Abstract
In the quest for an effective vaccine against human immunodeficiency virus (HIV), live attenuated virus vaccines have proven to be very effective in the experimental model system of simian immunodeficiency virus (SIV) in macaques. However, live attenuated HIV vaccines are considered unsafe for use in humans because the attenuated virus may accumulate genetic changes during persistence and evolve to a pathogenic variant. As an alternative approach, we earlier presented a conditionally live HIV-1 variant that replicates exclusively in the presence of doxycycline (DOX). Replication of this vaccine strain can be limited to the time that is needed to provide full protection through transient DOX administration. Since the effectiveness and safety of such a conditionally live AIDS vaccine should be tested in macaques, we constructed a similar DOX-dependent SIVmac239 variant in which the Tat-TAR (trans-acting responsive) transcription control mechanism was functionally replaced by the DOX-inducible Tet-On regulatory mechanism. Moreover, this virus can be used as a tool in SIV biology studies and vaccine research because both the level and duration of replication can be controlled by DOX administration. Unexpectedly, the new SIV variant required a wild-type Tat protein for replication, although gene expression was fully controlled by the incorporated Tet-On system. This result suggests that Tat has a second function in SIV replication in addition to its role in the activation of transcription.
Live attenuated virus vaccines have proven to be highly successful at inducing protective immunity against pathogenic viruses such as variola virus, poliovirus, and measles virus. Research on the development of a live attenuated human immunodeficiency virus (HIV) vaccine has focused on the experimental model system of the pathogenic simian immunodeficiency virus (SIV) and the infection of macaques. In most of these studies, several accessory functions have been deleted from the viral genome, either individually or in combination (reviewed in references 41, 48, 57, and 76). The majority of monkeys vaccinated with such deletion mutants of SIV can efficiently control the replication of pathogenic challenge virus strains. However, the attenuated virus could revert to virulence and cause disease over time in vaccinated animals (3, 4, 17, 75). Similarly, some of the long-term survivors of the Sydney Blood Bank Cohort infected with an HIV type 1 (HIV-1) variant in which nef and long terminal repeat (LTR) sequences were deleted eventually showed progression to AIDS (22), and an HIV-1 Δ3 variant with deletions in the vpr, nef, and LTR sequences regained substantial replication capacity in long-term cell culture infections by acquiring compensatory changes in the viral genome (13). These results highlight the genetic instability and evolutionary capacity of attenuated SIV/HIV strains, which pose a serious safety risk for any future experimentation with live attenuated HIV vaccines in humans.
The major problem with live attenuated HIV/SIV vaccines is the persistence of the attenuated virus and ongoing low-level replication. Due to the error-prone replication machinery of the virus, this may eventually lead to the appearance of fitter and more pathogenic virus variants. The pathogenicity of the vaccine strain can be further reduced by lowering the replication capacity through progressive deletion or mutation of accessory genes or regulatory elements. However, reduced replication may lower the efficacy of the vaccine (51, 77). As an alternative strategy to prevent the evolution of a pathogenic variant, replication of the vaccine virus should be limited to the extent that is needed to provide full protection. For instance, virus replication can be stopped upon vaccination by the administration of antiviral drugs (50). Whereas this may be a good strategy for in vitro studies, its application in humans seems problematic because long-term virus inhibition will require continuous drug administration, and the virus may develop drug resistance. Alternatively, a virus that can execute only a single round of replication can be used as a vaccine (31). However, due to the limited replication, such a single-cycle virus vaccine may be less appropriate for the induction of protective immunity.
We and others previously presented a unique genetic approach that exploits a conditionally live HIV-1 variant (11, 26, 27, 68, 73). In this HIV-rtTA variant, the Tat-TAR (trans-acting responsive) regulatory mechanism that controls viral gene expression and replication was inactivated by mutation of both the Tat gene and the TAR RNA structure and functionally replaced by the Tet-On system for inducible gene expression (5). The rtTA gene, encoding a man-made transcriptional activator, was inserted in place of the nef gene, and the tet operator (tetO) DNA binding sites were inserted into the LTR promoter. Since the rtTA protein can bind tetO and activate transcription only in the presence of doxycycline (DOX), the HIV-rtTA variant replicates exclusively when DOX is administered. Upon vaccination with this virus, replication can be temporarily activated and controlled to the extent needed for induction of the immune system by transient DOX administration. The initial HIV-rtTA variant has been improved significantly by virus evolution (28, 53, 54, 79-81), and we have shown efficient and DOX-dependent replication not only in vitro in T-cell lines but also ex vivo in human lymphoid tissue (47). In addition, we constructed an HIV-1 variant that depends not only on DOX for gene expression but also on the T20 peptide for cell entry (24). Replication of this virus can be limited to the level required to induce the immune system by the transient administration of DOX and T20. The subsequent withdrawal of these inducers efficiently blocks viral replication and prevents evolution.
The successful development of a conditionally live HIV-1 variant encouraged us to construct a similar DOX-dependent SIV variant that can be used to study the efficacy and safety of a conditionally live virus vaccine against AIDS in macaques. Furthermore, this SIV variant may be an attractive tool to study the correlates of immune protection upon vaccination, because the level and duration of replication can be controlled by DOX administration. Given our expertise with the HIV-rtTA design, the construction of SIV-rtTA seemed straightforward, but it turned out to be much more complicated. Surprisingly, SIV-rtTA was found to require a wild-type Tat protein, suggesting that Tat has a second function in SIV replication in addition to its role in the activation of transcription.
MATERIALS AND METHODS
Construction of SIV-rtTA.
For the construction of SIV-rtTA, we modified the SIVmac239 genome (62) (GenBank accession number M33262) by (i) mutation of TAR and introduction of tetO elements in both the 5′ and the 3′ LTRs, (ii) mutation of Tat, (iii) introduction of the rtTA gene at the site of the nef gene, and (iv) mutation of the original AUGNef start codon and three downstream AUG codons (AUGII-IV) that preceded the AUGrtTA start codon. The plasmid p239SpSp5′, which contains the 5′ half of the SIVmac239 genome (44), and pSIVmac239ΔNef (p239SpE3′/nef-del), which contains the 3′ half of the proviral genome with a deletion in the nef coding region (nucleotides [nt] 9507 to 9689) (33, 59), were used to construct 5′ and 3′ halves of SIV-rtTA, named pKP-5′SIV and pBS-3′SIV-rtTA, respectively. These plasmids were subsequently combined as described below to construct the complete pSIV-rtTA molecular clone.
Plasmids containing the 3′ end of the env gene, the nef gene, and the 3′ LTR (5′ and 3′ nucleotides corresponding to positions 8880 and 10535 in SIVmac239, respectively), and in which either TAR or AUG codons are mutated, were constructed by mutagenesis PCR (56) on the pSIVmac239ΔNef template with the forward mutagenic primers (primer M) MutTAR (5′-GCGGAGAGGCTGGCAGAAAGAGCCATTGGAGGTTCTCTCCAGCACTAGCAGGAAGAGCATTGGTGTTCCCTGCTAG-3′; nucleotides mismatching with the TAR bulge and loop sequences underlined), MutEnv1 (5′-GGGGAGACTTGTGGGAGACTCTTAGGAGAGGTGGAAGGTGGATACTCG-3′; nucleotides mismatching with AUGIII and AUGIV underlined), and MutEnv2 (5′-ACCTACAATACGGGTGGAGCTATTTCCACGAGGCGGTCC-3′; nucleotides mismatching with AUGNef and AUGII underlined); the forward primer SIV-LTR1 (5′-AGTACTGCGGCCGCAGGCATG8880CTGGGATGTGTTTGGCAATTG-3′; NotI and SphI sites underlined); and reverse primers SIV-LTR2 (5′-TATCTATAGCCCAGCACCGGCCAAGTG-3′; mismatching nucleotides underlined) and SIV-LTR3 (5′-ATCGGTACCGACGTCTCGAGT10535GCTAGGGATTTTCCTGCTTCG-3′; Asp718, AatII, and XhoI sites underlined). Briefly, PCRs were performed with primer M plus SIV-LTR3 and with SIV-LTR1 plus SIV-LTR2. The PCR products were purified, mixed, and PCR amplified with SIV-LTR1 and SIV-LTR3 (see reference 56 for details on the PCR strategy). The resulting mutated fragments were cloned as NotI-Asp718 fragments into pBluescript SK+ (Stratagene), resulting in the plasmids pBS-SIV-3′LTR-TAR, pBS-SIV-3′LTR-Env1, and pBS-SIV-3′LTR-Env2. The BglII-Asp718 fragment from pBS-SIV-3′LTR-Env1, containing the AUGNef and AUGII mutations, was used to replace the corresponding fragment in pBS-SIV-3′LTR-Env2, resulting in plasmid pBS-SIV-3′LTR-Env12, which contains all four AUG mutations.
For the insertion of tetO elements, the plasmid pBS-SIV-3′LTR-TAR was used as the template for PCR with primers Mut2Δ15tetO1 (5′-GAAACAGCAGGGACTTTCCACAGGATCCCTATCAGTGATAGAGAAATACCACTCCCTATCAGTGATAGAGAATTCAGGGGATGTTACGGGGAGGT-3′; the BamHI site is underlined, and tetO sites are in bold) and SIV-LTR3. The PCR product was digested with BamHI and Asp718 and ligated into the corresponding sites of pBluescript SK+, which resulted in plasmid pBS-SIV-3′LTR-2ΔtetO-TAR. In parallel, we introduced unique restriction enzyme recognition sites directly downstream of the env gene, which subsequently were used to insert the rtTA gene. For this, pSIVmac239ΔNef was PCR amplified with primers MutRtTA1 (5′-CACTCTCTTGTGAGGGACAGTCGACCATGTCTAGACTGGCCCGGGTAACTAAGTAAGGATCTCATTTTATAAAAGAAAAGGGGGGA-3′; SalI, XbaI, and SmaI sites are underlined, and the Env stop codon is in bold) and Mut2Δ15tetO2 (5′-TAGGGATCCTGTGGAAAGTCCCTGCTGTTTC-3′; BamHI site underlined). The PCR product was ligated into the pCR2.1-TOPO TA-cloning vector (Invitrogen), resulting in plasmid pCR-SIV-3′LTR-SB. Separately, we PCR amplified upstream pSIVmac239ΔNef sequences with primers SIV-LTR1 and MutRtTA2 (5′-CCCGGGCCAGTCTAGACATGGTCGACTGTCCCTCACAAGAGAGTGAGC-3′; SalI site underlined) and digested the PCR product with NotI and SalI. We subsequently ligated this fragment together with the SalI-BamHI fragment from pCR-SIV-3′LTR-SB into the NotI and BamHI sites of pBS-SIV-3′LTR-2ΔtetO-TAR, resulting in plasmid pBS-SIV-3′LTR-rtTA-2ΔtetO-TAR. The SacI-Asp718 fragment from this plasmid (SacI site present downstream of the env gene) and the NotI-SacI fragment from pBS-SIV-3′LTR-Env12 were jointly ligated into the NotI and Asp718 sites of pBluescript SK+, resulting in plasmid pBS-SIV-3′LTR. The rtTAF86Y A209T gene, isolated as an XbaI-SmaI fragment from pCMV-rtTAF86Y A209T (28), was subsequently ligated into the XbaI and SmaI sites of pBS-SIV-3′LTR, resulting in pBS-SIV-3′LTR-rtTA.
For the introduction of the Y55A mutation in Tat, pSIVmac239ΔNef was used as the template in a PCR with primers MutTatY55A (5′-AAGCTTGCATGCTATAACACATGCGCCTGTAAAAAGTGTTGCTA-3′; the SphI site is underlined, and mismatching nucleotides are in bold) and SIV-Env4 (5′-CCCTGTCATGTTGAATTTACAGCT-3′), and the product was digested with SphI and SpeI and subsequently ligated into the corresponding sites of pSIVmac239ΔNef, resulting in pSIVmac239ΔNef-Tat-Y55A. The SphI-NheI fragment from this plasmid was ligated into the SphI and NheI sites of pBS-SIV-3′LTR, resulting in pBS-SIV-3′LTR-Tat-Y55A. The NotI-SalI fragment of this plasmid was subsequently ligated into the corresponding sites of pBS-SIV-3′LTR-rtTA, resulting in pBS-3′SIV-rtTA, which contains the 3′ half of SIV-rtTA.
For the construction of the 5′ half of SIV-rtTA, we PCR amplified the tetO-containing and TAR-mutated LTR fragment of pBS-SIV-3′LTR with primers SIV-LTR4 (5′-AGCTCTAGAGCGGCCGCTGGAAGGGATTTATTACAGTGCA-3′; XbaI and NotI sites underlined) and SIV-LTR5 (5′-ATGGACGTCTCGAGTCGCATGCTAGGCGCCAATCTGCTAGGGATTTTCCTGCT-3′; AatII, XhoI, SphI, and NarI sites underlined). Upon digestion with XbaI and AatII, the PCR product was ligated into the corresponding sites of the pKP59 vector, a pBR322 derivative that was previously used for the construction of the HIV-1 LAI molecular clone (60), to generate pKP-SIV-5′LTR. We subsequently introduced the NarI-SphI fragment of p239SpSp5′ into pKP-SIV-5′LTR. However, since direct ligation of this fragment into the NarI and SphI sites of pKP-SIV-5′LTR failed for unknown reasons, we used a two-step approach. We first ligated the complete SphI-SphI insert of p239SpSp5′ into the SphI site of pKP-SIV-5′LTR, which resulted in a duplication of the LTR region, and subsequently removed the undesired LTR copy by NarI digestion and religation of the plasmid. The resulting plasmid, pKP-5′SIV, contains the 5′ half of the SIV-rtTA genome. The SphI-XhoI insert of pBS-3′SIV-rtTA was ligated into the corresponding sites of pKP-5′SIV to complete the construction of pSIV-rtTA, which contains the full-length SIV-rtTA genome.
For the introduction of wild-type TAR into SIV-rtTA, we PCR amplified the LTR region from p239SpSp5′ with primers Mut2Δ15tetO1 and SIV-Gag1 (5′-ATTTAATGTTCTCGGGCTTA-3′) and used the EcoRI-NarI-digested PCR product to replace the corresponding sequences in pKP5′SIV. Similarly, p239SpSp5′ was PCR amplified with primers Mut2Δ15tetO1 and SIV-LTR3, and the EcoRI-XhoI-digested PCR product was used to replace the corresponding sequences in pBS3′SIV-rtTA. The resulting plasmids, pKP5′SIV-TARwt and pBS3′SIV-rtTA-TARwt, were subsequently combined for the construction of pSIV-rtTA-TARwt as described for the construction of pSIV-rtTA.
For the construction of SIV-rtTA variants with wild-type env sequences, the env-nef region of pSIVmac239ΔNef was PCR amplified with forward primer SIV-LTR1 and reverse primer Mut-RtTA2 or MutRtTA3-Nef-stop (5′-ACATGGTCGACTATCCCTCACAAGAGAGTGAGC-3′; the SalI site is underlined, and the Env stop codon is in bold). The NheI-SalI-digested PCR product was subsequently used to replace the corresponding sequences in pSIV-rtTA, resulting in pSIV-rtTA-Envwt and pSIV-rtTA-Envwt-stop.
For the deletion of U3 sequences in SIV-rtTA, we used pBS-3′SIV-rtTA as the template in a PCR with primers MutEnv1 and DelSIV-U3-383 (5′-AGCGGATCCTGTGGAAAGTCCCTGCTGTTTCAGCGACGC▵GTAATAAATCCCTTCCAGTCC-3′; the BamHI site is underlined, and the position of deleted [▵] and replacing nucleotides is in bold) or DelSIV-U3-409 (5′-AGCGGATCCTACGC▵GTAATAAATCCCTTCCAGTCCC-3′). The PCR products were digested with SalI and BamHI and used to replace the SalI-BamHI fragment of pBS-3′SIV-rtTA. The NheI-XhoI fragments from the resulting plasmids pBS-3′SIV-rtTAΔU3-383 and pBS-3′SIV-rtTAΔU3-409 were used to substitute the corresponding sequences in pSIV-rtTA to construct pSIV-rtTAΔU3-383 and pSIV-rtTAΔU3-409.
For the construction of pSIV-rtTA-Tatwt, we exchanged the SphI-NheI fragment of pSIV-rtTA with the corresponding fragment of pSIVmac239ΔNef. We used standard molecular biology procedures for all manipulations, and plasmids were propagated in Escherichia coli TOP10 (for TA cloning; Invitrogen), DH5α (for the LTR mutation steps), or Stbl4 (all later steps; Invitrogen). All constructs were verified by sequence analysis.
Expression plasmids and reporter gene constructs.
For the construction of plasmids expressing wild-type or Y55A-mutated Tat of SIVmac239, we PCR amplified the first Tat exon of pSIV-rtTA-Tatwt and pSIV-rtTA with the primers SIV-Tat1 (5′-GGTAGTGGAGGTTCTGGAAGA-3′) and SIV-Tat-splice2 (5′-GTTGGATATGGGT▴TTGTTTGATGCAGAAGATGTATT-3′; ▴ indicates the splice site position) and the second Tat exon of pSIV-rtTA-Tatwt with the primers SIV-Tat-splice3 (5′-CTTCTGCATCAAACAA▴ACCCATATCCAACAGGAC-3′) and SIV-Tat-3 (5′-AGCGCTCGAGGATCCGGTATACTCTCGATAGCAA-3′; BamHI site underlined). PCR products were purified, and the first and second exon fragments were mixed (exon 1wt plus exon 2; exon 1Y55A plus exon 2) and used as the template in a subsequent PCR with primers SIV-Tat1 and SIV-Tat3. The PCR products comprise the complete Tat open reading frame, with a HindIII site located at 63 nt upstream of the translation start site and a BamHI site at 88 nt downstream of the Tat stop codon. PCR products were digested with HindIII and BamHI and ligated into the corresponding sites of expression plasmid pcDNA3 (Invitrogen), resulting in pcDNA3-SIV-Tatwt and pcDNA3-SIV-TatY55A.
For the construction of reporter plasmids in which the expression of firefly luciferase is driven by the SIV-rtTA promoter, we first inserted the HindIII-BamHI fragment of pGL3-Basic (Promega), which encompasses the luciferase-coding sequence and the simian virus 40 polyadenylation signal, in the corresponding sites of pBluescript SK+, resulting in pBS-luc. We subsequently ligated the NotI-XhoI LTR fragments from pBS-SIV-3′LTR and pBS-SIV-3′LTR-TARwt into the compatible Bsp120I and XhoI sites of pBS-luc, resulting in the SIV-rtTA and SIV-rtTA-TARwt LTR-luc constructs, respectively.
The expression of firefly luciferase is controlled by the wild-type HIV-1 promoter in the HIV-1 LTR-luc plasmid (pBlue3′LTR-luc in reference 40) and controlled by the HIV-rtTA promoter in the HIV-rtTA LTR-luc plasmid (pLTR-2ΔtetO-lucff in reference 28). We previously described the rtTA-expressing plasmid pCMV-rtTAF86Y A209T (28) and the HIV-1 Tat-expressing plasmid pcDNA3-Tat (72). Plasmid pRL-CMV (Promega), in which the expression of Renilla luciferase is controlled by a cytomegalovirus promoter, is cotransfected into the C33A cells to allow correction for differences in transfection efficiency.
Promoter activity assay.
To determine the DOX responsiveness levels of the promoter-luciferase constructs, C33A cells were transfected with 20 ng LTR-luc plasmid, 0.4 ng pCMV-rtTAF86Y A209T, and 0.5 ng pRL-CMV. The plasmid pBluescript was added as the carrier DNA to the transfection mix to a total of 1 μg of DNA. The cells were cultured after transfection for 48 h with 0 to 1,000 ng/ml DOX (D-9891; Sigma). Cells were lysed in passive lysis buffer, and firefly and Renilla luciferase activities were determined with the dual luciferase assay (Promega). The expression of firefly and Renilla luciferase was within the linear range, and no squelching effects were observed. The promoter activity was calculated as the ratio between the firefly and Renilla luciferase activities and corrected for between-session variation (66). To determine the Tat responsiveness levels of the promoter-luciferase constructs, C33A cells were transfected with 20 ng LTR-luc plasmid plus 0 to 50 ng Tat expression plasmid, 0 to 50 ng pcDNA3 (empty expression vector; the total amount of Tat expression plasmid and pcDNA3 was kept at 50 ng), 0.5 ng pRL-CMV, and 950 ng pBluescript. Cells were cultured for 48 h, and luciferase activities were subsequently measured.
Electrophoretic mobility shift assay (EMSA).
The SIV-rtTA and SIV-rtTA-TARwt LTR-luc plasmids were used for PCR amplification of the mutant and wild-type TAR regions, respectively, with the sense primer 5′-T7-PRO-TAR (5′-TAATACGACTCACTATAGGGAGTCGCTCTGCGGAGAG-3′), encoding the T7 promoter sequence (underlined) directly upstream of the +1 position, and the antisense primer 3′-T7-TAR (5′-GGAGTCACTCTGCCCAGCACCG-3′). DNA products were in vitro transcribed with the MEGAshortscript T7 transcription kit (Ambion). The transcripts were purified with a NucAway spin column (Ambion), and the RNA concentration was determined by spectrophotometry. RNA (25 pmol) was dephosphorylated with calf intestine alkaline phosphatase and 5′ end labeled with the KinaseMax kit (Ambion) in the presence of 1 μl [γ-32P]ATP (0.37 MBq/μl; Amersham Biosciences). TAR RNA was checked for integrity and purified on a denaturing 8% acrylamide gel. The RNA was resuspended in 100 mM KCl, 50 mM Tris-HCl (pH 8.0) and renatured by heating to 85°C for 2 min and slowly cooling to room temperature. His-tagged SIVmac-J5 Tat protein (22 kDa) was obtained from the Centralised Facility for AIDS Reagents at the National Institute for Biological Standards and Control, Potters Bar, United Kingdom (ARP685). 32P-labeled TAR RNA (200 counts/s) was incubated with 0 to 40 ng Tat protein in 50 mM Tris-HCl (pH 8.0), 20 mM KCl, 5 mM dithiothreitol, and 0.05% Triton X-100 for 15 min at room temperature. Calf liver tRNA (1 μg; Roche) was added as the competitor to minimize aspecific interactions when 40 ng Tat was used. After adding 4 μl nondenaturing loading buffer (30% glycerol, bromophenol blue), samples were loaded on a nondenaturing 4% acrylamide gel containing 45 mM Tris, 45 mM borate, and 0.1% Triton X-100. Electrophoresis was performed at 450 V at 4°C. The gel was subsequently dried and analyzed with a PhosphorImager (Molecular Dynamics).
Cells and viruses.
The PM1 T-cell line (52), a clonal derivative of HUT78, was cultured at 37°C and 5% CO2 in RPMI 1640 medium containing 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 units/ml streptomycin. To assay virus replication, cells were transfected with SIV-rtTA molecular clones or with the full-length SIVmac239 clone (34) by electroporation. Briefly, 5 × 106 cells were washed in RPMI 1640 with 20% FBS and mixed with 5 μg DNA in 250 μl RPMI 1640 with 20% FBS. Cells were electroporated in 0.4-cm cuvettes at 250 V and 960 μF and subsequently resuspended in 5 ml RPMI 1640 with 10% FBS. Upon the addition of 0.25 × 106 untransfected PM1 cells, cultures were maintained at 0 to 1,000 ng/ml DOX. The virus level in the culture medium was determined with a real-time PCR-based reverse transcriptase (RT) assay (55) in which avian myeloblastosis virus RT was used as the standard.
C33A cervix carcinoma cells (ATCC HTB31) (2) were cultured and transfected by calcium phosphate precipitation as previously described (28). C33A cells were cultured in 2-cm2 wells and transfected with 1 μg SIV-rtTA or SIVmac239 DNA. Virus production was measured after 48 h by RT assay and CA-p27 enzyme-linked immunosorbent assay (SIV core antigen kit; Beckman Coulter) on culture medium samples.
RESULTS
Design of a DOX-inducible SIV variant.
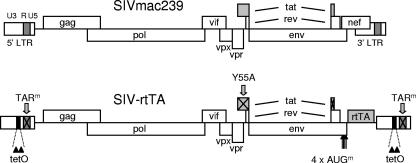
SIV gene expression and replication are naturally controlled by the viral Tat protein, which binds to the 5′ TAR region in the nascent RNA transcript and subsequently activates transcription (12). For the construction of a conditionally live SIVmac239 variant, this Tat-TAR regulatory mechanism was inactivated by mutation and functionally replaced by the DOX-inducible gene expression system (Tet-On system) (5). This E. coli-derived gene expression system is controlled by the rtTA protein. The binding of DOX to rtTA triggers a conformational switch in the protein that allows it to bind to tetO elements and activate transcription from the downstream-positioned promoter. To replace the Tat-TAR axis with the Tet-On system, we modified the SIVmac239 genome by (i) inactivation of TAR through mutation, (ii) introduction of tetO elements in the U3 promoter region, (iii) mutational inactivation of Tat, and (iv) introduction of the rtTA gene at the site of the nef gene (Fig. 1). Transcription of the resulting SIV-rtTA variant will thus no longer be activated by the binding of Tat to TAR but instead by the binding of the DOX-rtTA complex to the tetO-LTR promoter. As a result, replication will critically depend on the presence of DOX.
FIG. 1.
Design of the DOX-inducible SIV-rtTA variant. For the construction of a conditionally live SIVmac239 variant, we inactivated the Tat-TAR regulatory mechanism through mutation of TAR (TARm; loop and bulge mutations as shown in Fig. 2A) and Tat (Y55A substitution; Fig. 3B) and introduced the Tet-On regulatory mechanism through the insertion of two tetO elements in the U3 promoter region (Fig. 3A and 4C) and the rtTA gene at the site of the nef gene (Fig. 4). In addition, we mutated the original AUGNef translation start codon and three downstream AUG codons that precede the new AUGrtTA start codon on the spliced Nef transcript (AUGm; Fig. 4B).
Transforming the Tat-responsive SIV promoter into a DOX-inducible promoter.
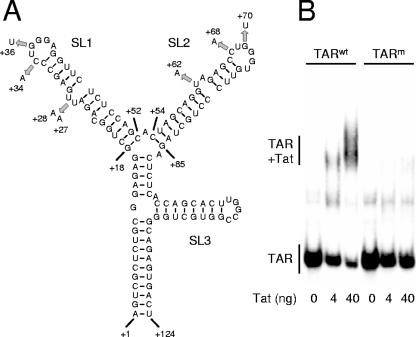
In the nascent SIV transcript, the TAR region from position +1 to position +124 forms a complex hairpin structure with three stem-loop domains (Fig. 2A). Both stem-loop 1 (+18/+52) and stem-loop 2 (+54/+85) resemble the HIV-1 TAR structure and are important for Tat-mediated activation of transcription (9, 32, 63). Whereas stem-loops 1 and 2 contain a 6-nt loop and a 2-nt bulge, HIV-1 TAR contains a 6-nt loop and a 3-nt bulge. HIV-1 studies demonstrated that the highly conserved pyrimidine nucleotides in the bulge are essential for the binding of the viral Tat transactivator protein (70), while the 6-nt loop binds the cyclin T1 subunit of the positive transcriptional elongation factor (pTEFb) in a Tat-dependent manner (65, 74). Upon binding, the kinase component of pTEFb, cyclin-dependent kinase 9 (CDK9), can phosphorylate the C-terminal domain of RNA polymerase II, which enhances the processivity of the elongating polymerase (15, 58). Furthermore, it was recently demonstrated that pTEFb also directs the recruitment of the TATA-box-binding protein (TBP) to the LTR promoter and thus stimulates the assembly of new transcription complexes (16, 61).
FIG. 2.
Inactivation of TAR. (A) The 5′ end of the nascent SIV transcript folds the TAR hairpin structure with three stem-loop regions (SL1 to SL3). Binding of Tat to TAR enhances transcription from the LTR promoter. To abolish Tat responsiveness, we mutated the bulge and loop sequences in SL1 and SL2. (B) Binding of SIV Tat to wild-type (TARwt) and mutant (TARm) TAR RNA was analyzed in an EMSA. TAR RNA was incubated with 0, 4, or 40 ng Tat protein and analyzed on a nondenaturing gel. The positions of unbound TAR RNA and the TAR-Tat complex are indicated.
The TAR structure of SIVmac239 is very similar to that of HIV-2ROD (8). It was previously demonstrated that mutation or deletion of the bulge nucleotides in HIV-2ROD TAR reduces Tat binding and trans activation (32, 63). Furthermore, the 6-nt loop sequences of SIVmac239 TAR are identical (stem-loop 1) or nearly identical (stem-loop 2) to the loop sequence in HIV-1 TAR, and it has been shown that mutation of the first and third loop nucleotides (corresponding to C+34/+68 and G+36/+70, respectively, in SIV TAR) abolishes Tat trans activation in HIV-1 (10). We therefore inactivated both stem-loop 1 and stem-loop 2 by mutation of the bulge and loop sequences to discontinue the Tat responsiveness of the SIVmac239 LTR promoter (Fig. 2A). We performed an EMSA to analyze the binding of the SIV Tat protein to wild-type and mutant TAR RNAs (Fig. 2B). In the absence of Tat, wild-type and mutant TAR RNAs migrate similarly on a nondenaturing polyacrylamide gel. Upon the incubation of the RNA with Tat, wild-type TAR efficiently binds Tat, resulting in the appearance of a slower-migrating Tat-TAR complex. This Tat-TAR complex is not formed with the mutant TAR RNA, demonstrating that the TAR mutations do effectively prevent the binding of Tat.
To turn this TAR-mutated LTR into a DOX-inducible SIV-rtTA promoter, we introduced two tetO elements between the NF-κB and Sp1 binding sites (Fig. 3A and 4C). This arrangement is effective in the DOX-dependent HIV-rtTA variant. We used the optimized 2ΔtetO configuration with a truncated spacer between the rtTA-binding sites (53), which was frequently observed during the evolution of HIV-rtTA and significantly improved virus replication (54). To test the DOX responsiveness of the SIV-rtTA promoter, a plasmid in which the new promoter controlled the expression of luciferase was constructed and cotransfected with an rtTA-expressing plasmid into C33A cells. After 2 days of culturing with 0 to 1,000 ng/ml DOX, we measured the intracellular luciferase level, which reflects promoter activity (Fig. 3C). This SIV-rtTA promoter was found to be inactive in the absence of DOX, and the activity gradually increased with an increasing DOX level. Similar promoter-reporter constructs with a wild-type TAR sequence (TARwt) or with the HIV-rtTA promoter, which were included as controls, show similar DOX-dependent activities (Fig. 3C). To demonstrate that the SIV-rtTA promoter is not responsive to Tat, we transfected C33A cells with the LTR-luciferase construct and 0 to 50 ng SIV Tatwt-expressing plasmid in which the Tat exon 1 and exon 2 sequences of SIVmac239 were fused and placed downstream of a cytomegalovirus promoter. Two days after transfection, the SIV-rtTA promoter activity was analyzed by measuring the intracellular luciferase activity (Fig. 3D). Whereas the SIV-rtTA construct did not respond to Tat, the control SIV-rtTA-TARwt construct showed increased activity with an increasing amount of Tat. An HIV-1 LTR-luciferase construct with a wild-type TAR hairpin similarly responded to SIV Tat. Cotransfection of the promoter constructs with an HIV-1 Tat expression plasmid revealed that the SIV-rtTA promoter is also not responsive to HIV-1 Tat, whereas the SIV-rtTA-TARwt and HIV-1 constructs are (Fig. 3E). This DOX-responsive, Tat-independent SIV-rtTA promoter was introduced in both the 5′ and 3′ LTRs of the SIVmac239 genome (Fig. 1), such that this promoter will be stably inherited in the viral progeny.
FIG. 3.
Expression from the SIV-rtTA LTR promoter is controlled by DOX. (A) To turn the SIV LTR into a DOX-inducible promoter, we mutated TAR and introduced two tetO elements between the NF-κB and Sp1 binding sites. This arrangement had previously been shown to be effective in the DOX-dependent HIV-rtTA variant. (B) Mutation of Tat. The SIVmac239 Tat protein consists of 130 amino acids and has a modular structure similar to that of the HIV-1 Tat protein, which consists of 86 to 101 amino acids (depending on the viral isolate). Both proteins have a transcription activation domain that can be subdivided in an N-terminal acidic domain, a cysteine-rich domain, a central core domain, and an RNA-binding domain consisting of a stretch of positively charged amino acids and therefore termed the basic domain. For the construction of HIV-rtTA, we previously inactivated HIV-1 Tat through the introduction of a tyrosine-to-alanine substitution at position 26 (Y26A) in the cysteine-rich domain. This cysteine-rich domain is highly conserved, and the tyrosine at position 26 in HIV-1 Tat corresponds to the tyrosine at position 55 in SIVmac239 Tat. We therefore introduced the Y55A mutation in the SIVmac239 Tat open reading frame. This mutation did not affect any other gene or known underlying sequence element. (C) C33A cells were transfected with a plasmid in which the expression of firefly luciferase was controlled by the LTR promoter of either SIV-rtTA, SIV-rtTA-TARwt, or HIV-rtTA and an rtTA-expressing plasmid. Transfected cells were cultured with 0 to 1,000 ng/ml DOX. (D to F) Cells were transfected with a plasmid in which the expression of firefly luciferase was controlled by the LTR promoter of either SIV-rtTA, SIV-rtTA-TARwt, or HIV-1 and 0 to 50 ng of a plasmid expressing wild-type SIV Tat (D), HIV-1 Tat (E), or Y55A-mutated SIV Tat (F). The intracellular luciferase level, which reflects promoter activity, was measured at 2 days after transfection. The error bars represent the standard deviations for two to five experiments (n.d., not determined).
FIG. 4.
Insertion of the rtTA gene and deletion of U3 sequences. In SIVmac239, the 5′ end of the nef open reading frame overlaps the env gene, while the 3′ end overlaps the U-box (U), the PPT, and the U3 region of the LTR promoter (including the attachment sequence required for integration; att). The SIV-rtTA modifications are shown schematically (A) and in detail (B and C). In SIV-rtTA, we replaced the nef sequences downstream of the env gene and upstream of the U-box with the rtTA gene. In addition, we mutated the original AUGNef translation start codon and three downstream AUG codons (AUGII-IV) that precede the new AUGrtTA start codon on the spliced Nef transcript (AUG codons are underlined in panel B). The four AUG mutations (boxed in gray in panel B) were chosen to be synonymous in the env open reading frame and thus not to affect the Env protein. The SIV-rtTA-Envwt variant does not carry these four AUG mutations and will produce a Nef-rtTA fusion protein in which 60 Nef amino acids are fused to the N terminus of the rtTA protein. In the SIV-rtTA-Envwt-stop variant, a translation stop codon was introduced between these Env and rtTA sequences (translation stop codons are indicated with *; mutations are boxed in gray). Accordingly, translation starting at AUGNef will result in the production of a 57-amino-acid Nef polypeptide, while the reinitiation of translation at the AUGrtTA will result in the production of rtTA. In SIV-rtTA-TARwt, the wild-type TAR sequence was reintroduced. In the SIV-rtTA-ΔU3 variants, the Nef-U3 sequences present between the att sequence and either the NF-κB site (SIV-rtTA-ΔU3-380) or the tetO elements (SIV-rtTA-ΔU3-405) were deleted. The numbering is according to the SIVmac239 proviral genome sequence in GenBank/EMBL (accession number M33262; gi 334647).
Inactivation of Tat.
Since the tat gene in the SIVmac239 genome overlaps with the vpr, rev, and env genes, we decided to minimally mutate the Tat open reading frame. For the construction of HIV-rtTA, we had previously introduced a tyrosine-to-alanine substitution at position 26 (Y26A) in the cysteine-rich domain of HIV-1 Tat, which effectively inactivated the transcription function (72). Despite differences in sequence and size between the HIV-1 and SIVmac239 Tat proteins, both proteins have similar modular structures (Fig. 3B). The cysteine-rich domain is highly conserved, and the tyrosine at position 26 in HIV-1 Tat corresponds to the tyrosine at position 55 in SIVmac239 Tat. We therefore introduced the Y55A mutation in the SIVmac239 Tat open reading frame. This mutation did not affect any other gene or any known underlying sequence element. To analyze the effect of this mutation on transcriptional activity, we made a Y55A-mutated SIVmac239 Tat expression plasmid (SIV Tatm), which was cotransfected with the SIV-rtTA-TARwt luciferase reporter construct into C33A cells. This reporter construct did not respond to SIV Tatm (Fig. 3F), whereas it was responsive to SIV Tatwt and HIV-1 Tat (Fig. 3D and E). Similarly, the HIV-1 luciferase construct was not activated by SIV Tatm (Fig. 3F), whereas it was activated by SIV Tatwt and HIV-1 Tat (Fig. 3D and E). These results demonstrate that the Y55A mutation effectively inactivated the transcription activation function of SIV Tat.
Insertion of the rtTA gene.
In SIV and HIV, the deletion of the accessory Nef function does not abolish replication, and the spontaneous deletion of nef sequences has been observed in vivo (22, 29, 45, 46, 49, 64). We therefore introduced the optimized rtTAF86Y A209T gene at the position of the nef gene (Fig. 1). This rtTA variant was isolated upon the evolution of HIV-rtTA and is more active and DOX sensitive than the original rtTA (28). In SIVmac239, the 5′ end of the nef open reading frame overlaps the env gene, while the 3′ end overlaps the U-box (39), the polypurine tract (PPT), and the U3 region of the LTR promoter (Fig. 4A). We replaced the nef sequences downstream of the env gene and upstream of the U-box/PPT/U3 region with the rtTA coding sequence. In addition, we mutated the original AUGNef translation start codon and three downstream AUG codons (AUGII-IV) that precede the new AUGrtTA start codon on the spliced Nef transcript (Fig. 4B). The four AUG mutations were chosen to be synonymous in the env open reading frame and thus do not affect the Env protein. As a result, the spliced RNA transcript that formerly encoded Nef now encoded rtTA. To demonstrate that SIV-rtTA can indeed express rtTA, we transfected the SIV-rtTA plasmid into HeLa X1/6 cells, which contain a chromosomally integrated copy of the DOX/rtTA-responsive tetO promoter/luciferase reporter construct (6). When cells were cultured for 2 days without DOX, no luciferase expression was detectable, while a high luciferase level was measured with DOX, which demonstrates that rtTA is properly expressed (data not shown).
Designed SIV-rtTA variant does not replicate in T cells.
Replication of the constructed SIV-rtTA variant was tested in the PM1 T-cell line, a clonal derivative of HUT78, which expresses CCR5 and allows the replication of the parental SIVmac239 strain (19, 52) (Fig. 5). Cells were transfected with the SIV-rtTA plasmid and cultured in the presence and absence of DOX. As intended, the virus did not replicate in the absence of DOX. However, the virus did also not show any replication in the presence of DOX. In contrast, the SIVmac239 control replicated efficiently and independent of DOX, and the HIV-rtTA control showed efficient replication only in the presence of DOX. Similarly, SIV-rtTA showed no replication in other T-cell lines (174xCEM, MT-2, MT-4, MOLT-CCR5, SupT1), whereas the SIVmac239 and HIV-rtTA controls did replicate in these cells (data not shown). These results are in strong contrast with our previous HIV-rtTA studies. The original HIV-rtTA construct did replicate, although considerably less efficiently than wild-type HIV-1. This poorly replicating HIV-rtTA could however evolve to a better-replicating variant through modifications in the tetO configuration and mutations in the rtTA gene (53, 54). The inability of SIV-rtTA to replicate came as a surprise, since we introduced the optimized tetO conformation and rtTA gene, and the modifications in TAR and Tat resemble the mutations that we introduced in HIV-rtTA. SIV-rtTA did also not show any replication in long-term cell cultures, indicating that one or several of the modifications affect replication so severely that evolution of the virus is prevented.
FIG. 5.
Replication of SIV-rtTA variants. PM1 T cells were transfected with plasmids encoding the SIV-rtTA variants SIVmac239 and HIV-rtTA. Cells were cultured in the presence (1 μg/ml) and absence of DOX, and viral replication was monitored by measuring the RT level in the culture supernatant. Similar DOX-dependent replication of SIV-rtTA-Tatwt and lack of replication of the other SIV-rtTA variants was observed in independent experiments.
SIV-rtTA requires Tat for replication.
To find out which SIV-rtTA modification obstructed viral replication, we made new SIV-rtTA variants (Fig. 4) in which we restored either the wild-type TAR sequence (SIV-rtTA-TARwt), the wild-type Tat sequence (SIV-rtTA-Tatwt), or the AUGNef and AUGII-IV codons in the nef-env overlap (SIV-rtTA-Envwt). The latter back-mutations will result in the production of a Nef-rtTA fusion protein in which 60 Nef amino acids are fused to the N terminus of the rtTA protein. Since the effect of such a fusion on rtTA activity is unknown, we made an additional variant in which a translation stop codon was introduced between the Env and rtTA sequences (SIV-rtTA-Envwt-stop). In this variant, translation starting at AUGNef will result in the production of a 57-amino-acid Nef polypeptide, while reinitiation of translation at AUGrtTA will result in the production of rtTA. Since the efficiency of this reinitiation step is expected to be relatively low, we realized that this variant might not produce sufficient rtTA to allow SIV-rtTA replication.
The introduction of rtTA and tetO elements in SIV-rtTA had increased the size of the viral RNA genome from 9637 to 10270 nt [excluding the poly(A) tail]. Since we could not exclude the possibility that this genome was too large to be efficiently packaged into virions, we made two additional SIV-rtTA variants from which we deleted the Nef-U3 sequences present between the attachment sequence required for integration (30) and either the NF-κB site (SIV-rtTA-ΔU3-380; Fig. 4C) or the tetO elements (SIV-rtTA-ΔU3-405; Fig. 4C), which reduced the genome size to 9890 or 9865 nt, respectively. Deletions in this part of the U3 region have previously been observed for Nef-deleted SIV and HIV variants in vivo (22, 29, 45, 46, 49, 64), which demonstrates that such deletions do not abolish viral replication.
We assayed the replication of all variants in PM1 cells (Fig. 5). Only the SIV-rtTA-Tatwt variant showed efficient replication in the presence of DOX and no replication in the absence of DOX. The other newly constructed variants did not show any replication with or without DOX. These results demonstrate that the Y55A mutation in Tat is responsible for the replication defect of the original SIV-rtTA. Restoration of the wild-type Tat sequence resulted in an SIV variant that replicated efficiently and exclusively in the presence of DOX.
Tat does not affect SIV-rtTA gene expression.
This SIV-rtTA-Tatwt variant carries the TAR mutations that abolish Tat-mediated activation of transcription (Fig. 3D). To demonstrate that the gene expression of this virus is controlled by the rtTA-tetO axis and therefore dependent on DOX, we transfected the SIV-rtTA-Tatwt molecular clone into C33A cells and monitored virus production by measuring the RT and CA-p27 levels in the culture supernatant after 2 days (Fig. 6). When cells were cultured without DOX, virus production was low, whereas high virus production was observed when cells were cultured with DOX. The SIVmac239 control showed high virus production both with and without DOX. This result demonstrates that the gene expression of SIV-rtTA-Tatwt is indeed dependent on DOX, which is in agreement with the strict DOX control of viral replication. The original SIV-rtTA (with the Y55A Tat mutation; Tatm in Fig. 6) showed a similarly high level of virus production with DOX, and a low level without DOX, compared with the SIV-rtTA-Tatwt variant. Consistent with this, Northern blot analysis of intracellular RNA revealed similar levels of spliced and unspliced viral RNA in the cells transfected with SIV-rtTA or SIV-rtTA-Tatwt and cultured with DOX (data not shown). These results demonstrate that wild-type Tat does not contribute to gene expression in the context of the TAR-mutated SIV-rtTA genome, in which transcription is controlled by DOX/rtTA. However, SIV-rtTA replication is dependent on wild-type Tat, suggesting that Tat has a second function in SIV replication other than the activation of transcription through TAR binding.
FIG. 6.
SIV-rtTA-Tatwt gene expression is controlled by DOX. C33A cells were transfected with the SIV-rtTA (Tatm), SIV-rtTA-Tatwt, or SIVmac239 molecular clone and cultured without and with 1 μg/ml DOX for 2 days. Virus production was monitored by measuring the RT (right) and CA-p27 (left) levels in the culture supernatant. The error bars represent the standard deviations from two experiments (n.d., not determined).
DISCUSSION
We here present a conditionally live SIVmac239 variant that replicates exclusively in the presence of DOX. The natural Tat-TAR transcription control mechanism in this SIV-rtTA variant was inactivated and functionally replaced by the rtTA and tetO components of the DOX-inducible Tet-On regulatory system. Since the rtTA protein can bind the tetO elements in the LTR promoter and activate transcription only in the presence of DOX, viral replication can be switched on and off by DOX administration and withdrawal, respectively. This DOX-controlled SIV variant will allow in vivo studies in macaques to determine the efficacy and safety of a conditionally live virus as an AIDS vaccine.
Although transcription from the SIV-rtTA promoter is fully controlled by DOX-rtTA, the virus was found to require a wild-type Tat gene for replication. This requirement indicates that Tat has a second essential function in SIV replication in addition to its role in transcription. Tat did not affect SIV-rtTA gene expression, suggesting that Tat is required in another step of the viral replication cycle. It seems unlikely that this second Tat function involves binding to TAR RNA, since the Tat-TAR binding study demonstrated that the mutations in the SIV-rtTA TAR element prevent Tat binding. Multiple additional roles for Tat have been suggested in HIV-1 biology; for example, Tat has been reported to affect mRNA capping, splicing, and translation (14, 18, 20, 21, 23, 67, 69, 78), to stimulate reverse transcription (1, 36, 37, 42, 43, 71), and to suppress RNA interference (7, 35). Moreover, Huang et al. demonstrated that HIV-1 variants regulated by the Gal4-VP16 trans-activator protein require Tat for infectivity (38), and we recently observed that the introduction of a frameshift mutation in the HIV-1 Tat gene, which prevents Tat production, abolished HIV-rtTA replication. These studies suggest that Tat has also a second, nontranscriptional function in HIV-1 replication. The introduction of a Y26A mutation in HIV-1 Tat, which corresponds to the Y55A mutation in SIV Tat, impaired the transcription function but did not hinder HIV-rtTA replication (73). These results suggest that the Y26A mutation affects the first but not the second function of Tat in HIV-1 biology.
SIV-rtTA, like wild-type SIV, is subject to spontaneous evolution during replication due to the error-prone reverse transcription process and a continuous selection pressure. We have extensively studied the evolutionary possibilities of HIV-rtTA in long-term cultures and demonstrated that the components of the Tet-On system, which are essential for virus replication, were stably maintained in the viral genome. In fact, we observed mutations in both the rtTA gene and the tetO elements that significantly improved virus replication (28, 53, 54, 80). We have also observed specific mutations in rtTA that reduced DOX control (81) and developed a novel rtTA variant that blocks this undesired evolutionary route (79, 81). This stabilized rtTA variant will similarly increase the genetic stability of SIV-rtTA. We never observed a reversion to the Tat-TAR mechanism of transcription control in the HIV-rtTA evolution experiments, and the virus stably maintained the introduced mutations in Tat and TAR. Unlike HIV-rtTA, the replicating SIV-rtTA variant expresses wild-type Tat protein, and the reversion of the TAR mutations during evolution would restore the Tat-TAR control mechanism. However, this evolution route would require multiple nucleotide substitutions in the TAR element, which is not likely to occur. Indeed, we never observed restoration of the Tat-TAR axis in multiple long-term cultures of SIV-rtTA. Nevertheless, we could further reduce the likelihood of this unwanted evolution route by introducing novel mutations in Tat that would inactivate its first function (activation of transcription) but not its second function. However, such Tat mutations remain to be identified. Alternatively, this evolution route can be blocked by the complete or partial deletion of TAR (e.g., only stem-loops 1 and 2), as we recently showed that the complete removal of TAR in HIV-rtTA does not significantly affect replication (25).
Why the live attenuated virus vaccines are so much more effective than other vaccine approaches remains to be determined. Whereas other vaccines contain or produce only one, or a subset, of the viral proteins, the live attenuated viruses express most of the viral antigens and may therefore elicit broad immune responses. Furthermore, since the live attenuated viruses replicate in a similar way and in the same cells as the wild-type virus, they may induce protective immune responses at the site of natural infection. Understanding the mechanism for protection conferred by live attenuated SIV will obviously facilitate the development of improved HIV vaccines (48, 76). The DOX-dependent SIV-rtTA variant seems to be an ideal tool to study the immune correlates of protection, since both the level and the duration of replication can be controlled by DOX administration. Such studies may reveal the critical information needed for the design of an HIV vaccine that is safe and equally effective as a live attenuated virus.
Acknowledgments
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: p239SpSp5′ and pSIVmac239ΔNef deletion mutant, from Ronald Desrosiers, and PM1, from Marvin Reitz. The full-length SIVmac239 clone was kindly provided by Yongjun Guan and Mark A. Wainberg (McGill University AIDS Centre, Montreal, Quebec, Canada). We thank FIT Biotech Oyj Plc, the Centralised Facility for AIDS Reagents supported by EU Programme EVA/MRC (contract QLKCT-1999-00609), and the UK Medical Research Council for the gift of the purified His-tagged SIVmac Tat protein.
This research was funded by the Dutch AIDS Foundation (Aids Fonds Netherlands grants 7007 and 2005022) and the Fondation pour la Recherche Medicale (postdoctoral fellowship to Mireille Centlivre).
Footnotes
Published ahead of print on 1 August 2007.
REFERENCES
- 1.Apolloni, A., C. W. Hooker, J. Mak, and D. Harrich. 2003. Human immunodeficiency virus type 1 protease regulation of Tat activity is essential for efficient reverse transcription and replication. J. Virol. 77:9912-9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auersperg, N. 1964. Long-term cultivation of hypodiploid human tumor cells. J. Natl. Cancer Inst. 32:135-163. [PubMed] [Google Scholar]
- 3.Baba, T. W., Y. S. Jeong, D. Penninck, R. Bronson, M. F. Greene, and R. M. Ruprecht. 1995. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science 267:1820-1825. [DOI] [PubMed] [Google Scholar]
- 4.Baba, T. W., V. Liska, A. H. Khimani, N. B. Ray, P. J. Dailey, D. Penninck, R. Bronson, M. F. Greene, H. M. McClure, L. N. Martin, and R. M. Ruprecht. 1999. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat. Med. 5:194-203. [DOI] [PubMed] [Google Scholar]
- 5.Baron, U., and H. Bujard. 2000. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 327:401-421. [DOI] [PubMed] [Google Scholar]
- 6.Baron, U., M. Gossen, and H. Bujard. 1997. Tetracycline-controlled transcription in eukaryotes: novel transactivators with graded transactivation potential. Nucleic Acids Res. 25:2723-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennasser, Y., S. Y. Le, M. Benkirane, and K. T. Jeang. 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22:607-619. [DOI] [PubMed] [Google Scholar]
- 8.Berkhout, B. 1992. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 20:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkhout, B., A. Gatignol, J. Silver, and K. T. Jeang. 1990. Efficient trans-activation by the HIV-2 Tat protein requires a duplicated TAR RNA structure. Nucleic Acids Res. 18:1839-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkhout, B., and K. T. Jeang. 1989. trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: a quantitative analysis. J. Virol. 63:5501-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berkhout, B., G. Marzio, and K. Verhoef. 2002. Control over HIV-1 replication by an antibiotic; a novel vaccination strategy with a drug-dependent virus. Virus Res. 82:103-108. [DOI] [PubMed] [Google Scholar]
- 12.Berkhout, B., R. H. Silverman, and K. T. Jeang. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. [DOI] [PubMed] [Google Scholar]
- 13.Berkhout, B., K. Verhoef, J. L. B. van Wamel, and B. Back. 1999. Genetic instability of live, attenuated HIV-1 vaccine strains. J. Virol. 73:1138-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berro, R., K. Kehn, C. de la Fuente, A. Pumfery, R. Adair, J. Wade, A. M. Colberg-Poley, J. Hiscott, and F. Kashanchi. 2006. Acetylated Tat regulates human immunodeficiency virus type 1 splicing through its interaction with the splicing regulator p32. J. Virol. 80:3189-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1999. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl. Acad. Sci. USA 96:7791-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brady, J., and F. Kashanchi. 2005. Tat gets the “green” light on transcription initiation. Retrovirology 2:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakrabarti, L. A., K. J. Metzner, T. Ivanovic, H. Cheng, J. Louis-Virelizier, R. I. Connor, and C. Cheng-Mayer. 2003. A truncated form of Nef selected during pathogenic reversion of simian immunodeficiency virus SIVmac239Δnef increases viral replication. J. Virol. 77:1245-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang, Y. N., D. J. Kenan, J. D. Keene, A. Gatignol, and K.-T. Jeang. 1994. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J. Virol. 68:7008-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, Z., A. Gettie, D. D. Ho, and P. A. Marx. 1998. Primary SIVsm isolates use the CCR5 coreceptor from sooty mangabeys naturally infected in West Africa: a comparison of coreceptor usage of primary SIVsm, HIV-2, and SIVmac. Virology 246:113-124. [DOI] [PubMed] [Google Scholar]
- 20.Chiu, Y. L., E. Coronel, C. K. Ho, S. Shuman, and T. M. Rana. 2001. HIV-1 Tat protein interacts with mammalian capping enzyme and stimulates capping of TAR RNA. J. Biol. Chem. 276:12959-12966. [DOI] [PubMed] [Google Scholar]
- 21.Chiu, Y. L., C. K. Ho, N. Saha, B. Schwer, S. Shuman, and T. M. Rana. 2002. Tat stimulates cotranscriptional capping of HIV mRNA. Mol. Cell 10:585-597. [DOI] [PubMed] [Google Scholar]
- 22.Churchill, M. J., D. I. Rhodes, J. C. Learmont, J. S. Sullivan, S. L. Wesselingh, I. R. Cooke, N. J. Deacon, and P. R. Gorry. 2006. Longitudinal analysis of human immunodeficiency virus type 1 nef/long terminal repeat sequences in a cohort of long-term survivors infected from a single source. J. Virol. 80:1047-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cullen, B. R. 1986. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell 46:973-982. [DOI] [PubMed] [Google Scholar]
- 24.Das, A. T., C. E. Baldwin, M. Vink, and B. Berkhout. 2005. Improving the safety of a conditional-live human immunodeficiency virus type 1 vaccine by controlling both gene expression and cell entry. J. Virol. 79:3855-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das, A. T., A. Harwig, M. M. Vrolijk, and B. Berkhout. 2007. The TAR hairpin of human immunodeficiency virus type 1 can be deleted when not required for Tat-mediated activation of transcription. J. Virol. 81:7742-7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das, A. T., K. Verhoef, and B. Berkhout. 2004. A conditionally replicating virus as a novel approach toward an HIV vaccine. Methods Enzymol. 388:359-379. [DOI] [PubMed] [Google Scholar]
- 27.Das, A. T., X. Zhou, M. Vink, B. Klaver, and B. Berkhout. 2002. Conditional live virus as a novel approach towards a safe live attenuated HIV vaccine. Expert Rev. Vaccines 1:293-301. [DOI] [PubMed] [Google Scholar]
- 28.Das, A. T., X. Zhou, M. Vink, B. Klaver, K. Verhoef, G. Marzio, and B. Berkhout. 2004. Viral evolution as a tool to improve the tetracycline-regulated gene expression system. J. Biol. Chem. 279:18776-18782. [DOI] [PubMed] [Google Scholar]
- 29.Deacon, N. J., A. Tsykin, A. Solomon, K. Smith, M. Ludford-Menting, D. J. Hooker, D. A. McPhee, A. L. Greenway, A. Ellett, C. Chatfield, V. A. Lawson, S. Crowe, A. Maerz, S. Sonza, J. Learmont, J. S. Sullivan, A. Cunningham, D. Dwyer, D. Dowton, and J. Mills. 1995. Genomic structure of an attenuated quasi species of HIV-1 from blood transfusion donor and recipients. Science 270:988-991. [DOI] [PubMed] [Google Scholar]
- 30.Du, Z., P. O. Ilyinskii, K. Lally, R. C. Desrosiers, and A. Engelman. 1997. A mutation in integrase can compensate for mutations in the simian immunodeficiency virus att site. J. Virol. 71:8124-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans, D. T., J. E. Bricker, H. B. Sanford, S. Lang, A. Carville, B. A. Richardson, M. Piatak, Jr., J. D. Lifson, K. G. Mansfield, and R. C. Desrosiers. 2005. Immunization of macaques with single-cycle simian immunodeficiency virus (SIV) stimulates diverse virus-specific immune responses and reduces viral loads after challenge with SIVmac239. J. Virol. 79:7707-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Martinez, L. F., G. Mavankal, P. Peters, F. Wu-Baer, and R. B. Gaynor. 1995. Tat functions to stimulate the elongation properties of transcription complexes paused by the duplicated TAR RNA element of human immunodeficiency virus 2. J. Mol. Biol. 254:350-363. [DOI] [PubMed] [Google Scholar]
- 33.Gibbs, J. S., D. A. Regier, and R. C. Desrosiers. 1994. Construction and in vitro properties of SIVmac mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retrovir. 10:607-616. [DOI] [PubMed] [Google Scholar]
- 34.Guan, Y., J. B. Whitney, K. Diallo, and M. A. Wainberg. 2000. Leader sequences downstream of the primer binding site are important for efficient replication of simian immunodeficiency virus. J. Virol. 74:8854-8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haasnoot, J., W. de Vries, E. J. Geutjes, M. Prins, P. de Haan, and B. Berkhout. 2007. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathogens 3:e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrich, D., C. Ulich, L. F. Garcia-Martinez, and R. B. Gaynor. 1997. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 16:1224-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hooker, C. W., J. Scott, A. Apolloni, E. Parry, and D. Harrich. 2002. Human immunodeficiency virus type 1 reverse transcription is stimulated by tat from other lentiviruses. Virology 300:226-235. [DOI] [PubMed] [Google Scholar]
- 38.Huang, L. M., A. Joshi, R. Willey, J. Orenstein, and K. T. Jeang. 1994. Human immunodeficiency viruses regulated by alternative trans-activators: genetic evidence for a novel non-transcriptional function of Tat in virion infectivity. EMBO J. 13:2886-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ilyinskii, P. O., and R. C. Desrosiers. 1998. Identification of a sequence element immediately upstream of the polypurine tract that is essential for replication of simian immunodeficiency virus. EMBO J. 17:3766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeeninga, R. E., M. Hoogenkamp, M. Armand-Ugon, M. de Baar, K. Verhoef, and B. Berkhout. 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J. Virol. 74:3740-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson, R. P., and R. C. Desrosiers. 1998. Protective immunity induced by live attenuated simian immunodeficiency virus. Curr. Opin. Immunol. 10:436-443. [DOI] [PubMed] [Google Scholar]
- 42.Kameoka, M., M. Morgan, M. Binette, R. S. Russell, L. Rong, X. Guo, A. Mouland, L. Kleiman, C. Liang, and M. A. Wainberg. 2002. The Tat protein of human immunodeficiency virus type 1 (HIV-1) can promote placement of tRNA primer onto viral RNA and suppress later DNA polymerization in HIV-1 reverse transcription. J. Virol. 76:3637-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kameoka, M., L. Rong, M. Gotte, C. Liang, R. S. Russell, and M. A. Wainberg. 2001. Role for human immunodeficiency virus type 1 Tat protein in suppression of viral reverse transcriptase activity during late stages of viral replication. J. Virol. 75:2675-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kestler, H., T. Kodama, D. Ringler, M. Marthas, N. Pedersen, A. Lackner, D. Regier, P. Sehgal, M. Daniel, N. King, and R. Desrosiers. 1990. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science 248:1109-1112. [DOI] [PubMed] [Google Scholar]
- 45.Kirchhoff, F., T. Greenough, D. B. Brettler, J. L. Sullivan, and R. C. Desrosiers. 1995. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 332:228-232. [DOI] [PubMed] [Google Scholar]
- 46.Kirchhoff, F., H. W. Kestler III, and R. C. Desrosiers. 1994. Upstream U3 sequences in simian immunodeficiency virus are selectively deleted in vivo in the absence of an intact nef gene. J. Virol. 68:2031-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiselyeva, Y., Y. Ito, R. G. Lima, J. C. Grivel, A. T. Das, B. Berkhout, and L. B. Margolis. 2004. Depletion of CD4 T lymphocytes in human lymphoid tissue infected ex vivo with doxycycline-dependent HIV-1. Virology 328:1-6. [DOI] [PubMed] [Google Scholar]
- 48.Koff, W. C., P. R. Johnson, D. I. Watkins, D. R. Burton, J. D. Lifson, K. J. Hasenkrug, A. B. McDermott, A. Schultz, T. J. Zamb, R. Boyle, and R. C. Desrosiers. 2006. HIV vaccine design: insights from live attenuated SIV vaccines. Nat. Immunol. 7:19-23. [DOI] [PubMed] [Google Scholar]
- 49.Kondo, M., T. Shima, M. Nishizawa, K. Sudo, S. Iwamuro, T. Okabe, Y. Takebe, and M. Imai. 2005. Identification of attenuated variants of HIV-1 circulating recombinant form 01_AE that are associated with slow disease progression due to gross genetic alterations in the nef/long terminal repeat sequences. J. Infect. Dis. 192:56-61. [DOI] [PubMed] [Google Scholar]
- 50.Lifson, J. D., J. L. Rossio, M. Piatak, Jr., T. Parks, L. Li, R. Kiser, V. Coalter, B. Fisher, B. M. Flynn, S. Czajak, V. M. Hirsch, K. A. Reimann, J. E. Schmitz, J. Ghrayeb, N. Bischofberger, M. A. Nowak, R. C. Desrosiers, and D. Wodarz. 2001. Role of CD8+ lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J. Virol. 75:10187-10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lohman, B. L., M. B. McChesney, C. J. Miller, E. McGowan, S. M. Joye, K. K. van Rompay, E. Reay, L. Antipa, N. C. Pedersen, and M. L. Marthas. 1994. A partially attenuated simian immunodeficiency virus induces host immunity that correlates with resistance to pathogenic virus challenge. J. Virol. 68:7021-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lusso, P., F. Cocchi, C. Balotta, P. D. Markham, A. Louie, P. Farci, R. Pal, R. C. Gallo, and M. S. Reitz, Jr. 1995. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 69:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marzio, G., K. Verhoef, M. Vink, and B. Berkhout. 2001. In vitro evolution of a highly replicating, doxycycline-dependent HIV for applications in vaccine studies. Proc. Natl. Acad. Sci. USA 98:6342-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marzio, G., M. Vink, K. Verhoef, A. de Ronde, and B. Berkhout. 2002. Efficient human immunodeficiency virus replication requires a fine-tuned level of transcription. J. Virol. 76:3084-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maudru, T., and K. W. Peden. 1998. Adaptation of the fluorogenic 5′-nuclease chemistry to a PCR-based reverse transcriptase assay. BioTechniques 25:972-975. [DOI] [PubMed] [Google Scholar]
- 56.Mikaelian, I., and A. Sergeant. 1992. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 20:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mills, J., R. Desrosiers, E. Rud, and N. Almond. 2000. Live attenuated HIV vaccines: proposal for further research and development. AIDS Res. Hum. Retrovir. 16:1453-1461. [DOI] [PubMed] [Google Scholar]
- 58.Parada, C. A., and R. G. Roeder. 1996. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature 384:375-378. [DOI] [PubMed] [Google Scholar]
- 59.Park, I. W., R. Steen, and Y. Li. 1991. Characterization of multiple mRNA species of simian immunodeficiency virus from macaques in a CD4+ lymphoid cell line. J. Virol. 65:2987-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peden, K., M. Emerman, and L. Montagnier. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661-672. [DOI] [PubMed] [Google Scholar]
- 61.Raha, T., S. W. Cheng, and M. R. Green. 2005. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 3:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Regier, D. A., and R. C. Desrosiers. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 6:1221-1231. [DOI] [PubMed] [Google Scholar]
- 63.Rhim, H., and A. P. Rice. 1994. Functional significance of the dinucleotide bulge in stem-loop1 and stem-loop2 of HIV-2 TAR RNA. Virology 202:202-211. [DOI] [PubMed] [Google Scholar]
- 64.Rhodes, D. I., L. Ashton, A. Solomon, A. Carr, D. Cooper, J. Kaldor, N. Deacon, et al. 2000. Characterization of three nef-defective human immunodeficiency virus type 1 strains associated with long-term nonprogression. J. Virol. 74:10581-10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Richter, S., Y. H. Ping, and T. M. Rana. 2002. TAR RNA loop: a scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl. Acad. Sci. USA 99:7928-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruijter, J. M., H. H. Thygesen, O. J. Schoneveld, A. T. Das, B. Berkhout, and W. H. Lamers. 2006. Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.SenGupta, D. N., B. Berkhout, A. Gatignol, A. M. Zhou, and R. H. Silverman. 1990. Direct evidence for translational regulation by leader RNA and Tat protein of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 87:7492-7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith, S. M., M. Khoroshev, P. A. Marx, J. Orenstein, and K. T. Jeang. 2001. Constitutively dead, conditionally live HIV-1 genomes. Ex vivo implications for a live virus vaccine. J. Biol. Chem. 276:32184-32190. [DOI] [PubMed] [Google Scholar]
- 69.Svitkin, Y. V., A. Pause, and N. Sonenberg. 1994. LA autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 RNA. J. Virol. 68:7001-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan, R., A. Brodsky, J. R. Williamson, and A. D. Frankel. 1997. RNA recognition by HIV-1 Tat and Rev. Semin. Virol. 8:186-193. [Google Scholar]
- 71.Ulich, C., A. Dunne, E. Parry, C. W. Hooker, R. B. Gaynor, and D. Harrich. 1999. Functional domains of Tat required for efficient human immunodeficiency type 1 reverse transcription. J. Virol. 73:2499-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Verhoef, K., M. Koper, and B. Berkhout. 1997. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology 237:228-236. [DOI] [PubMed] [Google Scholar]
- 73.Verhoef, K., G. Marzio, W. Hillen, H. Bujard, and B. Berkhout. 2001. Strict control of human immunodeficiency virus type 1 replication by a genetic switch: Tet for Tat. J. Virol. 75:979-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei, P., M. E. Garber, S.-M. Fang, W. H. Fisher, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 75.Whatmore, A. M., N. Cook, G. A. Hall, S. Sharpe, E. W. Rud, and M. P. Cranage. 1995. Repair and evolution of nef in vivo modulates simian immunodeficiency virus virulence. J. Virol. 69:5117-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Whitney, J. B., and R. M. Ruprecht. 2004. Live attenuated HIV vaccines: pitfalls and prospects. Curr. Opin. Infect. Dis. 17:17-26. [DOI] [PubMed] [Google Scholar]
- 77.Wyand, M. S., K. H. Manson, M. Garcia-Moll, D. Montefiori, and R. C. Desrosiers. 1996. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J. Virol. 70:3724-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou, M., L. Deng, F. Kashanchi, J. N. Brady, A. J. Shatkin, and A. Kumar. 2003. The Tat/TAR-dependent phosphorylation of RNA polymerase II C-terminal domain stimulates cotranscriptional capping of HIV-1 mRNA. Proc. Natl. Acad. Sci. USA 100:12666-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou, X., M. Vink, B. Berkhout, and A. T. Das. 2006. Modification of the Tet-On regulatory system prevents the conditional-live HIV-1 variant from losing doxycycline-control. Retrovirology 3:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou, X., M. Vink, B. Klaver, B. Berkhout, and A. T. Das. 2006. Optimization of the Tet-On system for regulated gene expression through viral evolution. Gene Ther. 13:1382-1390. [DOI] [PubMed] [Google Scholar]
- 81.Zhou, X., M. Vink, B. Klaver, K. Verhoef, G. Marzio, A. T. Das, and B. Berkhout. 2006. The genetic stability of a conditional-live HIV-1 variant can be improved by mutations in the Tet-On regulatory system that restrain evolution. J. Biol. Chem. 281:17084-17091. [DOI] [PubMed] [Google Scholar]