathway independent of the known receptors nectin1, HveA, and nectin2. J. Virol. 78:4720-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crump, C. M., C. Yates, and T. Minson. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 81:7380-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. [DOI] [PubMed] [Google Scholar]
- 19.Deng, L., C. Wang, E. Spencer, L. Yang, A. Braun, J. You, C. Slaughter, C. Pickart, and Z. J. Chen. 2000. Activation of the I B kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103:351-361. [DOI] [PubMed] [Google Scholar]
- 20.Fan, Z., M. L. Grantham, M. S. Smith, E. S. Anderson, J. A. Cardelli, and M. I. Muggeridge. 2002. Truncation of herpes simplex virus type 2 glycoprotein B increases its cell surface expression and activity in cell-cell fusion, but these properties are unrelated. J. Virol. 76:9271-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farnsworth, A., K. Goldsmith, and D. C. Johnson. 2003. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J. Virol. 77:8481-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farnsworth, A., T. W. Wisner, M. Webb, R. Roller, G. Cohen, R. Eisenberg, and D. C. Johnson. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. USA 104:10187-10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Favoreel, H. W., H. J. Nauwynck, H. M. Halewyck, P. Van Oostveldt, T. C. Mettenleiter, and M. B. Pensaert. 1999. Antibody-induced endocytosis of viral glycoproteins and major histocompatibility complex class I on pseudorabies virus-infected monocytes. J. Gen. Virol. 80:1283-1291. [DOI] [PubMed] [Google Scholar]
- 24.Favoreel, H. W., G. Van Minnebruggen, H. J. Nauwynck, L. W. Enquist, and M. B. Pensaert. 2002. A tyrosine-based motif in the cytoplasmic tail of pseudorabies virus glycoprotein B is important for both antibody-induced internalization of viral glycoproteins and efficient cell-to-cell spread. J. Virol. 76:6845-6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finley, D., S. Sadis, B. P. Monia, P. Boucher, D. J. Ecker, S. T. Crooke, and V. Chau. 1994. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 14:5501-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foster, T. P., J. M. Melancon, and K. G. Kousoulas. 2001. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology 287:18-29. [DOI] [PubMed] [Google Scholar]
- 27.Futter, C. E., A. Pearse, L. J. Hewlett, and C. R. Hopkins. 1996. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J. Cell Biol. 132:1011-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galan, J., and R. Haguenauer-Tsapis. 1997. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 16:5847-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 30.Gerdts, V., J. Beyer, B. Lomniczi, and T. C. Mettenleiter. 2000. Pseudorabies virus expressing bovine herpesvirus 1 glycoprotein B exhibits altered neurotropism and increased neurovirulence. J. Virol. 74:817-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gianni, T., L. Menotti, and G. Campadelli-Fiume. 2005. A heptad repeat in herpes simplex virus 1 gH, located downstream of the alpha-helix with attributes of a fusion peptide, is critical for virus entry and fusion. J. Virol. 79:7042-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gianni, T., R. Fato, C. Bergamini, G. Lenaz, and G. Campadelli-Fiume. 2006. Hydrophobic alpha-helices 1 and 2 of herpes simplex virus gH interact with lipids, and their mimetic peptides enhance virus infection and fusion. J. Virol. 80:8190-8198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harley, C. A., A. Dasgupta, and D. W. Wilson. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 75:1236-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartlieb, B., and W. Weissenhorn. 2006. Filovirus assembly and budding. Virology 344:64-70. [DOI] [PubMed] [Google Scholar]
- 36.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heineman, T. C., and S. L. Hall. 2001. VZV gB endocytosis and Golgi localization are mediated by YXXphi motifs in its cytoplasmic domain. Virology 285:42-49. [DOI] [PubMed] [Google Scholar]
- 40.Heineman, T. C., and S. L. Hall. 2002. Role of the varicella-zoster virus gB cytoplasmic domain in gB transport and viral egress. J. Virol. 76:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hicke, L., and R. Dunn. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19:141-172. [DOI] [PubMed] [Google Scholar]
- 42.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405-439. [DOI] [PubMed] [Google Scholar]
- 43.Hofmann, R. M., and C. M. Pickart. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96:645-653. [DOI] [PubMed] [Google Scholar]
- 44.Jasenosky, L. D., and Y. Kawaoka. 2004. Filovirus budding. Virus Res. 106:181-188. [DOI] [PubMed] [Google Scholar]
- 45.Jayakar, H. R., E. Jeetendra, and M. A. Whitt. 2004. Rhabdovirus assembly and budding. Virus Res. 106:117-132. [DOI] [PubMed] [Google Scholar]
- 46.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular body sorting. Nat. Rev. Mol. Cell. Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 48.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for Gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 50.Martin-Serrano, J., D. Perez-Caballero, and P. D. Bieniasz. 2004. Context dependent effects of L-domains and ubiquitination on viral budding. J. Virol. 78:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167-180. [DOI] [PubMed] [Google Scholar]
- 53.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9:423-429. [DOI] [PubMed] [Google Scholar]
- 54.Mettenleiter, T. C., T. Minson, and P. Wild. 2006. Egress of alphaherpesviruses. J. Virol. 80:1610-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mimnaugh, E. G., P. Bonvini, and L. Neckers. 1999. The measurement of ubiquitin and ubiquitinated proteins. Electrophoresis 20:418-428. [DOI] [PubMed] [Google Scholar]
- 56.Mimnaugh, E. G., and L. M. Neckers. 2005. Measuring ubiquitin conjugation in cells. Methods Mol. Biol. 301:223-241. [DOI] [PubMed] [Google Scholar]
- 57.Mitchell, B. M., and J. G. Stevens. 1996. Neuroinvasive properties of herpes simplex virus type 1 glycoprotein variants are controlled by the immune response. J. Immunol. 156:246-255. [PubMed] [Google Scholar]
- 58.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 59.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 61.Ott, D. E., L. V. Coren, R. C. Sowder, I. I., J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76:3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ott, D. E., L. V. Coren, R. C. Sowder, I. I. J. Adams, and U. Schubert. 2003. Retroviruses have differing requirements for proteasome function in the budding process. J. Virol. 77:3384-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patnaik, A., V. Chau, F. Li, R. C. Montelaro, and J. W. Wills. 2002. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J. Virol. 76:2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162:443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peng, T., M. Ponce de Leon, M. J. Novotny, H. Jiang, J. D. Lambris, G. Dubin, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1998. Structural and antigenic analysis of a truncated form of the herpes simplex virus glycoprotein gH-gL complex. J. Virol. 72:6092-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pereira, L. 1994. Function of glycoprotein B homologues of the family herpesviridae. Infect. Agents Dis. 3:9-28. [PubMed] [Google Scholar]
- 68.Pickart, C. M. 2000. Ubiquitin in chains. Trends Biochem. Sci. 25:544-548. [DOI] [PubMed] [Google Scholar]
- 69.Piper, R. C., and D. J. Katzmann. 2007. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. [Epub ahead of print.] doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed]
- 70.Pornillos, O., S. L. Alam, D. R. Davis, and W. I. Sundquist. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9:812-817. [DOI] [PubMed] [Google Scholar]
- 71.Pornillos, O., S. L. Alam, R. L. Rich, D. G. Myszka, D. R. Davis, and W. I. Sundquist. 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21:2397-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176:633-637. [DOI] [PubMed] [Google Scholar]
- 73.Radsak, K., M. Eickmann, T. Mockenhaupt, E. Bogner, H. Kern, A. Eis-Hubinger, and M. Reschke. 1996. Retrieval of human cytomegalovirus glycoprotein B from the infected cell surface for virus envelopment. Arch. Virol. 141:557-572. [DOI] [PubMed] [Google Scholar]
- 74.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Slagsvold, T., K. Pattni, L. Malerod, and H. Stenmark. 2006. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell. Biol. 16:317-326. [DOI] [PubMed] [Google Scholar]
- 76.Spear, P. G. 1993. Membrane fusion induced by herpes simplex virus, p. 201-232. In J. Bentz (ed.), Viral fusion mechanisms. CRC Press Inc., Ann Arbor, MI.
- 77.Spence, J., R. R. Gali, G. Dittmar, F. Sherman, M. Karin, and D. Finley. 2000. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102:67-76. [DOI] [PubMed] [Google Scholar]
- 78.Springael, J. Y., J. M. Galan, R. Haguenauer-Tsapis, and B. Andre. 1999. NH4+-induced downregulation of the Saccharomyces cerevisiae Gap1p permease involves its ubiquitination with lysine-63-linked chains. J. Cell Sci. 112:1375-1383. [DOI] [PubMed] [Google Scholar]
- 79.Strack, B., A. Calistri, M. A. Accola. G. Palù, and H. G. Göttlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Strack, B., A. Calistri, and H. G. Göttlinger. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 76:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Strack, B., A. Calistri, S. Craig, E. Popola, and H. G. Göttlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 82.Takimoto, T., and A. Portner. 2004. Molecular mechanism of paramyxovirus budding. Virus Res. 106:133-145. [DOI] [PubMed] [Google Scholar]
- 83.Urata, S., T. Noda, Y. Kawaoka, H. Yokosawa, and J. Yasuda. 2006. Cellular factors required for Lassa virus budding. J. Virol. 80:4191-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yuhasz, S. A., and J. G. Stevens. 1993. Glycoprotein B is a specific determinant of herpes simplex virus type 1 neuroinvasiveness. J. Virol. 67:5948-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]