Abstract
Alpha/beta interferon (IFN-α/β) produces antiviral effects through upregulation of many interferon-stimulated genes (ISGs) whose protein products are effectors of the antiviral state. Previous data from our laboratory have shown that IFN-α/β can limit Sindbis virus (SB) replication through protein kinase R (PKR)-dependent and PKR-independent mechanisms and that one PKR-independent mechanism inhibits translation of the infecting virus genome (K. D. Ryman et al., J. Virol. 79:1487-1499, 2005). Further, using Affymetrix microarray technology, we identified 44 genes as candidates for PKR/RNase L-independent IFN-induced antiviral activities. In the current studies, we have begun analyzing these gene products for antialphavirus activity using three techniques: (i) overexpression of the protein from SB vectors and assessment of virulence attenuation in mice; (ii) overexpression of the proteins in a stable tetracycline-inducible murine fibroblast culture system and assessment of effects upon SB replication; and (iii) small interfering RNA-mediated knockdown of gene mRNA in fibroblast cultures followed by SB replication assessment as above. Tested proteins included those we hypothesized had potential to affect virus genome translation and included murine ISG20, ISG15, the zinc finger antiviral protein (ZAP), viperin, p56, p54, and p49. Interestingly, the pattern of antiviral activity for some gene products was different between in vitro and in vivo assays. Viperin and ZAP attenuated virulence most profoundly in mice. However, ISG20 and ZAP potently inhibited SB replication in vitro, whereas and viperin, p56, and ISG15 exhibited modest replication inhibition in vitro. In contrast, p54 and p49 had little to no effect in any assay.
Members of the Alphavirus genus of the family Togaviridae are causes of arthropod-borne outbreaks of encephalitis, polyarthritis, and hemorrhagic fever in humans and domesticated animals (15). Furthermore, these viruses have potential for increased numbers of cases through more widespread natural emergence (15) or malicious release in a bioterrorism attack (6, 35). Understanding the alphavirus-specific antiviral activities of the innate immune system is important to development of antiviral therapies that either stimulate or artificially mimic natural antiviral mechanisms of the host and thereby suppress alphavirus replication.
Previous studies using mice that were deficient in alpha/beta interferon (IFN-α/β) signaling indicated that this pathway was a primary protective response after alphavirus infection (1, 14, 36, 46), preventing disseminated virus replication in dendritic cells (DCs) and macrophages which, when unrestrained, resulted in rapid mortality (36). Moreover, IFN responses in normal mice against adult mouse-avirulent alphaviruses, such as Sindbis virus (SB), were so potent that virus replication was curtailed to the point that disease signs were completely prevented (36). In vitro, growth of all tested alphaviruses can be greatly suppressed (although with differing sensitivity, depending upon the virus) by the antiviral effects of IFN-α/β when it is added to cells prior to infection (3, 9, 30, 39, 46).
The canonical pathway for IFN-α/β signaling involves interaction with the heterodimeric IFN-α/β receptor (IFNAR1 and IFNAR2 subunits), activation of Jak1 and Tyk2 kinases, phosphorylation, and heterodimerization of the STAT1 and STAT2 transcription factors. Subsequently, STAT1/2 heterodimers associate with IRF9, forming the ISGF3 complex, which translocates to the nucleus and binds to IFN-stimulated response elements, resulting in transcriptional upregulation of many genes, including some that possess antiviral activities (reviewed in reference 5). However, STAT1-independent effects of IFN-α/β have also been documented (13).
Historically, the primary IFN-upregulated antiviral responses have been thought to involve interaction of the double-stranded RNA (dsRNA)-dependent protein kinase R (PKR) and 2′-5′-oligoadenylate synthetase (2′-5′-OAS) with dsRNA. For PKR, the antiviral effects have been attributed to phosphorylation of the alpha subunit of eukaryotic initiation factor 2 (eIF2α) and consequent inhibition of host cell and viral translation initiation. Additionally, activated PKR plays a role in apoptosis of infected cells (47). The IFN-inducible 2′-5′-OAS binds dsRNA and is activated to synthesize 2′-5′-linked oligoadenylates that activate constitutive RNase L to cleave host and viral single-stranded RNA. Antiviral effects toward specific viruses have also been attributed to the IFN-α/β-inducible Mx proteins (17), a tryptophan-degrading enzyme (31), adenosine deaminase (ADAR1) (25), IFN-stimulated gene 20 (ISG20) (12), p56 (45), ISG15 (23, 33), mGBP2 (7), GBP-1 (2), the APOBEC proteins (41), viperin (8), and other, as-yet-undescribed factors (39, 48).
With alphaviruses, we previously demonstrated that, while RNase L appeared to have little effect, PKR severely limited SB replication in DCs prior to IFN signaling. However, IFN signaling was sufficient to protect PKR−/− or PKR/RNase L−/− mice from lethal infection by upregulating the expression of other antiviral proteins (39). Therefore, the most potent IFN-induced protective factors are independent of PKR or RNase L and have yet to be identified. Further investigation of the mechanism of IFN-mediated inhibition of alphavirus growth in PKR/RNase L-deficient cells revealed that virus replication was inhibited at the point of translation of infecting genomes either through degradation or mislocalization of viral mRNA or through direct inhibition of translation initiation (38). In these studies, we also used a genetic approach (Affymetrix Genechip microarray) to identify IFN-induced genes as candidates for antiviral effectors in IFN-treated PKR/RNase L−/− cells, resulting in the designation of 44 candidate genes.
In the current work we have assessed the antiviral effects of a subset of the 44 genes previously identified in our microarray studies as candidates for a non-PKR/RNase L-mediated antiviral activity (38). The ISG20, ISG49, ISG54, ISG56, and viperin genes encoding the murine zinc finger antiviral protein (ZAP), the ISG20 exonuclease (10), p49, p54, and p56 putative translation inhibitors (42, 44), and viperin, a gene of unknown action that blocks viral protein production (8), were chosen due to the likelihood that they might comprise or contribute to the translation inhibition induced by IFN treatment of PKR/RNase L−/− DCs. Controls included IFN-induced proteins shown to possess antialphavirus activity (murine ZAP and ISG15) when overexpressed in vitro or in vivo (4, 23, 33, 38). Assays utilized included expression of the genes from SB double subgenomic promoter viruses, overexpression of the genes in a tetracycline (Tet)-inducible mouse embryonic fibroblast (MEF) system, and small interfering RNA (siRNA)-mediated knockdown of candidate gene mRNA abundance in MEF cells with or without IFN priming. Results indicated that ISG20, p56, ZAP, virperin, and ISG15 contributed, to varying degrees, to the antiviral state in vitro, suggesting direct antiviral roles; however, in vitro results did not always correlate with the effects of each protein upon virus disease in mice.
MATERIALS AND METHODS
Cell culture.
The Tet-Off MEF cell lines were purchased from BD Biosciences Clontech and maintained in Dulbecco's modified Eagle's medium (DMEM; Mediatech), supplemented with 10% heat-inactivated fetal bovine serum, 200 mM l-glutamine (Sigma), 10,000 units/ml penicillin G sodium (Sigma), 10 mg/ml streptomycin sulfate (Sigma), and 100 μg/ml G418 (Invitrogen) (DMEM complete). The Tet-Off MEF antiviral protein-expressing cell lines were maintained in DMEM complete further supplemented with 100 μg/ml hygromycin sulfate (BD Biosciences Clontech) and 2 μg/ml doxycycline (Dox; BD Biosciences Clontech) to suppress transcription of target genes. For target gene induction (Dox withdrawal), cell monolayers were washed with phosphate-buffered saline (PBS) three times before replacing with DMEM complete supplemented with hygromycin. Growth medium was replaced at 24 h after withdrawal of Dox, and gene induction was allowed to continue for 1 to 2 days, depending upon the experiment. BHK and L929 cells were maintained in AlphaMEM (Mediatech) supplemented with 10% donor calf serum, 200 mM l-glutamine (Sigma), 10,000 units/ml penicillin G sodium (Sigma), and 10 mg/ml streptomycin sulfate (Sigma). Cell lines were grown at 37°C in a humidified chamber with 5% CO2.
Establishing Tet-Off MEF cell lines expressing FLAG-tagged candidate proteins.
A cassette containing the FLAG tag was removed from the pCMVTag-1 (Stratagene) vector using NotI and EcoRV and was inserted into the multiple cloning site of a similarly digested Tet-inducible pTRE2hyg vector (BD Biosciences Clontech), generating the pTRE2hyg-FLAG vector. This allowed FLAG epitope tagging of proteins inserted via NotI sites into the pTRE2hyg-FLAG vector. Amplicons encoding generic controls (enhanced green fluorescent protein [eGFP] and firefly luciferase [fLuc]) or tested genes (murine ISG20, ISG15, ISG49 [murine ortholog of human ISG60], ISG56, ISG54, ZAP, and viperin) with NotI ends were produced either by PCR of 39MCS viruses (described below) or reverse transcription-PCR (RT-PCR) of total mRNA from IFN-treated Raw 264.7 macrophages or bone marrow-derived dendritic cells (Table 1) with primers designed against GenBank sequences. All PCR products were initially placed into the PCR Blunt cloning vector (Invitrogen) and then cloned into the NotI site of pTRE2hyg-FLAG. In this configuration, the antiviral protein gene inserts are under the control of the Tet-Off expression system, allowing the induction of target gene expression after withdrawal of repressor (Dox). Tet-Off MEF cells expressing the Dox-responsive cytomegalovirus promoter transactivator (BD Biosciences Clontech) were electroporated with each construct and immediately placed in the presence of Dox. The transfected cells were selected on growth medium containing hygromycin and then were cloned from single cells by limiting dilution, always in the presence of Dox. Depending upon the construct, approximately 30 Geneticin 418-resistant clones were isolated. Among Hygr colonies, positive clones were identified by Dox withdrawal, lysis, and Western blotting using anti-FLAG antibody (Sigma). Two separately selected clones of each gene that exhibited substantial induction of FLAG-positive protein after Dox withdrawal were used in subsequent studies.
TABLE 1.
Sequences of primers used to amplify mRNAsa
| Gene | Sense primer | Antisense primer |
|---|---|---|
| β-Actin internal | 5′-GGATCCTCAGAAGGACTCCTATGTGG-3′ | 5′-GGATCCATGAGGTAGTCTGTCAGGTC-3′ |
| IFN-β internal | 5′-AGGGCGGACTTCAAGATC-3′ | 5′-CTCATTCCACCCAGTGCT-3′ |
| Cyclophilin B internal | 5′-AACGATAAGAAGAAGGGACCTA-3′ | 5′-CAGGCTCTCTACTCCTTGGC-3′ |
| ISG20 | 5′-ATGGCAGGCATCCCAGAGGTGGTGG-3′ | 5′-GTCTGACGTCCCAGGGCAAGGCAGCCCTCG-3′ |
| ISG54 | 5′-ATGAGTACAACGAGTAAGGAGTCACTGGAG-3′ | 5′-AGTATTCAGCACCTGCTTCATCC-3′ |
| ISG56 | 5′-GCTATGCAGTCGTAGCCTATCGCC-3′ | 5′-CCTGCTCTATGTGAGCCACGAGGG-3′ |
| ISG49 | 5′-ATGAGTGAGGTCAACCGGGAATCTC-3′ | 5′-CTATGTTTGCTCTTTAACCTCTTCC-3′ |
| Viperin | 5′-ATGGGGATGCTGGTGCCCACTGCTCTA-3′ | 5′-CCAGTCCAGCTTCAGGTCAGC-3′ |
| ZAP | 5′-ATGACGGATCCCGAGGTATTCTGTTTCATCACC-3′ | 5′-CTCTGGACCTCTTCTCTTCTGCTCCACATCC-3′ |
| ZAP internal | 5′-ACCAGGCCGGGATCACTCGGTCGGTGGTGG-3′ | 5′-AGACACATCCTCCAGGGGATCCTTACAGCC-3′ |
| ISG15 | 5′-ATGGCCTGGGACCTAAAGGTGAAG-3′ | 5′-TTAGGCACACTGGTCCCCTCCCCC-3′ |
| MVH | 5′-ATGGGAGATGAAGATTGGGAGGCAG-3′ | 5′-TCAGTCCCATGATTCGTCATCAACTGG-3′ |
Includes only the target mRNA sequence. Primers amplify the full-length gene unless otherwise indicated (“internal”).
Viruses, virus vectors, and stocks.
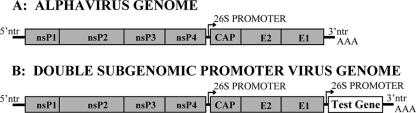
Construction of the consensus sequence pTR339 plasmid has been described previously (22). Infectious viral RNA was generated by in vitro transcription using the Message Machine kit (Ambion) with XhoI-linearized pTR339 DNA temple and transfected into BHK-21 cells by electroporation. Virus particles were harvested from the supernatant 18 to 20 h after electroporation, clarified by centrifugation, and stored at −70°C in single-use aliquots, and titers were determined by standard BHK cell plaque assay. Virus plasmids derived from TR339 and expressing either eGFP (virus designated 39MCS-eGFP) or fLuc (virus designated 39MCS-Luc) from a second copy of the subgenomic promoter were described previously (37, 38). cDNA clones of murine ISG20, ISG15, ISG49, ISG56, ISG54, ZAP, and viperin genes containing NotI sites at both ends were generated by RT-PCR from IFN-treated Raw264.7 or bone marrow-derived DC mRNA and cloned into the virus plasmid downstream of the 26S subgenomic promoter (Fig. 1A). Infectious viral RNA was generated as described above from each expression virus template linearized with either XhoI or FseI. Protein expression from virus vectors was confirmed by either (i) fluorescence microscopy, (ii) a luciferase assay, (iii) Western blotting, or (iv) a radiolabeling assay (data not shown). For radiolabeling of expressed proteins, BHK cells were electroporated with 10 to 20 μg of a transcription reaction mixture for each virus followed by replacement of growth medium between 5 and 8 h postinfection (hpi) with methionine-free/cysteine-free DMEM (1% fetal bovine serum) (Invitrogen) and addition of 10 μCi/ml of [35S]Cys and [35S]Met at 8 hpi and continued incubation for 18 h. Whole-cell lysates were then prepared, and radiolabeled proteins were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels followed by exposure to film (Kodak) and visualization of expressed proteins.
FIG. 1.
Diagrams of the genomes of wild-type Sindbis virus strain TR339 (A) and the 39MCS double subgenomic promoter virus (B) used for in vivo expression of candidate genes.
Mice.
Pregnant (13 to 15 days of gestation) CD1 mice were purchased from Charles River Laboratories. Mice were housed in the Animal Resources Center (Louisiana State University Health Sciences Center, Shreveport) under specific-pathogen-free conditions. All procedures were carried out in accordance with federal and institutional guidelines for animal care and use.
Mortality studies.
Five micrograms of RNA from in vitro transcriptions of Sindbis viruses encoding the candidate antiviral proteins eGFP, fLuc, or the mouse vasa homolog (MVH) gene were inoculated subcutaneously into CD1 mice using a 30-gauge needle and Hamilton syringe. Virus-infected mice were observed at 12-hour intervals, and the percent mortality and average survival time were calculated as described by Ryman et al. (39). Experiments were performed at least twice with similar results. Three weeks postinfection, all surviving mice were challenged intracranially with 1,000 PFU SAAR 86 clone S55, a Sindbis virus strain that causes 100% mortality in naive intracranially inoculated adult mice (18). Mouse mortality curves were compared using a Mantel-Cox log rank test with GraphPad Prism software.
IFN-α/β treatment and virus infection.
Tet-Off MEF cells were primed by the addition of IFN-α/β (Access Biomolecular) to supernatants for either 3 or 6 h prior to infection. After removal of the medium, cells were infected with TR339 or 39MCS-fLuc diluted in RPMI medium containing HEPES buffer and 1% donor calf serum (cRPMI) at a low multiplicity of infection (MOI; 0.1) for 1 h at 37°C. After infection, cells were washed three times with Dulbecco's PBS (Mediatech) supplemented with 10% calf serum, and growth medium was replaced. Biological interferon assays were performed using L929 cells and encephalomyocarditis virus as previously described (39).
Virus growth curves and determination of luciferase activity.
Tet-Off MEF cell lines expressing different genes (two separate cloned lines per gene) or nontransfected Tet-Off cells either treated with IFN-α/β or mock treated were infected in triplicate as described above in 24-well plates. For progeny virus particle titration, 10 μl of supernatant was collected at 12 and 24 hpi into 90 μl of cRPMI followed by storage at −80°C and subsequent BHK cell plaque assay titration. For the luciferase assay as a measurement, infected and uninfected cell lysates were harvested at 8 and 24 hpi with 1× passive luciferase lysis buffer (Promega), and luciferase activity was determined by using the single luciferase reporter system (Promega) on a PolarStar microplate reader (BMG Technologies) and expressed as relative light units. Expression of eGFP in infected cells was monitored on a Nikon TE300 fluorescence microscope and recorded photographically at constant shutter speed to facilitate comparison of GFP fluorescence intensity between samples. All experiments were performed at least twice with similar results. Significance of antiviral effects in in vitro assays was determined by a one-tailed Student's t test for samples of equal variance using GraphPad Prism software.
Western blot analysis.
Cellular extracts were subjected to immunoblot analysis as described previously (38). Briefly, whole-cell extract lysates were prepared by washing cells twice with PBS and lysing them in whole-cell extract lysis buffer containing protease and phosphatase inhibitor cocktails (Sigma). Protein concentrations were determined by Bradford assay (Bio-Rad), and equal protein was loaded. Proteins were fractionated by 7.5%, 12%, or 15% SDS-PAGE (dependent upon the size of loaded proteins) and transferred onto a polyvinylidene difluoride membrane (Bio-Rad). The membranes were blocked with 5% nonfat milk in PBS and incubated with anti-FLAG M2 monoclonal antibody (Sigma). Protein bands were visualized using ECL Plus Western blotting detection reagents (Amersham Biosciences), followed by exposure to Kodak Bio-Max film.
siRNA transfection, gene knockdown, and luciferase activity determination.
ISG20 (catalog no. L-063834-00), ZAP (catalog no. L-045974-00), ISG15 (catalog no. L-040962-00), ISG56 (catalog no. L-046524-00), viperin (catalog no. L-059375-1), and positive (targeting mouse cyclophilin B; catalog no. D-001820-20-05) and negative (nontargeting siRNA; catalog no. D-001810-10-05) control synthetic siRNA duplexes were purchased from Dharmacon RNA Technologies (SMARTpool ON-Targetplus siRNA system) and resuspended in RNase-free H2O. Transfection of siRNAs targeting each mRNA was carried out according to the manufacturer's instructions with some modifications. MEF/3T3 cells were plated in 24-well plates for 18 to 24 h before transfection. On the day of transfection, RNA-lipid complexes were introduced into each well of cells (10 to 200 nM RNA) by using DharmaFECT3 transfection reagent (Dharmacon RNA Technologies). For cotransfection of multiple siRNAs, two different pools of siRNA duplexes were introduced into each well of cells (50 nM each; 100 nM siRNA total) using DharmaFECT3 as above. Optimum transfection efficiency was determined at 48 h posttransfection using the fluorescent positive control siRNA duplex (siGLO cyclophilin B siRNA), which is a dual-purpose silencing control targeting cyclophilin B that can be visualized by fluorescence microscopy. The effect of specific siRNAs on target gene mRNA abundance was assessed by RT-PCR for the target gene, alongside RT-PCR for β-actin as a loading control. Eighty-one hours after transfection, siRNA-treated and nontreated control cells were either left untreated or exposed to 10 IU/ml IFN-α/β for 3 h. All cells were then infected as described above with 39MCS-Luc followed by lysis at 8 hpi for luciferase assay processing as described above.
RNA isolation and RT-PCR.
For semiquantitative RT-PCR, total cellular RNA was isolated using the RNeasy kit (QIAGEN). Five micrograms of total RNA per sample was prepared, treated with DNase I (Promega) to remove genomic DNA contamination, and reverse transcribed using an oligo(dT)18 primer as previously described (38). Equal volumes of each cDNA were then subjected to PCR amplification with gene-specific primers (Integrated DNA Technologies) (Table 1) designed against GenBank sequences of each gene (ISG20, ISG15, ISG49, ISG56, ISG54, ZAP, viperin, cyclophilin B, IFN-β, and β-actin). The volume of the cDNA template included in these reaction mixtures and the number of amplification cycles were optimized to ensure that reactions were stopped during the linear phase of product amplification, permitting semiquantitative comparisons of mRNA abundance between different RNA preparations. To exclude the possibility of contaminating DNA, control reactions were performed in parallel in the absence of reverse transcriptase. RT-PCR products were resolved by agarose gel electrophoresis and visualized on a VersaDoc 4000 imaging system (Bio-Rad).
RESULTS
Overexpression of proteins from TR339-based Sindbis virus vectors and assessment of virulence attenuation in mice.
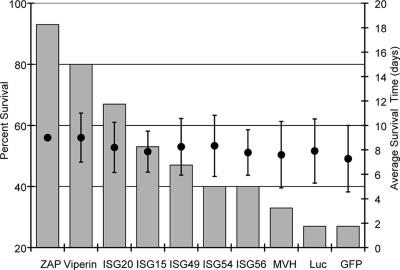
To characterize the potential of candidate proteins to inhibit SB replication, we first tested the effect of expression of genes in infected cells using the TR339-based double subgenomic promoter virus (Fig. 1). Initial experiments using adult IFNAR1−/− mice infected subcutaneously (s.c.) failed to show any significant effect of ISG15 or several other proteins in average survival time or percent mortality versus a GFP-expressing control (data not shown), suggesting that our SB vector system was not as sensitive to antiviral activities as those used by other investigators in which cell culture-adapted, attenuated laboratory strains of SB were used for coexpression of antiviral proteins (23). For this reason, we infected IFN response-competent, 1-day-old CD1 mice. Our previous studies suggested that these mice may be partially, but not completely, defective in IFN-α/β antiviral activities (21, 37), providing a more natural context for determination of protein antiviral activity. Furthermore, to avoid the possibility that expression of genes with strong antiviral activities would be disproportionately attenuated by mutation or deletion during in vitro packaging reactions, we immunized mice directly with transcription reaction mixtures. We previously found that similar immunization of mice with TR339 transcripts resulted in 100% mortality with a slight increase in survival time over similar immunizations with virus particles (data not shown). Presumably, entry of viral RNA into susceptible cells (e.g., DCs or macrophages) was sufficient to initiate the virus life cycle in vivo. Controls for these experiments were designed to meet two criteria: (i) a control protein was actually expressed from the second subgenomic promoter, and (ii) the gene encoding the protein was of similar size to each of the candidate genes. We used eGFP as a control for ISG15 and ISG20 (gene length, <1.0 kb), firefly luciferase as a control for ISG56, ISG49, ISG54, and viperin (gene length, ∼1.6 kb), and the mouse MVH gene as a control for ZAP (gene length, ∼3.0 kb). We had previously found that these proteins did not exhibit antiviral activity in vitro (data not shown).
Mice inoculated with the eGFP-, fLuc-, or MVH-expressing control viruses exhibited an extended average survival time indicative of the attenuating effect of the second subgenomic promoter upon viruses inoculated by the s.c. route, causing mortality in approximately 75% of animals (Fig. 2). The percent mortality caused by expression of MVH was slightly lower, perhaps reflecting an effect of expressed gene size upon the virulence of the vector. Virulence data for the tested genes reflected a range of effects upon mouse mortality, with ZAP exhibiting the greatest protection versus the control (P < 0.005), followed by viperin (P < 0.005) and ISG20 (P < 0.05) (Fig. 2). A hierarchy of protection from mortality was observed among the other genes (ISG15 > ISG49 > ISG54/ISG56); however, these results were not significant in comparison with controls (P > 0.05). None of the genes had a strong effect upon average survival time in the animals that succumbed in comparison with controls (Fig. 2). All mice receiving virus RNA were protected from intracranial challenge with SA.AR86 (naïve controls succumbed to the challenge), indicating that RNA inoculation initiated infection effectively.
FIG. 2.
Neonatal mouse mortality and average survival time data from infections with viruses. Mice were inoculated subcutaneously in the axial region at exactly 24 h of age with 5 μg of RNA from an in vitro transcription reaction mixture of each expression virus. Two separate pooled litters totaling 15 to 20 mice were used for each virus. Percent mortality and average survival times were calculated as described in Materials and Methods.
Establishment and evaluation of Tet-Off MEF cell clones expressing potential antiviral proteins.
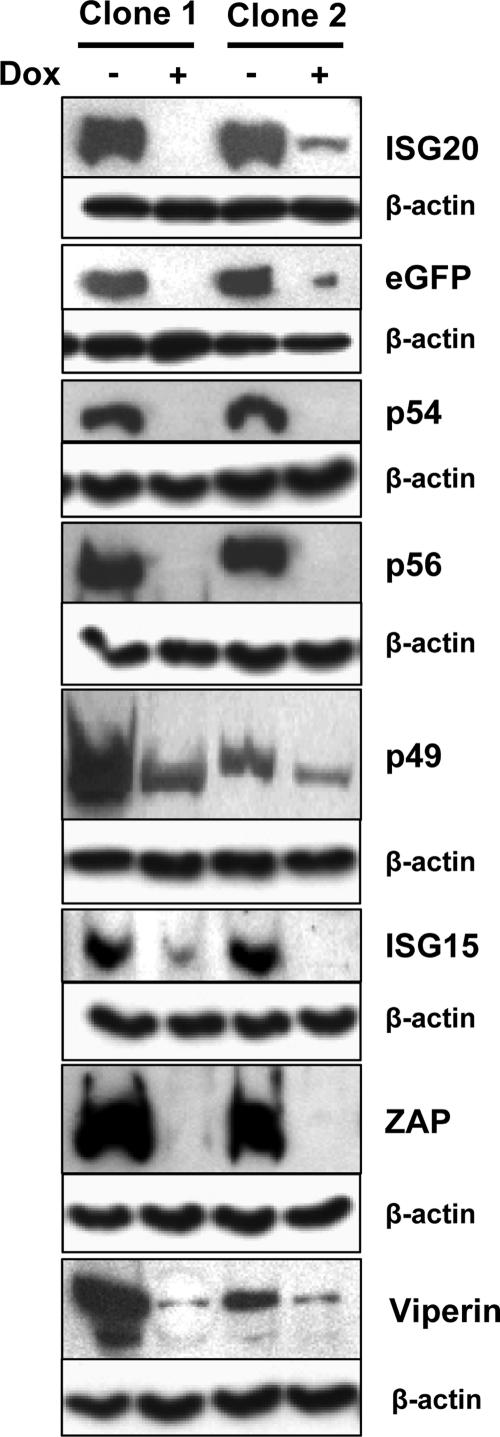
To begin analysis of the IFN-stimulated gene products for direct antialphavirus activity in vitro, we derived a series of stably transfected Tet-inducible MEF cell clones expressing the proteins. Two cell clones for each gene were selected, and inducibility and protein expression were confirmed by anti-FLAG immunostaining of proteins before and after induction by Dox withdrawal (Fig. 3). It was evident that low levels of some target proteins were also observed in uninduced cells, indicating that shutoff was incomplete in some cell lines (Fig. 3). Staining for β-actin in companion samples confirmed equal protein loading between the different samples.
FIG. 3.
Basal and induced expression of tested proteins in Tet-OFF MEF cultures. Cultures of two separately selected clones expressing each protein were either incubated in doxycycline (+) or incubated without doxycycline (−) for 48 to 72 h followed by lysis, SDS-PAGE separation, and Western blotting for FLAG tag as described in Materials and Methods. Fifty micrograms of protein was loaded per well. Staining for β-actin in the samples confirmed equal loading.
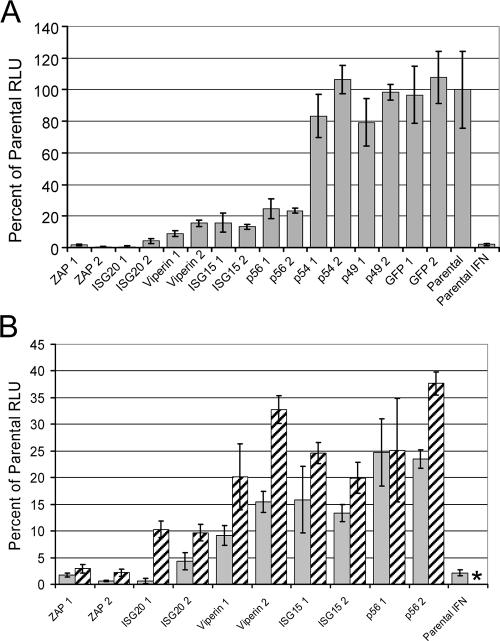
To assess the antialphaviral activity of overexpressed proteins, we began by determining titers of progeny virus from growth assays with one cloned cell line of each candidate gene infected with TR339 at a low MOI. Virus production in this experiment revealed the effects of candidate protein overexpression upon replication within individual cells and spread between cells (Fig. 4). In the growth assays, ISG20 and ZAP exhibited significant inhibition of virus release versus GFP-expressing cells at both 12 hpi (P < 0.005 for both) and 24 hpi (P = 0.05 and P < 0.005, respectively), but the other proteins did not. However, ISGS15 did produce inhibition at 24 h only (P = 0.05). As expected, IFN-α/β treatment of the untransfected parental cells was more potent than isolated expression of either gene, individually, at 12 hpi but was similarly effective to ZAP at 24 hpi. This may reflect waning of the IFN-mediated antiviral effect in the normal cells.
FIG. 4.
Growth of TR339 at 12 (solid bars) or 24 (hatched bars) hours postinfection (MOI, 0.01) of GFP- or candidate gene-expressing Tet-OFF MEFs or mock-treated (parental) or IFN-α/β-treated, untransfected parental cells (1,000 IU for 6 h). Samples titers were determined by standard plaque assay on BHK cells.
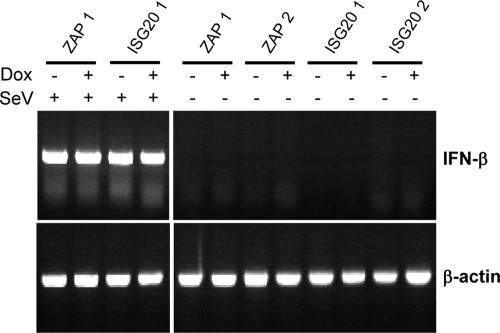
To eliminate the possibility that overexpression of ZAP or ISG20 resulted in IFN-α/β induction that might account for any antiviral effects observed, we performed RT-PCR for IFN-β mRNA on RNA harvested from uninduced and induced ZAP- or ISG20-expressing cells compared with a similar analysis of RNA harvested 8 hpi of the cells with Sendai virus, a known IFN inducer (Fig. 5). IFN-β mRNA was not detected in any of the samples from uninfected MEFs; however, it was clearly upregulated by PCR (Fig. 5) or the biological IFN assay (data not shown) after infection with Sendai virus. Finally, we also performed a biological IFN-α/β assay on supernatants from induced, uninfected cells and did not detect any IFN-α/β production (data not shown).
FIG. 5.
RT-PCR for IFN-β mRNA in ISG20- and ZAP-expressing Tet-Off MEF clones 1 and 2 in the presence or absence of DOX and either infected or mock infected with Sendai virus (MOI, 3).
We have found that detection of an indicatory protein produced from the subgenomic mRNA of SB is a more sensitive and direct measure of replication inhibition than titration of particles produced (data not shown). Therefore, we infected the cells with a TR339-based double promoter virus expressing fLuc from the second subgenomic promoter (39MCS-fLuc) and utilized a luciferase assay of infected cell lysates as a measure of replication. This approach could detect effects of individual proteins upon attachment and entry, genome translation, minus-strand synthesis, subgenomic mRNA synthesis, or translation of the subgenomic mRNA, but not upon structural protein sorting or particle egress. We considered a protein antiviral in this system if (i) growth of SB was inhibited by a statistically significant amount in the overexpressing cells versus an eGFP-expressing control, (ii) the increase in protein expression achieved by removal of Dox was associated with an increase in the degree of inhibition versus uninduced cells, and (iii) these effects were consistent for both cloned cell lines.
Similar to particle titer data, SB virus replication was significantly inhibited by overexpression of ISG20 (160-fold and 20-fold for clones 1 and 2, respectively; for combined data from both clones, P < 0.005) or ZAP (50-fold or 160-fold for clones 1 and 2, respectively; for combined data from both clones, P < 0.005) in induced Tet-Off MEF cells compared to control untransfected cells or GFP-expressing cells at 8 hpi (Fig. 6A), while GFP-transfected and untransfected parental cells were not different (combined data for both clones, P > 0.05). The inhibition by ISG20 and, in particular, ZAP was similar in magnitude to IFN-α/β priming of parental, untransfected cells. In contrast to the failure of other genes to inhibit virus particle production significantly at this time (Fig. 4), production of luciferase was inhibited 4- to 10-fold by viperin, ISG15, or p56 overexpression (combined data for both clones of each protein, P < 0.005). There was no significant and consistent difference in SB virus replication between controls and cells overexpressing p54 or p49 (combined data for either clone, P > 0.05). Similar patterns of results were obtained when samples were taken at 24 hpi (data not shown).
FIG. 6.
Luciferase expression from Tet-OFF MEF cultures at 8 hpi with 39MCS-fLuc. (A) Comparison of all clones with GFP or untransfected (parental) clones or parental cells treated with IFN (1,000 IU/ml for 6 h). (B) Comparison of clones exhibiting an antiviral effect in experiments of panel A either in the presence (hatched bars) or absence (gray bars) of Dox. *, not done.
We further compared the luciferase expression in the presence (suppressed) or absence (induced) of Dox for each of the clones that showed a consistent effect in the previous assay to determine if candidate protein induction as measured by Western blotting (Fig. 3) was associated with an increase in the inhibition of replication (Fig. 6B). In repeated experiments, a decrease in virus replication was associated with withdrawal of Dox (induction of gene expression) for Zap, ISG20, viperin, and ISG15 (Fig. 6B) but not the eGFP control, p54, or p49 (data not shown). However, the presence of Dox in the medium did not, in any case, restore virus replication to control levels. This was presumably due to low-level expression of proteins even in the repressed state (as detected for several genes in Fig. 3). In the experiment presented in Fig. 6B, p56 clone 1 did not show a decrease in virus-mediated luciferase expression associated with induction of gene expression; however, a decrease in luciferase in the induced clone was observed in this experiment at 24 hpi, and repeated experiments showed such a decrease at this time point (data not shown). Therefore, based upon the criteria outlined above, we consider ZAP, ISG20, viperin, ISG15, and p56 to have an antiviral effect against Sindbis virus when overexpressed in vitro. Furthermore, data are suggestive that ZAP and ISG20 are the most potent of these antiviral proteins when expressed prior to virus infection.
siRNA-mediated reduction of ISG20, ZAP, ISG56, ISG15, and viperin mRNA is correlated with increased permissivity of Tet-Off MEF parental cells to SB replication.
Overexpression of proteins likely does not recreate physiological conditions of gene induction following IFN-α/β stimulation. To further confirm the results from overexpression studies, we used synthetic siRNAs targeting mRNAs from these genes followed by infection of nontransfected Tet-Off MEF parental cells with the 39MCS-fLuc virus and quantitation of luciferase activity as described above. In these experiments, a nontargeting negative control siRNA, an siRNA targeting the cyclophilin B gene, or untreated cells served as controls.
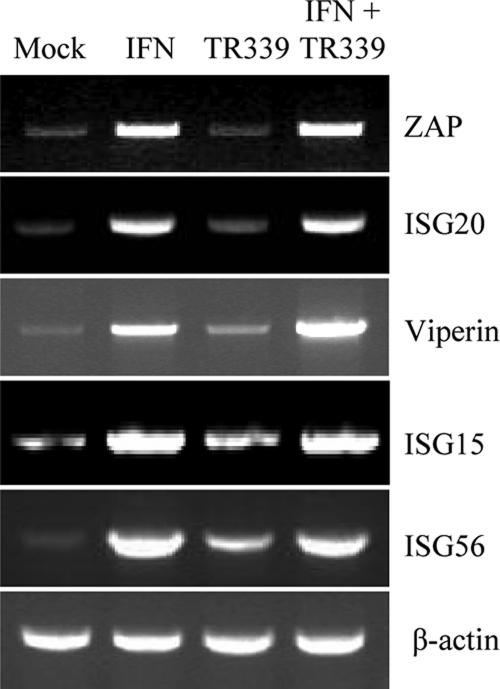
To first confirm that each of these genes was IFN inducible in the Tet-Off MEF cells, we performed RT-PCR using gene-specific primers (Table 1) on RNA harvested at 6 h post-IFN treatment (1,000 IU/ml IFN-α/β) (Fig. 7). Interestingly, some constitutive expression of each gene was observed prior to IFN treatment. In addition, stimulation with IFN-α/β resulted in an increase of ISG20, ZAP, p56, ISG15, and viperin mRNA but not β-actin mRNA abundance in the cells (Fig. 7). Furthermore, infection with TR339 alone was less stimulating of their transcription than IFN treatment, consistent with criteria used in our original microarray studies with murine DCs to classify genes as candidates for a PKR-RNase L-independent activity (39) and with reports describing IFN inducibility of each of these genes in other cell types (8, 26, 27, 38, 43).
FIG. 7.
Semiquantitative RT-PCR demonstrating the induction profiles of ZAP, ISG20, viperin, ISG15, and ISG56 mRNAs in Tet-OFF MEF cells after IFN treatment (1,000 IU/ml for 6 h) or TR339 infection (MOI, 3).
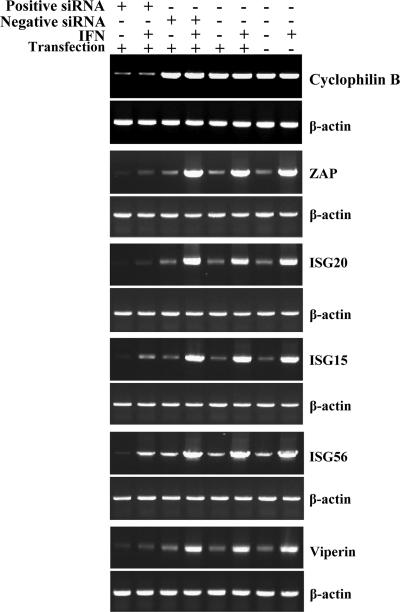
In Tet-Off MEF parental cells transfected with the siGLO mouse cyclophilin B siRNA (Dharmacon) positive control siRNA duplexes, the abundance of target cylophilin B mRNA was reduced compared to mock-transfected cells (Fig. 8), indicating that the siRNA transfection was effective at reducing target mRNA abundance. Transfection of a negative control nontargeting siRNA pool, treatment of the cell with transfection reagent alone, or mock treatment of the cells confirmed that the decrease in the cyclophilin B gene expression levels observed with gene-specific siRNAs was related to sequence-specific targeting of mRNAs (Fig. 8). Similarly, constitutive expression and IFN-α/β-induced expression of ZAP, ISG20, ISG15, viperin, and p56 mRNAs was greatly reduced after transfection of specific targeting siRNA pools (Fig. 8).
FIG. 8.
Semiquantitative RT-PCR for mRNAs targeted by siRNA at 48 h post-siRNA transfection. Positive siRNA refers to transfection with the gene-specific RNA listed on the right of the figure in each pair with β-actin. Negative siRNA refers to the presence or absence of a nontargeting control siRNA. In IFN+ samples, cells were treated (at 45 h post-siRNA transfection) with 10 IU of IFN-α/β for 3 h at 37°C followed by sample analysis. “Transfection” refers to the presence or absence of the lipid-based transfection reagent. For each gene, PCR cycles were adjusted such that the maximum signal for a given gene (for example, IFN-treated cells with ZAP, ISG20, ISG15, and p56) was not saturating. Cyclophilin B was included as a control for a non-IFN-inducible mRNA. The β-actin loading control was performed separately for each treatment. PCR primers used for each gene are listed in Table 1.
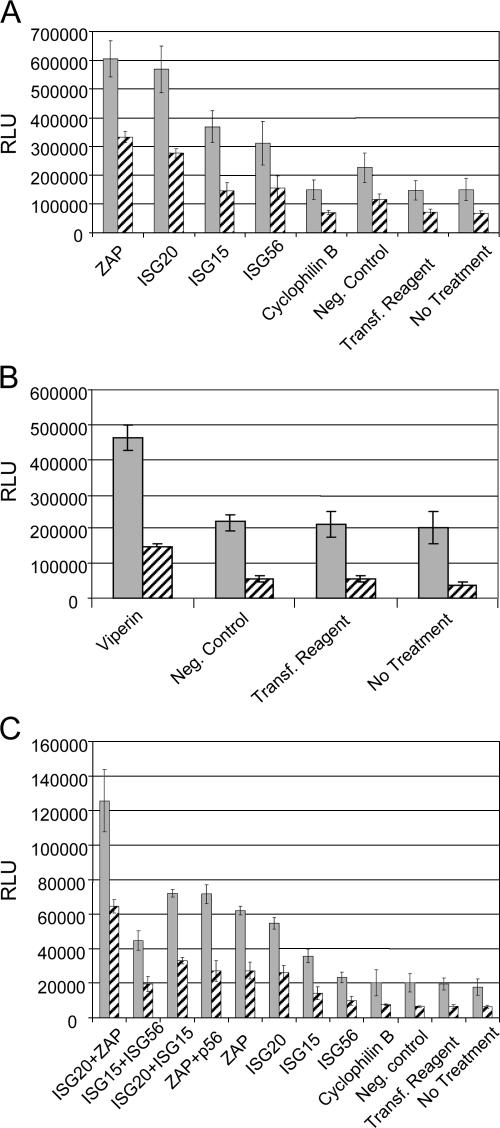
Interestingly, in the absence of IFN-α/β treatment, siRNA-mediated knockdown of constitutive levels of ZAP (P < 0.005), ISG20 (P < 0.005), viperin (P < 0.01), ISG15 (P < 0.01), and ISG56 (P < 0.05) mRNAs resulted in a significant increase in luciferase activity expressed by the 39MCSLuc virus compared with the negative control siRNA-treated cells (Fig. 9A and B). For all genes, an increase in virus replication was also observed in comparison with the cyclophilin B positive control, the nontargeting negative control, or no treatment (Fig. 9A and B). Similar to results following overexpression of the proteins, reduction of the abundance of the ISG20 or ZAP mRNAs had a larger effect on replication than reduction of viperin, ISG15, or p56. However, in cells pretreated with 1,000 IU of IFN-αβ for 6 h prior to infection, the siRNA transfections had minimal effect upon SB replication (data not shown). Presuming that this was due to strong upregulation of multiple antiviral mediators by the high dose of IFN-α/β, we subsequently used 10 IU/ml of IFN-αβ and only 3 h of pretreatment prior to infection. With this treatment, luciferase expression in control cells was reduced two- to threefold (compared with >100-fold after 1,000 IU/ml for 6 h). Using this regimen, a reduction in the efficacy of the antiviral state was clearly detectable in the cultures transfected with ISG20 (P < 0.005), ZAP (P < 0.005), viperin (P < 0.01), ISG15 (P < 0.01), or p56 (P < 0.05) siRNAs (Fig. 9A and B).
FIG. 9.
Luciferase expression in Tet-OFF MEF cultures that were mock treated (gray bars) or treated with 10 IU/ml of IFN-α/β for 3 h (hatched bars) prior to infection. Cells were harvested at 8 hpi with 39MCS-fLuc. (A) Comparison of specific siRNA pools targeting ZAP, ISG20, ISG15, and ISG56 with controls, including a cyclophilin B-specific siRNA pool, a nontargeting negative control pool, transfection reagent alone, or no treatment. (B) Comparison of specific siRNA pools targeting viperin with a nontargeting negative control pool, transfection reagent alone, or no treatment. (C) Comparison of individual siRNA pools targeting ZAP, ISG20, ISG15, or ISG56 with combinations of two siRNA pools and control treatments described for panel A above.
Finally, to determine if the antiviral state induced by IFN-α/β priming of the Tet-Off MEF parental cells was comprised of multiple independent antiviral activities, we cotransfected the cells with different combinations of several siRNAs that exhibited activity in previous experiments and examined luciferase production 8 h after infection with 39MCS-fLuc (Fig. 9C). The combination of ISG20 siRNA and ZAP siRNA reversed the inhibition of SB virus replication more than either ISG20 or ZAP individually in both the presence and absence of added IFN (P < 0.005 for all). As anticipated, other combinations were less effective, only exhibiting significant inhibition versus individual siRNAs if ZAP or ISG20 siRNA was present. Interestingly, when cells were cotransfected with all four siRNAs (including ISG20, ISG15, p56, and ZAP), SB replication was lower than either individual ISG20 or ZAP siRNA-transfected cells (data not shown). Recent reports have suggested that duplexed RNAs of the size of siRNAs can activate PKR (40) or the IFN induction system (32). Since the Tet-Off MEF cells were capable of producing and responding to IFN, we sought to determine if siRNA-induced IFN played any role in our results. First, we completed a biological IFN assay using the supernatant from the cells 24 h after transfecting the cells with siRNAs but did not detect any IFN production (data not shown). Second, induction of IFN-inducible genes was not observed at this time after transfection with the siRNA concentration (100 nmol) used in all experiments (Fig. 9) except for the four-gene combination experiment, in which 200 nmol was used. Finally, virus replication as measured by luciferase production was very similar in untreated cells versus the negative control siRNA-transfected or cyclophilin B siRNA-transfected cells, suggesting no differences in the antiviral state. However, use of 200 nmol of any of the siRNAs resulted in nonspecific upregulation at 48 h posttransfection of mRNA abundance for the IFN-inducible genes but not the non-IFN-inducible genes cyclophilin B or β-actin (data not shown), suggestive of production of IFN-α/β.
DISCUSSION
With alphaviruses, PKR, but not RNase L, has been shown to exert substantial antiviral activity after infection of DCs (38). We identified inhibition of translation (occurring either directly or indirectly) as a major IFN-α/β-induced antiviral activity that occurs in the absence of PKR (38). This activity reduces translation of transfected reporter mRNAs 10- to 20-fold in PKR−/− DCs or MEFs (38; M. Tesfay, J. Yin, K. Ryman, and W. Klimstra, unpublished data). In contrast, we have observed up to 1,000-fold inhibition of virus particle production after IFN-α/β treatment of these cells, implying the existence of additional antiviral activities that inhibit alphavirus growth. In the current studies, we have identified ISG20, p56, and viperin as IFN-induced proteins with antialphavirus activity and confirmed the activities of ZAP and ISG15 previously identified by others (4, 23, 24, 33). In contrast, p54 and p49 showed little to no antiviral effect in any assay. These are the first studies to compare directly the efficacy of antiviral effects between different proteins. In our studies, ZAP and ISG20 both showed potent antiviral activity when overexpressed or knocked down by siRNA in vitro, while the effects of viperin, ISG15, and p56 were similar to each other and less potent. However, these conclusions are tempered by the fact that overexpression or siRNA knockdown levels may not have been equal between the different proteins.
In in vivo studies, these hierarchies were maintained in general; however, viperin was more potent in protection from virus-induced mortality than ISG20, while p56 was of limited effect. Possibly affecting these results is the fact that in vivo virus-based expression of candidate antiviral genes differs substantially from the in vitro systems in that target proteins are produced significantly after the initiation of the virus replication cycle as opposed to preinfection manipulation of protein levels as occurs in vitro. Furthermore, expression of a particular protein may also influence the development of innate or adaptive immune responses unrelated to the IFN system in mice infected with expression vectors. Therefore, it is possible that the antiviral effect of a given protein is dramatically different in the two situations. Finally, our data also provide evidence that the activities of individual antiviral proteins (e.g., ZAP and ISG20) are additive, indicating that the IFN-induced antiviral state is comprised of multiple, independent antiviral activities that likely act at different points in the virus replication cycle.
It is currently unclear whether or not any members of this group of genes whose protein products have antialphavirus activity account for the IFN-α/β-inducible translation-inhibitory activity we previously identified in PKR−/− cells (38). ISG20 is an RNA exonuclease that exhibits inhibitory activity towards human immunodeficiency virus (11), vesicular stomatitis virus, influenza virus, and encephalomyocarditis virus (12). Viperin is an IFN-induced cytoplasmic protein of unknown function that inhibits production of human cytomegalovirus structural proteins (8), hepatitis C virus replicon replication (19), and human immunodeficiency virus type 1 replication (34). The murine ortholog of human p56 appears to inhibit translation via bonding to the eIF3 c subunit and inhibiting eIF4F interaction with the 40S ribosomal subunit (20). ZAP specifically binds to regions within the SB genome and a subset of other mRNAs and recruits the RNA to the exosome, where it may be degraded (4, 16). ISG15 is a ubiquitin ligase-like molecule that may enhance proteasome targeting and degradation of certain proteins (28).
Data from our other studies indicate that inhibition of translation of viral mRNAs electroporated into IFN-α/β-treated PKR-deficient cells is not associated with degradation of the RNA (Tesfay et al., unpublished). Therefore, ISG20 and ZAP activities that result in mRNA degradation may not be involved, although silencing of translation by mislocalization of the RNA may still occur. However, it is also unlikely that ZAP is primarily responsible, as IFN-α/β-treated PKR−/− cells suppressed the translation of the identical mRNA used by Bick and coworkers (4) as a negative control for ZAP activity (38). It is possible that viperin or p56 suppresses initial translation of electroporated SB and host-mimicking mRNAs or that ISG15 activity results in rapid proteolysis of viral polypeptides such that virus replication is inhibited. However, we do not observe significant global inhibition of host cell mRNA translation in IFN-α/β-primed PKR−/− cells, suggesting that a broadly active mechanism is not operating (Tesfay et al., unpublished). We are currently examining the point in the alphavirus replication cycle at which each of these proteins acts to inhibit virus replication, for comparison with the translation-inhibiting activity observed in PKR−/− cells.
In contrast with some of the previous studies that examined the antialphavirus activities of host proteins (23), we have utilized cell lines and mouse virulence test systems capable of producing and responding to IFN-α/β. ISG15 has been shown, in certain circumstances and with particular SB strains, to exert significant antiviral activity in vivo (23, 24, 33). In preliminary experiments, we attempted to screen the antiviral proteins by expression from TR339-based viruses in IFNAR1−/− mice but were unable to detect a phenotype for most of the proteins (including ISG15) in neonatal or adult mice (data not shown). However, our laboratory strain of SB, TR339, kills these mice in less than 72 h (21, 22, 36); therefore, we attribute at least some of the differences between our data and others to the enhanced virulence of TR339 versus SB laboratory strains in these model systems or to the s.c. route of inoculation used in our studies. We speculate that the use of attenuated SB strains or use of SB expression vectors which we have found to be attenuated by the presence of the second subgenomic promoter (data not shown) may magnify the antiviral effects of certain proteins that may not be strong inhibitors of wild-type, fully virulent alphaviruses. Indeed, only a minor antiviral effect could be attributed to ISG15 after SB infection of wild-type and ISG15−/− MEF cultures (a gift from Klaus-Peter Knobeloch) (29) in the presence or absence of IFN-α/β pretreatment (data not shown). Ultimately, our approach likely gives a more realistic measure of the relative potencies of antiviral proteins against naturally circulating alphaviruses in a natural context; however, it does raise the possibility of indirect effects of protein expression or inhibition upon induction and activity of endogenous IFN-α/β. In the in vitro system, we addressed this possibility by examining the induction of IFN-α/β mRNA after protein overexpression or siRNA knockdown, but it remains possible that altered induction of IFN influenced the outcome of mouse experiments.
Interestingly, our siRNA data suggest that constitutive expression of antiviral proteins in cells that have not been exposed to IFN may significantly inhibit alphavirus replication. This result may, in part, explain the cell and tissue tropisms observed following SB infection of mice deficient in IFN-α/β or IFN-γ receptors (36, 37). In these mice, virus replication was poorly restrained, particularly in DCs and macrophages, but nevertheless, certain tissues (e.g., skeletal muscle, visceral organ parenchymal cells, endothelium, and most epithelia) remained refractory to virus replication in the absence of all IFN responses. It is possible that differential constitutive expression of antiviral proteins such as ZAP or ISG20 as well as appropriate expression of attachment/entry receptors account for these results.
Currently, there is a great deal of research effort focused on the effects of virus infection upon the inductive phase of the IFN-α/β-mediated antiviral system, specifically with regard to viral antagonism of IFN gene induction and IFN receptor signaling. These studies have revealed numerous points at which this component of the vertebrate host innate immune response is vulnerable to viral pathogens. However, the IFN response has evolved such that it is generally very effective at protecting uninfected cells from infection and replication if they are exposed to IFN prior to contact with viruses and expression of viral antagonist elements. This condition occurs during natural infections of vertebrate hosts as soon as paracrine or systemic IFN is produced from the initially infected cells. Understanding of the antiviral activities stimulated by IFN treatment of cells will be vital to development of antiviral drugs by distinguishing viral replicative processes from cellular processes and identifying mechanisms of inhibition that eventually can be mimicked pharmacologically.
Acknowledgments
Janice Anderson, Michael Farmer, Tanya Debenport, and Danielle Gonzalez provided excellent technical assistance.
This work was supported by NIH/NCRR grants R01AI22186 (W.B.K.), R21 AI069158-01 (K.D.R), and P20 RR 018724-01 and an Alliance for Cancer Gene Therapy Young Investigator Award.
Footnotes
Published ahead of print on 8 August 2007.
REFERENCES
- 1.Aguilar, P. V., S. Paessler, A. S. Carrara, S. Baron, J. Poast, E. Wang, A. C. Moncayo, M. Anishchenko, D. Watts, R. B. Tesh, and S. C. Weaver. 2005. Variation in interferon sensitivity and induction among strains of eastern equine encephalitis virus. J. Virol. 79:11300-11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, S. L., J. M. Carton, J. Lou, L. Xing, and B. Y. Rubin. 1999. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology 256:8-14. [DOI] [PubMed] [Google Scholar]
- 3.Anishchenko, M., S. Paessler, I. P. Greene, P. V. Aguilar, A. S. Carrara, and S. C. Weaver. 2004. Generation and characterization of closely related epizootic and enzootic infectious cDNA clones for studying interferon sensitivity and emergence mechanisms of Venezuelan equine encephalitis virus. J. Virol. 78:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bick, M. J., J. W. Carroll, G. Gao, S. P. Goff, C. M. Rice, and M. R. Macdonald. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 77:11555-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boxel-Dezaire, A. H., M. R. Rani, and G. R. Stark. 2006. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 25:361-372. [DOI] [PubMed] [Google Scholar]
- 6.Burgess, T. H., K. E. Steele, B. A. Schoneboom, and F. B. Grieder. 2001. Clinicopathologic features of viral agents of potential use by bioterrorists. Clin. Lab. Med. 21:475-493. [PubMed] [Google Scholar]
- 7.Carter, C. C., V. Y. Gorbacheva, and D. J. Vestal. 2005. Inhibition of VSV and EMCV replication by the interferon-induced GTPase, mGBP-2: differential requirement for wild-type GTP binding domain. Arch. Virol. 150:1213-1220. [DOI] [PubMed] [Google Scholar]
- 8.Chin, K. C., and P. Cresswell. 2001. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA 98:15125-15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Despres, P., J. W. Griffin, and D. E. Griffin. 1995. Antiviral activity of alpha interferon in Sindbis virus-infected cells is restored by anti-E2 monoclonal antibody treatment. J. Virol. 69:7345-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espert, L., G. Degols, C. Gongora, D. Blondel, B. R. Williams, R. H. Silverman, and N. Mechti. 2003. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J. Biol. Chem. 278:16151-16158. [DOI] [PubMed] [Google Scholar]
- 11.Espert, L., G. Degols, Y. L. Lin, T. Vincent, M. Benkirane, and N. Mechti. 2005. Interferon-induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J. Gen. Virol. 86:2221-2229. [DOI] [PubMed] [Google Scholar]
- 12.Espert, L., C. Gongora, and N. Mechti. 2003. Interferon: antiviral mechanisms and viral escape. Bull. Cancer 90:131-141. [PubMed] [Google Scholar]
- 13.Gil, M. P., E. Bohn, A. K. O'Guin, C. V. Ramana, B. Levine, G. R. Stark, H. W. Virgin, and R. D. Schreiber. 2001. Biologic consequences of Stat1-independent IFN signaling. Proc. Natl. Acad. Sci. USA 98:6680-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grieder, F. B., and S. N. Vogel. 1999. Role of interferon and interferon regulatory factors in early protection against Venezuelan equine encephalitis virus infection. Virology 257:106-118. [DOI] [PubMed] [Google Scholar]
- 15.Griffin, D. E. 2001. Alphaviruses, p. 917-962. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, PA.
- 16.Guo, X., J. Ma, J. Sun, and G. Gao. 2007. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl. Acad. Sci. USA 104:151-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hefti, H. P., M. Frese, H. Landis, C. Di Paolo, A. Aguzzi, O. Haller, and J. Pavlovic. 1999. Human MxA protein protects mice lacking a functional alpha/beta interferon system against Lacrosse virus and other lethal viral infections. J. Virol. 73:6984-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heise, M. T., D. A. Simpson, and R. E. Johnston. 2000. A single amino acid change in nsP1 attenuates neurovirulence of the Sindbis-group alphavirus S.A.AR86. J. Virol. 74:4207-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helbig, K. J., D. T. Lau, L. Semendric, H. A. Harley, and M. R. Beard. 2005. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology 42:702-710. [DOI] [PubMed] [Google Scholar]
- 20.Hui, D. J., F. Terenzi, W. C. Merrick, and G. C. Sen. 2005. Mouse p56 blocks a distinct function of eukaryotic initiation factor 3 in translation initiation. J. Biol. Chem. 280:3433-3440. [DOI] [PubMed] [Google Scholar]
- 21.Klimstra, W. B., K. D. Ryman, K. A. Bernard, K. B. Nguyen, C. A. Biron, and R. E. Johnston. 1999. Infection of neonatal mice with Sindbis virus results in a systemic inflammatory response syndrome. J. Virol. 73:10387-10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klimstra, W. B., K. D. Ryman, and R. E. Johnston. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 72:7357-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenschow, D. J., N. V. Giannakopoulos, L. J. Gunn, C. Johnston, A. K. O'Guin, R. E. Schmidt, B. Levine, and H. W. Virgin. 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 79:13974-13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenschow, D. J., C. Lai, N. Frias-Staheli, N. V. Giannakopoulos, A. Lutz, T. Wolff, A. Osiak, B. Levine, R. E. Schmidt, A. Garcia-Sastre, D. A. Leib, A. Pekosz, K. P. Knobeloch, I. Horak, and H. W. Virgin. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. USA 104:1371-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, Y., C. X. George, J. B. Patterson, and C. E. Samuel. 1997. Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J. Biol. Chem. 272:4419-4428. [DOI] [PubMed] [Google Scholar]
- 26.Loeb, K. R., and A. L. Haas. 1992. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J. Biol. Chem. 267:7806-7813. [PubMed] [Google Scholar]
- 27.Mattei, M. G., C. Tissot, C. Gongora, and N. Mechti. 1997. Assignment of ISG20 encoding a new interferon-induced PML nuclear body-associated protein, to chromosome 15q26 by in situ hybridization. Cytogenet. Cell. Genet. 79:286-287. [DOI] [PubMed] [Google Scholar]
- 28.Narasimhan, J., J. L. Potter, and A. L. Haas. 1996. Conjugation of the 15-kDa interferon-induced ubiquitin homolog is distinct from that of ubiquitin. J. Biol. Chem. 271:324-330. [DOI] [PubMed] [Google Scholar]
- 29.Osiak, A., O. Utermohlen, S. Niendorf, I. Horak, and K. P. Knobeloch. 2005. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol. Cell. Biol. 25:6338-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perri, S., C. E. Greer, K. Thudium, B. Doe, H. Legg, H. Liu, R. E. Romero, Z. Tang, Q. Bin, T. W. Dubensky, Jr., M. Vajdy, G. R. Otten, and J. M. Polo. 2003. An alphavirus replicon particle chimera derived from Venezuelan equine encephalitis and Sindbis viruses is a potent gene-based vaccine delivery vector. J. Virol. 77:10394-10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfefferkorn, E. R. 1984. Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc. Natl. Acad. Sci. USA 81:908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynolds, A., E. M. Anderson, A. Vermeulen, Y. Fedorov, K. Robinson, D. Leake, J. Karpilow, W. S. Marshall, and A. Khvorova. 2006. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA 12:988-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritchie, K. J., C. S. Hahn, K. I. Kim, M. Yan, D. Rosario, L. Li, J. C. de la Torre, and D. E. Zhang. 2004. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat. Med. 10:1374-1378. [DOI] [PubMed] [Google Scholar]
- 34.Rivieccio, M. A., H. S. Suh, Y. Zhao, M. L. Zhao, K. C. Chin, S. C. Lee, and C. F. Brosnan. 2006. TLR3 ligation activates an antiviral response in human fetal astrocytes: a role for viperin/cig5. J. Immunol. 177:4735-4741. [DOI] [PubMed] [Google Scholar]
- 35.Rotz, L. D., A. S. Khan, S. R. Lillibridge, S. M. Ostroff, and J. M. Hughes. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryman, K. D., W. B. Klimstra, K. B. Nguyen, C. A. Biron, and R. E. Johnston. 2000. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J. Virol. 74:3366-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryman, K. D., K. C. Meier, C. L. Gardner, and W. B. Klimstra. Nonpathogenic Sindbis virus causes viral hemorrhagic fever in the absence of alpha/beta and gamma interferons. Virology, in press. [DOI] [PubMed]
- 38.Ryman, K. D., K. C. Meier, E. M. Nangle, S. L. Ragsdale, N. L. Korneeva, R. E. Rhoads, M. R. Macdonald, and W. B. Klimstra. 2005. Sindbis virus translation is inhibited by a PKR/RNase L-independent effector induced by alpha/beta interferon priming of dendritic cells. J. Virol. 79:1487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryman, K. D., L. J. White, R. E. Johnston, and W. B. Klimstra. 2002. Effects of PKR/RNase L-dependent and alternative antiviral pathways on alphavirus replication and pathogenesis. Viral Immunol. 15:53-76. [DOI] [PubMed] [Google Scholar]
- 40.Sledz, C. A., M. Holko, M. J. de Veer, R. H. Silverman, and B. R. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5:834-839. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka, Y., H. Marusawa, H. Seno, Y. Matsumoto, Y. Ueda, Y. Kodama, Y. Endo, J. Yamauchi, T. Matsumoto, A. Takaori-Kondo, I. Ikai, and T. Chiba. 2006. Anti-viral protein APOBEC3G is induced by interferon-alpha stimulation in human hepatocytes. Biochem. Biophys. Res. Commun. 341:314-319. [DOI] [PubMed] [Google Scholar]
- 42.Terenzi, F., D. J. Hui, W. C. Merrick, and G. C. Sen. 2006. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J. Biol. Chem. 281:34064-34071. [DOI] [PubMed] [Google Scholar]
- 43.Terenzi, F., S. Pal, and G. C. Sen. 2005. Induction and mode of action of the viral stress-inducible murine proteins, P56 and P54. Virology 340:116-124. [DOI] [PubMed] [Google Scholar]
- 44.Wacher, C., M. Muller, M. J. Hofer, D. R. Getts, R. Zabaras, S. S. Ousman, F. Terenzi, G. C. Sen, N. J. King, and I. L. Campbell. 2007. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J. Virol. 81:860-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, C., J. Pflugheber, R. Sumpter, Jr., D. L. Sodora, D. Hui, G. C. Sen, and M. Gale, Jr. 2003. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J. Virol. 77:3898-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White, L. J., J. G. Wang, N. L. Davis, and R. E. Johnston. 2001. Role of alpha/beta interferon in Venezuelan equine encephalitis virus pathogenesis: effect of an attenuating mutation in the 5′ untranslated region. J. Virol. 75:3706-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeung, M. C., J. Liu, and A. S. Lau. 1996. An essential role for the interferon-inducible, double-stranded RNA-activated protein kinase PKR in the tumor necrosis factor-induced apoptosis in U937 cells. Proc. Natl. Acad. Sci. USA 93:12451-12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]