Abstract
Six amino acid substitutions in the shared N-terminal region of the P subunit of the viral polymerase and the accessory V protein convert the noncytopathic paramyxovirus simian virus 5 (SV5), which is a poor inducer of host cell responses, into a P/V mutant (P/V-CPI-) that induces high levels of apoptosis, interferon-beta (IFN-beta), and proinflammatory cytokines. In this study, we addressed the question of whether these new mutant phenotypes are due to the presence of an altered P protein or of an altered V protein or of both proteins. By the use of the P/V-CPI- mutant as a backbone, new mutant viruses were engineered to express the wild-type (WT) V protein (+V-wt) or WT P protein (+P-wt) from an additional gene inserted between the HN and L genes. In human epithelial cell lines, the +V-wt virus showed reduced activation of apoptosis and lower secretion of IFN-beta and proinflammatory cytokines compared to the parental P/V-CPI- virus. The presence of a V protein lacking the C-terminal cysteine-rich domain (corresponding to the SV5 I protein) did not reduce these host cell responses to P/V-CPI- infection. Unexpectedly, the +P-wt virus, which expressed a WT P subunit of the viral polymerase, also induced much lower levels of host cell responses than the parental P/V-CPI- mutant. For both +V-wt and +P-wt viruses, reduced levels of IFN-beta synthesis correlated with reduced IRF-3 dimerization and nuclear localization of IRF-3 and NF-κB, suggesting that the WT P and V proteins acted at an early stage in antiviral pathways. Host cell responses induced by the various P/V mutants directly correlated with levels of viral mRNA accumulation but not with steady-state levels of genomic RNA. Our results support the hypothesis that WT P and V proteins limit induction of antiviral responses by controlling the production of key viral inducers. A model is presented for the mechanism by which both the P subunit of the viral polymerase and the V accessory protein contribute to the ability of a paramyxovirus to limit activation of antiviral responses.
Host cell responses to viral infection are important factors that can contribute to viral pathogenesis and to tropism for specific tissues or cells (4). These antiviral responses can include the secretion of proinflammatory cytokines such as interleukin-6 (IL-6) and IL-8, stimulation of type I interferon (IFN) pathways (26, 46), and induction of apoptosis (45). The overall goal of the work described here was to determine the individual contributions of the paramyxovirus phosphoprotein P and the V protein to limiting these host cell antiviral responses.
Members of the paramyxovirus family of negative-strand RNA viruses employ a diverse range of mechanisms to circumvent host cell antiviral responses, including limiting cytokine induction and blocking IFN signaling pathways or both of these processes (reviewed in reference 9; 15, 18, 27). Many of these mechanisms for counteracting IFN have been attributed to products of the P/V (or sometimes P/V/C) gene which encodes both the phosphoprotein P subunit of the RNA-dependent RNA polymerase and the accessory V protein (13, 17, 35, 38). For simian virus 5 (SV5), accurate transcription of the P/V gene results in an mRNA that codes for the V protein. The P mRNA is identical to the V mRNA except for the addition of two nontemplated G residues that are inserted by the viral polymerase at a precise location in the P/V transcript (51). Thus, the SV5 P and V proteins are identical for the 164 amino-terminal residues (the shared P/V region) but differ in their C-terminal sequences. The P and V proteins have unique C-terminal domains, with the V protein encoding a highly conserved cysteine-rich (cys-rich) zinc-binding domain that is required for many V-associated functions (21, 41).
The paramyxovirus phosphoprotein P is an essential subunit of the viral RNA-dependent RNA polymerase (36). In a complex with the L catalytic subunit, the P protein plays multiple roles during mRNA transcription and during replication of genomic and antigenomic RNAs. The paramyxovirus V protein is also thought to function in the regulation of viral RNA synthesis (11, 24, 37, 39) but has additional roles in counteracting host cell antiviral responses (reviewed in reference 9; 15, 18).
A major function of the SV5 V protein is the inhibition of IFN signaling (13). In infected cells or cells transfected with V-expressing plasmids, V protein forms a cytoplasmic complex that directs the ubiquitylation and targeting of STAT1 (signal transducer and activator of transcription 1) for degradation (1, 52). Recently, the SV5 V protein was also shown to block activation of the IFN-beta promoter by transfected double-stranded RNA (dsRNA) (43) or following infection with a recombinant SV5 (rSV5) encoding a mutant V protein that lacked the cys-rich region (VΔC virus; 21). The inhibition of IFN-beta induction is thought to be due to V protein targeting the IFN-inducible RNA helicase mda-5 (7) by binding through the cys-rich region (2). Thus, the multifunctional V protein counteracts IFN responses at two steps, resulting in both limited induction of IFN synthesis and a block in IFN signaling.
In addition to IFN pathways, apoptosis of virus-infected cells can be a major antiviral response that limits virus growth (45). The factors that contribute to whether a particular paramyxovirus induces or limits apoptosis are not completely understood. Wild-type (WT) SV5 has the unusual property among paramyxoviruses of being largely noncytopathic in most epithelial and fibroblast cell types (8, 20, 40). The V protein has been implicated in limiting cell death in SV5-infected cells, since apoptosis is induced following infection with the VΔC rSV5 (21, 48).
In addition to the cys-rich C-terminal domain, the N-terminal P/V region of V protein contributes to counteracting the activity of host cell antiviral pathways (6, 54, 55). This is evident from the results seen with the naturally occurring CPI- strain of SV5, which is defective in inducing STAT1 degradation and blocking type I IFN signaling (6). Mutational analyses have identified amino acid differences in the P/V region between WT rSV5 and CPI- that are responsible for this loss of function in targeting STAT1 for degradation (6, 47). We have previously engineered an rSV5 mutant (rSV5-P/V-CPI-) to encode these same six CPI- P/V substitutions in the background of the WT rSV5 genome (54). These P/V gene substitutions converted WT rSV5 into a mutant that failed to target STAT1 for degradation as expected, but the chimeric virus was also found to be a potent inducer of IFN and proinflammatory cytokines (54, 59). The most striking contrast between rSV5-P/V-CPI- and WT SV5 was seen in their activation of a cytopathic effect (CPE), since the rSV5-P/V-CPI- mutant induced extensive cell death by apoptosis that is not seen with WT SV5 (54, 56).
Since the rSV5-P/V-CPI- mutant contained substitutions in the N-terminal domains of both P and V, the contribution of an altered P or altered V to these new phenotypes of the P/V-CPI- mutant was not known. Given the roles of V protein in counteracting host antiviral responses and the P protein in RNA synthesis, we hypothesized that alterations to the V protein were responsible for the striking differences between the host antiviral responses to infection with WT rSV5 and the P/V-CPI- mutant. In the present study, we tested this hypothesis by generating novel rSV5 viruses that expressed either WT V or WT P protein as a result of the presence of an additional gene within the rSV5-P/V-CPI- virus. Consistent with our hypothesis, expression of the WT V protein limited activation of IFN, proinflammatory cytokines, and apoptosis by the P/V-CPI- mutant. Unexpectedly, however, expression of the WT P subunit of the viral polymerase also limited the induction of these same host cell responses. Decreased host cell responses induced by the novel V- or P-expressing viruses correlated with reduced activation of IRF-3 and NF-κB and with reduced levels of viral mRNA, suggesting that WT V and WT P proteins acted at an early stage in activation of antiviral pathways. We propose a model to explain the effect of the N-terminal P/V mutations on SV5 gene expression and the roles of both the accessory protein V and the P subunit of the viral polymerase in limiting host cell responses.
MATERIALS AND METHODS
Cells, viruses, and growth analysis.
Monolayer cultures of cells were grown in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS). An rSV5 expressing green fluorescent protein (GFP) (19) was recovered as described previously (40) from cDNA plasmid kindly provided by Robert Lamb (Northwestern University) and Biao He (Pennsylvania State University). This virus has growth properties indistinguishable from those of WT rSV5 lacking the GFP-encoding gene (19). rSV5-P/V-CPI- was recovered from cDNA and grown in Vero cells as described previously (54). The rSV5-P/V-CPI- viruses with the extra WT V (+V-wt), truncated WT V (+V-Δcys), or WT P (+P-wt) gene inserted between the HN and L genes were constructed using standard molecular biology techniques (40; details available upon request) by insertion of an HpaI-SalI fragment containing the HN-L gene end-gene start sequence into the pBH311 plasmid (19) followed by insertion of an open reading frame encoding V only or P only. Editing of the V and P genes was disrupted through changes in the nucleotide sequence upstream of and including the editing site of the P/V gene (see Fig. 1) such that lysine 162 was converted to an arginine (51). The gene encoding the extra WT V protein was truncated so that the mRNA could be distinguished from the mRNA encoding the CPI- V protein expressed from the authentic P/V gene. The truncated WT V protein in the +V-Δcys virus was created by insertion of two translational stop codons in place of amino acids 169 and 170 in the +V-wt open reading frame. The +V-wt, +V-Δcys, and +P-wt viruses were recovered as described previously (54) and were grown in Vero cells. The presence of the inserted V and P genes was confirmed by sequencing reverse transcription-PCR products derived from infected-cell RNA. Virus infections and plaque assays for infectivity were performed as described previously (40, 54).
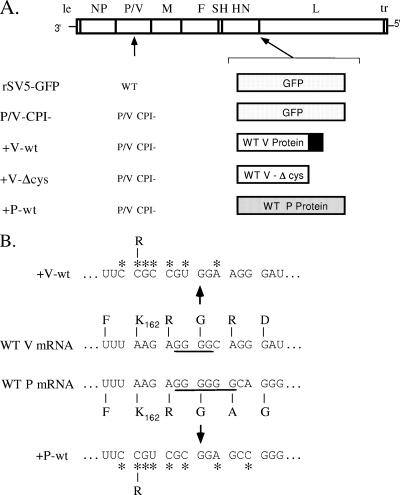
FIG. 1.
(A) Schematic diagram of SV5 viruses used in this study. Genome structures are shown schematically as negative-sense RNA, with an additional gene (encoding GFP, WT V protein, V-Δcys, or WT P protein) inserted between HN and L. Extra 5′-end P or V genes have an altered editing site, as described in Materials and Methods, such that only the P or V protein is expressed. The black box denotes the cys-rich C-terminal region of V protein which is missing in the case of the V-Δcys open reading frame. With the exception of rSV5-GFP, which has a WT P/V gene, all viruses encoded a P/V gene with a substitution of the CPI- shared P/V N-terminal region as described previously (54). le, leader; tr, trailer. (B) Sequence at the SV5 editing site. The nucleotide and amino acid sequence at the SV5 P/V gene-editing site (51) is shown, with underlined nucleotides denoting the sites of four (V mRNA) or six (P mRNA) G residues. Asterisks denote the locations of substitutions used to disrupt RNA editing.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis Western blotting.
For Western blotting, six-well dishes of cells were infected with viruses as described in each figure legend. The cells were washed with phosphate-buffered saline (PBS) and incubated in DMEM containing 2% FBS. At each time point, cells were washed with PBS and lysed in 1% sodium dodecyl sulfate. The protein concentrations of cell lysates were determined by bicinchoninic acid assay (Pierce Chemicals), and equivalent amounts of protein were analyzed with rabbit antisera against cellular STAT-1 (Santa Cruz Biotechnology) or the individual SV5 NP, P, and V proteins (40, 41). Alternatively, lysates were analyzed using a mouse monoclonal antibody against cellular actin or the common region of the P and V proteins (V5 antibody; Invitrogen). Samples were analyzed using horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (Pierce Chemicals).
IRF-3 dimerization assay.
The assay for dimerization of cellular IRF-3 was carried out as described previously (29). Briefly, cells were washed two times with cold PBS and then lysed in buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 5 μg/ml aprotinin and leupeptin, 1 mM phenylmethylsulfonyl fluoride, 100 nM okadaic acid, 0.1 mM NaF, 1 mM sodium orthovanadate, and 100 nM microcystin. After incubation on ice for 10 min, samples were centrifuged at 4°C for 15 min at 13,000 × g. Protein (50 μg) was analyzed by electrophoresis on 7.5% nondenaturing acrylamide gels (Ready Gels; Bio-Rad), with 1% deoxycholate in the cathode buffer. IRF-3 monomers and dimers were detected by Western blot analysis with monoclonal anti-IRF-3 SL-12.1 antibody (BD PharMingen) diluted 1:500.
Analysis of viral RNA.
To measure viral replication products, six-well dishes of Vero cells were either mock infected or virus infected at a multiplicity of infection (MOI) of 10. At various times postinfection (p.i.), cell lysates were treated with micrococcal nuclease to digest unencapsidated viral RNA as described previously (31, 42), and RNA was purified by TRIzol extraction. To measure the accumulation of positive-sense viral mRNA, total RNA was isolated from mock-infected or virus-infected cells at the indicated times p.i. by use of TRIzol (Invitrogen).
RNase protection assays (RPA) were performed using an Ambion RPAIII kit according to the manufacturer's instructions and an RNase A-RNase T1 digestion step followed by analysis on 6% polyacrylamide gels containing 9 M urea and autoradiography. Riboprobes were generated using in vitro transcription of linearized pGem plasmids encoding SV5-specific sequences in the presence of [32P]CTP as described previously (44). To detect viral genomic RNA, a positive-sense riboprobe was used that corresponded to SV5 genomic RNA bases 1 to 171 spanning the le-NP junction. Negative-sense riboprobes that would detect positive-sense viral RNAs specific for NP (genomic RNA bases 1454 to 1680) and M (bases 4320 to 4488) have been described previously (44).
Cell viability, microscopy, and apoptosis assays.
Cell viability was measured by using a One Solution cell proliferation assay (Promega) according to the manufacturer's instructions. Briefly, cells in 96-well dishes were mock infected or virus infected at an MOI of 10, washed with PBS, and replaced with DMEM containing 2% FBS. At the indicated times p.i., 20 μl of CellTiter 96 AQueous One Solution reagent was added to each sample well. Absorbance at 450 nm was recorded and normalized to wells containing only media (no cells) to compensate for background reading.
Microscopy was carried out as described previously (3) using a Nikon Eclipse fluorescence microscope and a ×20 lens. Images were captured using a QImaging digital camera and processed using QCapture software. Exposure times were manually set to be constant between samples.
For annexin V staining, 6-cm dishes of cells were mock infected or virus infected at an MOI of 5 and replaced in DMEM containing 2% FBS. Infections were staggered such that the annexin V staining at the indicated times p.i. occurred at one time. At the indicated times p.i., cells were trypsinized, pelleted, and resuspended in binding buffer at an approximate concentration of 1 × 106 cells/ml. Samples were mixed with annexin V-PE (BD Biosciences) according to the manufacturer's instructions and analyzed using a FACSCalibur flow cytometer (BD Biosciences). Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining was performed as described previously (54) using an in situ death detection kit (Roche Molecular Biochemicals).
Nuclear localization assays.
Cells were infected with the indicated viruses and stained for IRF-3 or the NF-κB p65 subunit at 20 h p.i. Cells were fixed in 4% paraformaldehyde for 6 min at room temperature and then permeabilized with 0.1% Triton X-100-PBS for 10 min at room temperature. After 35 min of blocking with 10% bovine serum albumin at room temperature, cells were washed once with PBS and incubated with primary antibody against IRF-3 at 1:400 (BD PharMingen clone SL-12.1) or NF-κB p65 at 1:100 (Santa Cruz Biotechnology clone C20) for 1 h at 4°C. Then, cells were washed four times with PBS for 5 min each time, incubated with secondary antibody (IRF-3, 1:1,000; NF-κB, 1:2,000) for 40 min at room temperature in the dark, and analyzed by microscopy.
ELISA.
Enzyme-linked immunosorbent assays (ELISA) were performed by using OptEIA human interleukin-6 (IL-6) and IL-8 sets (BD PharMingen) and Human IFN-beta kits (PBL) following the instructions of the manufacturers. A parallel sample of uninfected cells was counted at 24 h p.i. for use in normalizing the secretion of cytokines to 106 cells.
RESULTS
Expression of the WT V protein from the genome of rSV5-P/V-CPI-.
In our initial approach to address the role of the P/V-CPI- V and P proteins in activation of host cell responses, we engineered genomic cDNA clones such that the P and V proteins were encoded as two separate nonedited genes. Infectious virus was never recovered from these cDNA clones, suggesting that RNA editing of the P/V gene is an important aspect of SV5 replication. The alternative approach described below was based on our previous studies showing that cells coinfected with WT rSV5 and the P/V-CPI- mutant displayed much lower activation of IFN pathways, CPE, and apoptosis (55). This result suggested that the WT gene products were dominant over those of P/V-CPI-. Based on the known functions of V, we hypothesized that expression of the WT V protein as an additional gene in the P/V-CPI- genome would complement defects from the original P/V-CPI- mutant.
To test this hypothesis, we utilized a variation on the approach previously described by Brzózka et al. (5) in which the +V-wt virus was generated by inserting a copy of the WT V gene in place of the GFP gene in the P/V-CPI- genome as shown in Fig. 1. Within the V coding region, the editing site was disrupted (Fig. 1B) as described in Materials and Methods such that RNA editing could not occur and only the V mRNA was transcribed from this gene. For these studies, WT rSV5-GFP (19) was used as the proper control for the P/V-CPI- mutant and variants such that all viruses in this study carried an additional gene inserted between HN and L (Fig. 1).
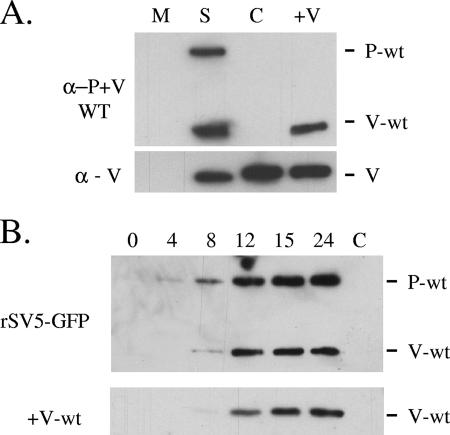
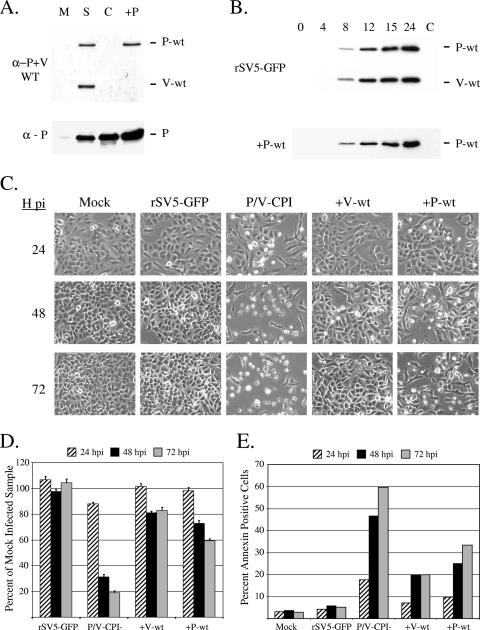
To confirm expression of the WT V protein by the +V-wt virus, A549 cells were mock infected or infected at an MOI of 10 with WT rSV5-GFP, P/V-CPI-, or +V-wt and cell lysates were harvested at 24 h p.i. for analysis by Western blotting (Fig. 2A). All viruses expressed a V protein (Fig. 2A, bottom panel), as shown by reactivity with a monoclonal anti-V antibody that is specific for the cys-rich C-terminal domain found in both WT and CPI- V proteins (41). By contrast, the V5 antibody, which is specific for only the WT P and V proteins (47), recognized both P and V proteins in the WT rSV5-GFP sample but not in the case of the P/V-CPI- sample (Fig. 2A, top panel, S and C lanes). As expected, the V5 antibody recognized V protein but not P protein in lysates from cells infected with the +V-wt virus. As shown in Fig. 2B, the kinetics of accumulation of WT V protein in cells infected with the +V-wt virus were slightly delayed in comparison to WT rSV5-GFP results (Fig. 2B), which is consistent with the presence of a viral gene encoded in a more 3′ promoter-distal position. Total WT V protein levels produced by rSV5-GFP and +V-wt were comparable at late times p.i. The P/V-CPI- virus grows to ∼10-fold-higher titer than WT rSV5-GFP in single-step assays (54). By contrast, the +V-wt virus had high MOI growth kinetics that were very similar to those of WT rSV5-GFP (data not shown).
FIG. 2.
Protein expression for the +V-wt virus. (A) Analysis of V protein expression. A549 cells were mock infected (M lane) or infected with rSV5-GFP (S lane), P/V-CPI- (C lane), or +V-wt (+V lane) at an MOI of 10. Cell lysates were harvested at 24 h p.i. and analyzed by Western blotting with V5 antibody, which recognizes WT P and V proteins (α−P+V WT; top panel), or a monoclonal antibody which recognizes both WT and CPI- V proteins (α - V; bottom panel). (B) Time course of WT V protein expression. A549 cells were infected with rSV5-GFP or +V-wt at an MOI of 10. Cell lysates were harvested at the indicated h p.i. and analyzed by Western blotting using the V5 antibody. P/V-CPI- infected cell lysates were loaded as an antibody specificity control (lane C).
The +V-wt virus induces lower levels of apoptosis compared to P/V-CPI-.
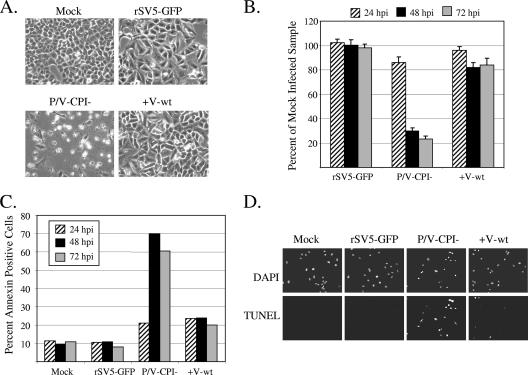
To test the hypothesis that the V protein can limit apoptosis induced by P/V-CPI-, A549 cells were mock infected or infected at an MOI of 10 with rSV5-GFP, P/V-CPI-, or +V-wt virus and cells were examined by microscopy at 72 h p.i. As shown in Fig. 3A, cells infected with the P/V-CPI- mutant showed high levels of CPE not evident in WT rSV5-GFP-infected cells. In the case of the +V-wt virus, some infected cells had rounded up by 72 h p.i., but the overall level of CPE closely matched that seen with the WT rSV5-GFP. Using a cell proliferation assay (Fig. 3B), high MOI infection with the P/V-CPI- virus resulted in a time-dependent loss of cell viability that was not seen with rSV5-GFP infection. Consistent with the microscopy pictures in panel A, the +V-wt virus-infected samples showed a loss in viability of only ∼20% and retained ∼80% viability even out to 72 h p.i. (Fig. 3B).
FIG. 3.
The +V-wt virus induces less apoptotic cell death than the parental P/V-CPI- virus. (A) Microscopy of cells infected with +V-wt virus. A549 cells were examined with a ×20 lens at 72 h p.i. with rSV5-GFP, P/V-CPI-, or +V-wt at an MOI of 10. (B) Cell viability. A549 cells were infected with the indicated viruses at an MOI of 10 and analyzed by a cell proliferation assay at 24, 48, and 72 h p.i. Values are expressed as a percentage of mock-infected sample values at the respective times p.i. with error bars denoting standard deviations from the mean. (C) Annexin staining. A549 cells infected with the indicated viruses at an MOI of 5 were harvested at 24, 48, or 72 h p.i. and analyzed by flow cytometry for cell surface annexin V staining. Data are representative of the results of two independent experiments. (D) TUNEL staining. A549 cells were mock infected or infected with the indicated viruses at an MOI of 10. At 48 h p.i., cells were analyzed for DAPI (4′,6′-diamidino-2-phenylindole) staining or TUNEL staining as described in Materials and Methods.
To determine the level of apoptosis induced by the +V-wt virus, A549 cells were mock infected or infected at an MOI of 5 with rSV5-GFP, P/V-CPI-, or +V-wt virus, and cells were analyzed for annexin V staining at 24, 48, and 72 h p.i. As shown in Fig. 3C, the P/V-CPI- virus induced a time-dependent increase in the percentage of annexin V-positive cells whereas WT rSV5-GFP infected cell results were similar to mock-infected control cell results. Infection with the +V-wt virus resulted in a small increase in annexin V-positive cell numbers at 24 h p.i., but this percentage did not increase further over time. Similarly, mock- and WT rSV5-GFP-infected cells were largely negative with respect to TUNEL staining, which measures late apoptotic stages, while P/V-CPI- virus induced high levels of TUNEL-positive cells (Fig. 3D). Infection with the +V-wt virus resulted in a small fraction of the cell population showing TUNEL-positive results. Taken together, these data indicate that expression of the WT V protein from the P/V-CPI- genome leads to increased cell viability and decreased apoptotic markers relative to the parental P/V-CPI- virus results.
The C-terminal cys-rich domain is required for V protein to limit apoptosis induced by the P/V-CPI mutant.
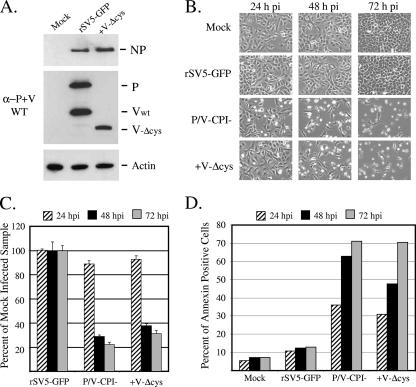
Most functions of the V protein are dependent on the presence of the cys-rich C-terminal domain (36). To determine whether a V protein lacking the cys-rich C-terminal domain could limit P/V-CPI-induced apoptosis, an rSV5 virus was constructed that expressed a truncated V protein from the genome of P/V-CPI- (+V-Δcys; see Fig. 1). The additional gene encoding the truncated V protein was engineered to be identical to that encoded by the +V-wt virus but contained two translational stop codons immediately following the RNA editing site. The resulting +V-Δcys virus directed the synthesis of a truncated V protein that reacted with the V5 antibody specific for the WT N-terminal domain (Fig. 4A).
FIG. 4.
A V protein which lacks the cys-rich C-terminal domain cannot limit apoptotic cell death induced by the parental P/V-CPI- virus. (A) Analysis of V protein expression. A549 cells were mock infected or infected with rSV5-GFP or +V-Δcys at an MOI of 10. Cell lysates were harvested at 24 h p.i. and analyzed by Western blotting for levels of NP (top panel), WT P and V proteins (α−P+V WT; middle panel), or actin (bottom panel). (B) Microscopy of cells infected with +V-Δcys virus. A549 cells were examined by microscopy at 24, 48, and 72 h after infection with the indicated viruses at an MOI of 10. (C and D) Cell viability and Annexin V staining. A549 cells were infected with the indicated viruses at an MOI of 10 (panel C) or 5 (panel D) and analyzed by an MTS cell proliferation assay (panel C) or for annexin V staining (panel D) as described in the legend to Fig. 3. Data in panel D are representative of the results of two independent experiments.
Cells infected with the +V-Δcys virus showed CPE that was very similar to that seen with the parental P/V-CPI- virus. This is evident in the time course of CPE for infected cells shown in Fig. 4B, where the kinetics and level of cell rounding and appearance of cell debris were very similar between +V-Δcys and P/V-CPI- samples. Similarly, time courses of MTS-based cell viability (Fig. 4C) and the percentage of annexin V-positive cells (Fig. 4D) showed only slight differences in kinetics and extent of cell killing and apoptosis between +V-Δcys and the parental P/V-CPI- virus. These data indicate that the cys-rich C-terminal domain of V protein is required in order to limit CPE and apoptosis induced by the P/V-CPI- virus.
Expression of wild-type P protein limits P/V-CPI- induction of CPE and apoptosis.
The induction of host cell responses by the P/V-CPI- virus could also be the result of an altered P protein. To test this hypothesis, we engineered an rSV5 that was analogous to the +V-wt virus except that the WT P protein was expressed from an additional nonedited gene in the genome of P/V-CPI- virus (+P-wt; see Fig. 1). To confirm WT P expression from this virus, A549 cells were mock infected or infected at an MOI of 10 with rSV5-GFP, P/V-CPI-, or +P-wt virus. At 24 h p.i., cell lysates were analyzed by Western blotting using the V5 antibody that recognizes only WT P and V proteins (anti-P+V WT) or with a polyclonal antibody that recognizes both WT and CPI- P proteins (anti-P). As shown in the top panel of Fig. 5A, WT P protein was detected in lysates from cells infected with rSV5-GFP and the +P-wt viruses (lanes S and +P) but not in those from cells infected with the P/V-CPI- virus (lane C). The WT V protein was detected only in the case of the WT rSV5-GFP samples. In time course experiments, Western blotting with the V5 antibody specific for WT P and V showed that the kinetics of WT P protein expression were similar between rSV5-GFP and the +P-wt virus (Fig. 5B). In single-step growth assays, the P/V-CPI- virus grows to ∼10-fold-higher titers than WT rSV5-GFP (54). By contrast, the +P-wt virus grew with kinetics and final yields that were intermediate between WT rSV5-GFP and the P/V-CPI- parental mutant results (data not shown).
FIG. 5.
The +P-wt virus induces less apoptotic cell death than the parental P/V-CPI- virus. (A and B) Analysis and time course of P protein expression. A549 cells were mock infected (M lane) or infected with rSV5-GFP (S lane), P/V-CPI- (C lane), or +P-wt (+P lane) at an MOI of 10. Cell lysates were harvested at 24 h p.i. (panel A) or at the indicated times p.i. (panel B) and analyzed by Western blotting with V5 antibody, which recognizes WT P and V proteins (α-P+V WT; top panels), or a polyclonal P-specific antibody which recognizes both WT and CPI- P proteins (α - P; bottom panels). (C) Microscopy of cells infected with +P-wt virus. A549 cells were infected with the indicated viruses at an MOI of 10. Cells were examined by microscopy at 24, 48, and 72 h p.i. (D and E) Cell viability and annexin V staining. A549 cells were infected at an MOI of 10 (panel D) or 5 (panel E) with the indicated viruses and analyzed at 24, 48, or 72 h p.i. by an MTS cell proliferation assay (panel D) or for annexin V staining (panel E) as described in the legend to Fig. 3. Data in panel E are representative of the results of two independent experiments.
To determine whether the WT P protein could limit CPE induced by P/V-CPI-, A549 cells were infected at an MOI of 10 with rSV5-GFP, P/V-CPI-, +V-wt, or +P-wt virus. At 24, 48, and 72 h p.i., levels of CPE were analyzed by microscopy (Fig. 5C). As shown previously, P/V-CPI- caused extensive CPE that was not evident in cells infected with either WT rSV5-GFP or the +V-wt virus. Unexpectedly, the level of CPE induced by the +P-wt virus was significantly lower than that seen with P/V-CPI- parental virus. This is most evident at the 48 and 72 h time points shown in Fig. 5C, where some rounded cells and some cellular debris were evident in +P-wt-infected cultures, but the majority of cells did not show the massive CPE seen following infection with the parental P/V-CPI- virus.
Cell viability and annexin staining were analyzed to confirm the reduced CPE in cells infected with the +P-wt virus. As shown in the time course of cell viability in Fig. 5D, infection with P/V-CPI- resulted in a dramatic loss in cell viability down to approximately 20% of that of mock-infected samples by 72 h p.i., while +V-wt-infected cells had a viability that was ∼80% of that of mock-infected cells. For the +P-wt virus, there was a small time-dependent decrease in cell viability, but levels were still at ∼60% of mock-infected cell results at 72 h p.i. Similarly, cells infected with the +P-wt virus showed a slow time-dependent increase in annexin-positive cells that was higher than the level seen with control cells infected with WT rSV5-GFP (Fig. 5E). Importantly, however, the percentage of annexin V-positive cells was much lower than that induced by the parental P/V-CPI- virus. Taken together, these data indicate that expression of either the WT P protein or WT V protein can limit CPE and apoptosis induced by the P/V-CPI- virus, with the +P-wt virus activating slightly higher levels of apoptosis than those seen with the +V-wt virus.
The +V-wt and +P-wt viruses induce lower IFN-beta synthesis and IRF-3 activation compared to the parental P/V-CPI- virus.
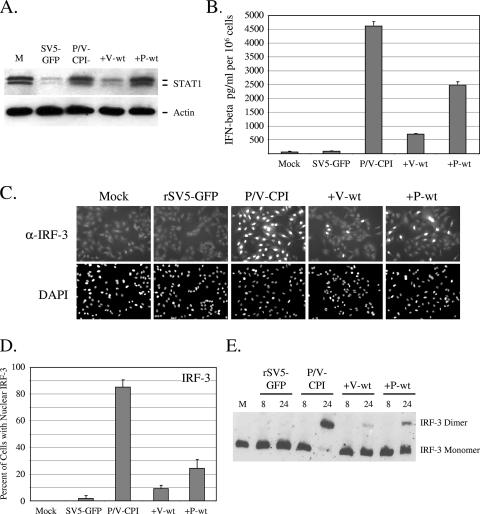
The parental P/V-CPI- virus is defective in targeting STAT1 for degradation and in blocking IFN signaling (54). To determine whether STAT1 levels were altered by infection with the +V-wt or +P-wt viruses, A549 cells were infected at an MOI of 10 with the viruses indicated in Fig. 6A, and levels of STAT1 were analyzed by Western blotting at 8 h p.i. This time point was chosen in order to detect any small differences in the STAT1 levels that may not be evident at later times p.i. Cells infected with WT rSV5-GFP showed a loss of STAT1 which was not seen in cells infected with P/V-CPI- (6, 12, 54). The +V-wt virus induced STAT1 degradation to levels that were similar to those seen with WT rSV5-GFP, while infection with +P-wt did not induce loss of STAT1. These results are consistent with previous data showing that the V protein is sufficient to target STAT1 degradation even in the context of a virus that is defective in this function (55).
FIG. 6.
The +V-wt and +P-wt viruses induce lower IFN-beta synthesis and IRF-3 activation compared to the parental P/V-CPI- virus. (A) STAT1 levels. A549 cells were infected at an MOI of 10 with the indicated viruses. Cell lysates were prepared at 8 h p.i., and levels of STAT1 and actin were determined by Western blotting. (B) IFN-beta synthesis. A549 cells were mock infected or infected at an MOI of 10 with rSV5-GFP, P/V-CPI-, +V-wt, or +P-wt. At 24 h p.i., media were analyzed by ELISA for levels of IFN-beta. Error bars denote standard deviations from the mean. (C) IRF-3 nuclear translocation. A549 cells were infected at an MOI of 10 with the indicated viruses. At 24 h p.i., cells were permeabilized and stained for IRF-3 by use of a monoclonal antibody and for the nucleus by use of DAPI (4′,6′-diamidino-2-phenylindole) as described in Materials and Methods. (D) Quantitation of nuclear IRF-3. Samples from the experiment displayed in panel C were used to determine the number cells displaying intense nuclear staining as a percentage of the population. For each sample, four random fields were counted and averaged, with error bars denoting standard deviations. (E) Dimerization of IRF-3. A549 cells were infected at an MOI of 10 with the indicated viruses, and cell lysates prepared at either 8 or 24 h p.i. were analyzed on nondenaturing gels followed by Western blotting with an IRF-3-specific antibody.
To determine whether expression of the WT V or P proteins could limit IFN induction by P/V-CPI-, A549 cells were infected at an MOI of 10 with rSV5-GFP, P/V-CPI-, +P-wt, or +V-wt virus and media collected at 24 h p.i. were analyzed by ELISA for secreted IFN-beta. As shown in Fig. 6B, WT rSV5-GFP was a poor inducer of IFN-beta whereas cells infected with the P/V-CPI- virus secreted ∼4,500 pg/ml of IFN-beta. Cells infected with the +V-wt virus showed markedly (eightfold) decreased IFN-beta production compared to those infected with P/V-CPI-, which is consistent with our previous results from coinfection studies (55). Unexpectedly, infection with the +P-wt virus also reproducibly induced less IFN-beta secretion than that seen with the parental P/V-CPI- parental virus, and levels of secreted IFN-beta were intermediate between those seen with WT rSV5-GFP and the parental P/V-CPI- virus.
Synthesis of IFN-beta requires the phosphorylation and homodimerization of IRF-3, which then translocates to the nucleus to initiate transcription of the IFN-beta gene (22, 49, 58). To determine the mechanism by which WT V and WT P proteins limit IFN-beta production, infected A549 cells were examined by immunofluorescence for changes in translocation of IRF-3 to the nucleus. As shown in Fig. 6C, the diffuse cytoplasmic IRF-3 location in WT SV5-GFP-infected cells was indistinguishable from mock-infected cell results. By contrast, nearly all cells in cultures infected with the P/V-CPI- virus showed intense staining for IRF-3 in the nucleus, which is consistent with this virus inducing high levels of IFN-beta. As seen with the mock-infected cells, the majority of cells infected with +V-wt or with +P-wt viruses showed IRF-3 staining largely confined to the cytoplasm, with a few cells showing bright nuclear staining. Quantitation from multiple microscopic fields showed that the percentage of cells with intense IRF-3 nuclear staining was reduced from >80% for cultures infected with the parental P/V-CPI- virus to ∼10% and ∼25% for cultures infected with the +V-wt and +P-wt viruses, respectively (Fig. 6D).
To further test the hypothesis that the +V-wt and +P-wt viruses limit IFN-beta synthesis at an early step in the induction pathway, virus-infected cells were assayed for activation of IRF-3 dimerization. A549 cells were infected with rSV5-GFP, P/V-CPI-, +P-wt, or +V-wt viruses at an MOI of 10, and cell lysates were harvested at 8 and 24 h p.i. IRF-3 homodimerization was examined through use of nondenaturing polyacrylamide gel electrophoresis coupled to Western blot analysis (29). At 8 h p.i. there were no detectable IRF-3 dimers for any samples (Fig. 6E). However, by 24 h p.i. the majority of IRF-3 was found as a dimer in lysates from cells infected with P/V-CPI- but not in those from the WT rSV5-GFP sample. Reduced levels of dimeric IRF-3 were seen for both +V-wt and +P-wt samples, which is consistent with the above data with respect to IRF-3 nuclear localization. Taken together, these data indicate that expression of either the WT V or WT P protein can limit IFN-beta induction by the parental P/V-CPI- mutant by acting at a step prior to IRF-3 dimerization and nuclear translocation.
The +V-wt and +P-wt viruses induce lower proinflammatory cytokine synthesis and NF-κB activation compared to the parental P/V-CPI- virus.
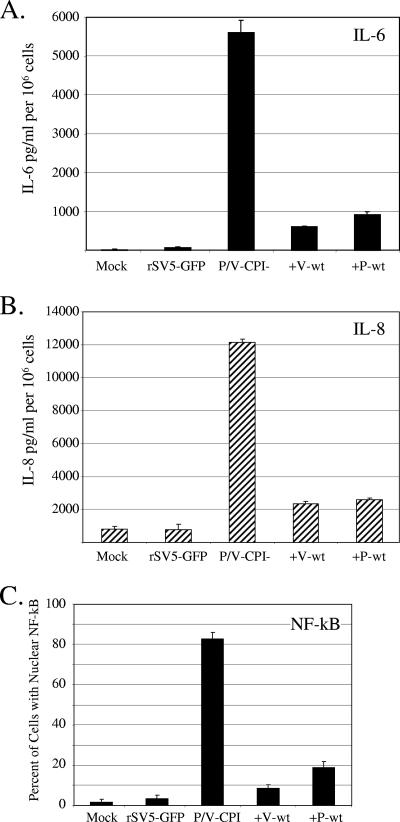
To determine whether expression of the WT V protein or WT P protein could limit proinflammatory cytokine synthesis, A549 cells were infected at an MOI of 10 with the viruses indicated in Fig. 7A, and media collected at 24 h p.i. were analyzed by ELISA for levels of secreted IL-6 (Fig. 7A) or IL-8 (Fig. 7B). WT rSV5-GFP induced IL-6 and IL-8 to levels similar to those seen with mock-infected control samples, while the P/V-CPI- mutant induced high levels of both cytokines as described previously (59). In the case of both +V-wt and +P-wt viruses, infected cells showed reduced levels of IL-6 and IL-8 secretion relative to the parental P/V-CPI- virus.
FIG. 7.
The +V-wt and +P-wt viruses induce lower proinflammatory cytokine synthesis and NF-κB activation than the parental P/V-CPI- virus. (A and B) Samples prepared as described for panel B of Fig. 6 were analyzed by ELISA for levels of IL-6 (panel A) and IL-8 (panel B). Error bars denote standard deviations from the mean. (C) Cells infected with the indicated viruses were permeabilized and stained for NF-κB p65. The number of cells displaying intense nuclear staining was determined as a percentage of the population as described for panel D of Fig. 6.
IL-6 and IL-8 synthesis is dependent on activation of NF-κB (30). To determine whether WT V or WT P protein affected P/V-CPI- induction of NF-κB, infected A549 cells were examined by immunofluorescence for nuclear translocation of NF-κB p65. As shown in Fig. 7C, the percentage of cells with intense NF-κB p65 nuclear staining at 24 h p.i. was reduced from the ∼80% seen with cultures infected with the parental P/V-CPI- to ∼10% and ∼20% for cultures infected with the +V-wt and +P-wt viruses, respectively. These data indicate that expression of either the WT V or WT P protein can also limit activation of proinflammatory cytokines and nuclear translocation of NF-κB compared to parental P/V-CPI- mutant results.
Replication and transcription levels for +V-wt and +P-wt viruses.
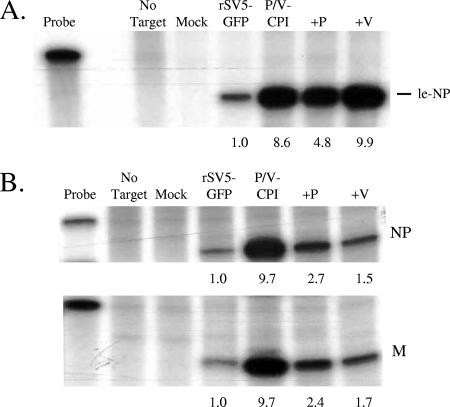
Induction of IFN-beta and apoptosis by the P/V-CPI- virus is sensitive to UV treatment (55), suggesting that activation of these host cell responses is dependent on viral gene expression. RPA were carried out to determine whether changes in viral RNA synthesis correlated with the limited host responses seen with +V-wt and +P-wt viruses. Vero cells were infected at an MOI of 10 with WT rSV5-GFP, P/V-CPI-, +V-wt, or +P-wt virus, and nucleocapsid-associated RNA that was harvested at 14 h p.i. was analyzed using a riboprobe specific for the leader-NP junction as described in Materials and Methods. As shown in Fig. 8A, accumulation of genomic RNA for the P/V-CPI- mutant was much higher than that seen with WT rSV5-GFP. Levels of genomic RNA for the +V-wt virus were very similar to those seen with the P/V-CPI- mutant, while the +P-wt virus reproducibly showed slightly reduced levels compared to P/V-CPI-. Similar results were seen with RNA harvested at 24 h p.i. (data not shown). These results indicate that the differential induction of host cell responses by P/V-CPI- versus +V-wt and +P-wt viruses does not directly correlate with levels of genomic RNA synthesis for these viruses.
FIG. 8.
RNA synthesis in cells infected with the +V-wt and +P-wt viruses. Vero cells were mock infected or infected at an MOI of 10 with the indicated viruses. At 14 h p.i., nucleocapsid RNA (panel A) or total RNA (panel B) was harvested as described in Materials and Methods. (A) RNA was analyzed by RPA using a 32P-labeled riboprobe that annealed to the leader-NP junction in genomic RNA. The position of the le-NP protected fragment is indicated. Data are representative of the results of four independent experiments. (B) Total RNA was analyzed by RPA using 32P-labeled riboprobe that annealed to positive-sense NP or M mRNA. The position of the virus-specific protected fragment is indicated. Data are representative of the results of three independent experiments. For panels A and B, the amount of RNA in the probe lane represents 1/120 of the amount used to hybridize with samples. Numbers below the lanes denote quantitation of the severalfold increase over the level of radioactivity in the WT rSV5-GFP samples set at 1.0.
To measure accumulation of mRNA transcripts, total RNA was harvested at 14 h p.i. from infected cells and analyzed by RPA using radiolabeled probes specific for viral NP or M mRNAs. As seen in Fig. 8B, levels of NP and M mRNA were much higher for the P/V-CPI- mutant than for WT rSV5-GFP, as shown previously (54). Importantly, however, both +P-wt and +V-wt samples showed a decrease in the accumulation of viral mRNA compared to the parental P/V-CPI- virus samples. Similar results were seen with HN mRNA (not shown), but the differences between samples were not as dramatic as those seen for NP and M. Cells infected with the +V-wt showed slightly lower levels of mRNAs than those infected with the +P-wt virus, and this correlated with +V-wt inducing host responses at a lower level than +P-wt virus. These data demonstrate a correlation between reduced viral mRNA accumulation for both the +V-wt and +P-wt viruses and the reduced host cell responses compared to the parental P/V-CPI- mutant responses.
DISCUSSION
Six naturally occurring P/V gene substitutions convert the noncytopathic WT rSV5, which is a poor inducer of host cell responses, into the highly cytopathic P/V-CPI- mutant, which is a potent inducer of IFN-beta, proinflammatory cytokine synthesis, and apoptosis (54, 59). The goal of the work described here was to determine the individual contributions of altered P and altered V proteins to these new phenotypes of the P/V-CPI- mutant virus. As is consistent with our original hypothesis, expression of the WT V protein in the context of the P/V-CPI- virus resulted in dramatic decreases in the secretion of IFN-beta and proinflammatory cytokines and in apoptotic cell death. The most striking result of our work, however, was our finding that expression of the WT P subunit of the viral polymerase also resulted in a clear reduction in apoptosis and in the secretion of proinflammatory cytokines and IFN-beta. Together, our results support the conclusion that the CPI- substitutions in both P and in V contribute to differences in activation of these host cell responses by WT rSV5 and the P/V-CPI- mutant.
Transcription of the SV5 P/V gene produces mRNAs encoding three proteins that share an N-terminal domain but differ in their C-terminal regions: the V protein, which contains the cys-rich domain, the P protein, which contains a longer and distinct C-terminal domain, and the I or W protein, which contains only a few unique C-terminal residues (36, 51). In the case of Sendai virus (SeV), the W protein is abundantly expressed in infected cells and is thought to play a role in controlling viral RNA synthesis (11, 24). In SV5-infected cells, the I protein is very difficult to detect, and the functions of I protein during a virus infection are not known. Our engineered V-Δcys protein is essentially the same as the I protein. Our results obtained with the +V-Δcys virus suggest that I protein, at least in the context of the P/V-CPI- mutant infection, cannot limit apoptosis induction or activation of host cell cytokine responses.
Our results with the +V-wt virus are consistent with the known roles of the paramyxovirus V protein in limiting IFN-beta induction (43) but raise the question of why the CPI- V protein cannot serve a similar function during P/V-CPI- infection. The paramyxovirus V protein has been shown to inhibit activation of IFN-beta synthesis in response to dsRNA through binding of the RNA helicase mda-5 (2, 7, 43), and this inhibition occurs when the V protein C-terminal cys-rich domain is expressed without the N-terminal domain. Consistent with this, the CPI- V protein has an intact cys-rich domain (6) and has been shown to block dsRNA-induced IFN-beta synthesis when expressed from a transfected plasmid (43). Importantly, however, the CPI- V protein cannot block IFN-beta induction in the context of the P/V-CPI- mutant virus, since this virus is a potent inducer of IFN-beta (55). One possibility to explain this discrepancy is that the activation of IFN-beta responses by P/V-CPI- virus is not through induction by dsRNA. This proposal would seem to be supported by recent work showing that viral components other than dsRNA can activate IFN synthesis (50) and that negative-strand RNA viruses do not generate dsRNA to levels that can be detected by use of a monoclonal antibody (57). The P/V-CPI- virus may induce IFN-beta through the RIG-I pathway, which is activated by RNA containing an uncapped 5′-end triphosphate (25). Consistent with this, it has been proposed that RIG-I is not blocked by the paramyxovirus V protein (7).
Many paramyxoviruses cause extensive CPE and induce apoptotic cell death (14, 28, 33). In the case of SeV, the gene products that modulate apoptosis are not completely defined, but the C proteins and trailer RNA have been identified as factors that can contribute to the efficiency of cell killing (16, 28, 34). Here we show that expression of the WT V protein in the context of the P/V-CPI- mutant infection leads to higher cell viability and decreased apoptosis compared to the results seen with the parental P/V-CPI- virus infection. While it has been shown that the measles virus V protein is sufficient to block apoptosis induced by exogenous activators of p53-related pathways (10), it remains to be seen whether the SV5 V protein is a general inhibitor of apoptosis or whether it is specific for virus-induced apoptosis. Since the V protein can inhibit viral RNA synthesis (23, 37), an alternative hypothesis is that the SV5 V protein indirectly reduces cell killing by altering the apoptosis-inducing potential of the virus by altering RNA synthesis rather than by directly acting as an antagonist of cellular death pathways.
The most striking finding from our work is that expression of the WT P protein from the genome of the P/V-CPI- virus leads to lower activation of apoptosis, IFN-beta synthesis, and proinflammatory cytokine secretion. While both the +V-wt and the +P-wt infections showed greatly reduced host cell responses compared to the parental P/V-CPI- responses, they were not completely reduced to the low levels seen with WT rSV5-GFP. We interpret these data as indicating that in the original P/V-CPI- mutant, mutations in both V and P proteins contribute to the potent activation of antiviral responses and that these responses are partially reduced when one or the other of the WT proteins is expressed.
Given the known functions of the P protein in viral RNA synthesis, how does the WT P protein limit these host cell responses to P/V-CPI- infection? The response of a host cell to virus infection can be viewed as a function of (i) the presence of a viral suppressor of antiviral pathways and (ii) the level or type of viral inducing component that is produced during the infection. Thus, the level at which a virus activates host responses can be determined by changes in the effectiveness of a viral antagonist as well as in the levels or types of key activating components that are synthesized. In terms of suppressing antiviral pathways, the P protein of both rabies virus and Borna disease virus can inhibit induction of the IFN-beta promoter by dsRNA, resulting in a block in IRF-3 activation (5, 53). In general, however, the paramyxovirus P protein is thought to function primarily as a subunit for the viral RNA-dependent RNA polymerase. Consistent with this, expression of the SV5 P protein from plasmid DNA is not able to counteract the induction of IFN by dsRNA (43).
Based on the known function of the SV5 P protein in RNA synthesis and the finding that early phases of cytokine response pathways (e.g., IRF-3 and NF-κB activation) are limited, we hypothesize that the WT P protein limits host cell responses by limiting the synthesis of key viral components that trigger antiviral pathways. This is supported by our finding that host responses to P/V-CPI- are sensitive to UV treatment (55). Thus, in this hypothesis the CPI- P mutations, in combination with the WT L protein, result in a polymerase complex which synthesizes levels or types of RNA different from those seen in the context of a WT rSV5 infection, and this results in activation of host antiviral pathways. In the case of the +P-wt virus, we hypothesize that polymerase complexes containing the WT P protein are dominant over those of the mutant P and that this results in lower synthesis of viral activating components.
P protein functional domains have been identified in the case of SeV by mutagenesis and structural analyses (reviewed in reference 32). The shared N-terminal P/V domain contains regions for chaperoning soluble NPo to the nascent viral RNA during the encapsidation stage of genome replication (23) as well as regions that are dispensable for RNA synthesis. By contrast, the unique C-terminal region of the SeV P protein can function by itself during transcription in vitro and is involved in P protein multimerization, binding to the polymerase catalytic subunit L, and forming stable interactions of the L-P polymerase complex with the nucleocapsid template (32). Future work should determine whether these six CPI- mutations reside in domains that are critical for these P functions or for other functions such as binding soluble NPo.
While the mechanism by which the WT P and V proteins limit host responses will require further analyses, our results with respect to the accumulation of viral RNA suggest the hypothesis that the level of viral transcription may be an important factor. The level of accumulation of viral genomes in infected cells did not correlate with activated host cell responses to infection with a particular rSV5 virus. This was made evident by our finding that cells infected with both the +V-wt and +P-wt viruses showed genomic RNA levels that were higher than that of WT rSV5-GFP and that +V-wt genomic RNA was detected at slightly higher levels than +P-wt but induced lower overall host cell responses. In the case of mRNA levels, however, both the +P-wt and +V-wt viruses directed the synthesis of NP and M mRNAs to levels that were lower than those seen with the P/V-CPI- mutant and that more closely matched that of the WT rSV5-GFP virus. Thus, one possibility is that the higher level of synthesis of viral mRNA by the P/V-CPI- mutant increases the chances of producing aberrant mRNAs that trigger antiviral responses and that expression of the WT P or V protein suppresses aberrant transcription. One such pathway that could be triggered by aberrant transcription involves RIG-I, a cellular RNA helicase that recognizes single-stranded RNA lacking a 5′ cap structure (25). Work is in progress to test this hypothesis and determine the mechanism by which WT P and V proteins limit the transcription activity of the viral polymerase.
Acknowledgments
We thank members of the Parks laboratory and Doug Lyles for helpful comments on the manuscript. We are grateful to Biao He and Robert A. Lamb (Northwestern University) for the original gift of the rSV5 infectious clones and to Robert Lamb and Reay Paterson for the kind gift of the anti-V monoclonal antibodies.
This work was supported by National Institutes of Health grant AI-42023.
Footnotes
Published ahead of print on 8 August 2007.
REFERENCES
- 1.Andrejeva, J., D. F. Young, S. Goodbourn, and R. E. Randall. 2002. Degradation of STAT1 and STAT2 by the V proteins of SV5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrejeva, J., K. S. Childs, D. F. Young, R. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arimilli, S., M. A. Alexander-Miller, and G. D. Parks. 2006. A simian virus 5 (SV5) P/V mutant is less cytopathic than wild-type SV5 in human dendritic cells and is a more effective activator of dendritic cell maturation and function. J. Virol. 80:3416-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biron, C. A., and G. C. Sen. 2007. Innate responses to viral infections, p. 249-278. In B. Fields, D. Knipe, and P. Howley. (ed.), Fields virology, 5th ed. Lippincott Williams and Wilkins Publishers, Philadelphia, PA.
- 5.Brzózka, K., S. Finke, and K.-K. Conzelmann. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 79:7673-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatziandreou, N., D. Young, J. Andrejeva, S. Goodbourn, and R. E. Randall. 2002. Differences in interferon sensitivity and biological properties of two related isolates of simian virus 5: a model for virus persistence. Virology 293:234-242. [DOI] [PubMed] [Google Scholar]
- 7.Childs, K. S., N. Stock, C. Ross, J. Andrejeva, L. Hilton, M. Skinner, R. E. Randall, and S. Goodbourn. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190-200. [DOI] [PubMed] [Google Scholar]
- 8.Choppin, P. W. 1964. Multiplication of myxovirus (SV5) with minimal cytopathic effects and without interference. Virology 23:224-233. [DOI] [PubMed] [Google Scholar]
- 9.Conzelmann, K. K. 2005. Transcriptional activation of alpha/beta interferon genes: interference by nonsegmented negative-strand RNA viruses. J. Virol. 79:5241-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz, C. D., H. Palosaari, J. P. Parisien, P. Devaux, R. Cattaneo, T. Ouchi, and C. M. Horvath. 2006. Measles virus V protein inhibits p53 family member p73. J. Virol. 80:5644-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curran, J., J. B. Marq, and D. Kolakofsky. 1995. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J. Virol. 69:849-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and SV5 block activation of IFN-responsive genes: importance of virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of SV5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elankumaran, S., D. Rockemann, and S. K. Samal. 2006. Newcastle disease virus exerts oncolysis by both intrinsic and extrinsic caspase-dependent pathways of cell death. J. Virol. 80:7522-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 16.Garcin, D., G. Taylor, K. Tanebayashi, R. Compans, and D. Kolakofsky. 1998. The short Sendai virus leader region controls induction of programmed cell death. Virology 243:340-353. [DOI] [PubMed] [Google Scholar]
- 17.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signaling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 19.He, B., R. G. Paterson, C. D. Ward, and R. A. Lamb. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249-260. [DOI] [PubMed] [Google Scholar]
- 20.He, B., G. Y. Lin, J. E. Durbin, R. K. Durbin, and R. A. Lamb. 2001. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J. Virol. 75:4068-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 22.Hiscott, J., P. Pitha, P. Genin, H. Nguyen, C. Heylbroeck, Y. Mamane, M. Algarte, and R. Lin. 1999. Triggering the interferon response: the role of IRF-3 transcription factor. J. Interferon Cytokine Res. 19:1-13. [DOI] [PubMed] [Google Scholar]
- 23.Horikami, S. M., J. Curran, D. Kolakofsky, and S. A. Moyer. 1992. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horikami, S. M., S. Smallwood, and S. A. Moyer. 1996. The Sendai virus V protein interacts with the NP protein to regulate viral genome RNA replication. Virology 222:383-390. [DOI] [PubMed] [Google Scholar]
- 25.Hornung, V., J. Ellengast, S. Kim, K. Brzózka, A. Jung, H. Kato, H. Poeck, S. Akira, K.-K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 26.Horvath, C. M. 2000. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem. Sci. 25:496-502. [DOI] [PubMed] [Google Scholar]
- 27.Horvath, C. M. 2004. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur. J. Biochem. 271:4621-4628. [DOI] [PubMed] [Google Scholar]
- 28.Iseni, F., D. Garcin, M. Nishio, N. Kedersha, P. Anderson, and D. Kolakofsky. 2002. Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J. 21:5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwamura, T., M. Yoneyama, K. Yamaguchi, W. Suhara, W. Mori, K. Shiota, Y. Okabe, H. Namiki, and T. Fujita. 2001. Induction of IRF-3/-7 kinase and NF-κB in response to double-stranded RNA and virus infection: common and unique pathways. 6:375-388. [DOI] [PubMed]
- 30.Karin, M., and Y. Ben-Narieh. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 31.Keller, M. A., S. K. Murphy, and G. D. Parks. 2001. RNA replication from the simian virus 5 antigenomic promoter requires three sequence-dependent elements separated by sequence-independent spacer regions. J. Virol. 75:3993-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolakofsky, D., P. Le Mercier, F. Iseni, and D. Garcin. 2004. Viral polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology 318:463-473. [DOI] [PubMed] [Google Scholar]
- 33.Kotelkin, A., E. A. Prikhodko, J. I. Cohen, P. L. Collins, and A. Bukreyev. 2003. Respiratory syncytial virus infection sensitizes cells to apoptosis mediated by tumor necrosis factor-related apoptosis-inducing ligand. J. Virol. 77:9156-9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koyama, A. H., H. Irie, A. Kato, Y. Nagai, and A. Adachi. 2003. Virus multiplication and induction of apoptosis by Sendai virus: role of the C proteins. Microbes Infect. 5:373-378. [DOI] [PubMed] [Google Scholar]
- 35.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C-terminal cys-rich region of mumps virus structural V protein correlates with block of interferon α and γ signal transduction pathway through decrease of STAT1. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 36.Lamb, R. A., and G. Parks. 2007. Paramyxoviridae: the viruses and their replication, p. 1449-1496. In B. Fields, D. Knipe, and P. Howley (ed.), Fields virology, 5th ed. Lippincott Williams and Wilkins Publishers, Philadelphia, PA.
- 37.Lin, Y., F. Horvath, J. A. Aligo, R. Wilson, and B. He. 2005. The role of simian virus 5 V protein on viral RNA synthesis. Virology 338:270-280. [DOI] [PubMed] [Google Scholar]
- 38.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 39.Parks, C. L., S. E. Witko, C. Kotash, S. L. Lin, M. S. Sidhu, and S. A. Udem. 2006. Role of V protein RNA binding in inhibition of measles virus minigenome replication. Virology 348:96-106. [DOI] [PubMed] [Google Scholar]
- 40.Parks, G. D., V. A. Young, C. Koumenis, E. K. Wansley, J. L. Layer, and K. M. Cooke. 2002. Controlled cell killing by a recombinant nonsegmented negative-strand RNA virus. Virology 293:192-203. [DOI] [PubMed] [Google Scholar]
- 41.Paterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. Paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208:121-131. [DOI] [PubMed] [Google Scholar]
- 42.Peeples, M. E., and P. L. Collins. 2000. Mutations in the 5′ trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J. Virol. 74:146-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 44.Rassa, J. C., and G. D. Parks. 1998. Molecular basis for naturally occurring elevated readthrough transcription across the M-F junction of the paramyxovirus SV5. Virology 247:274-286. [DOI] [PubMed] [Google Scholar]
- 45.Roulston, A., R. C. Marcellus, and P. E. Branton. 1999. Viruses and apoptosis. Annu. Rev. Microbiol. 53:577-628. [DOI] [PubMed] [Google Scholar]
- 46.Sen, G. C. 2000. Novel functions of interferon-induced proteins. Semin. Cancer Biol. 10:93-101. [DOI] [PubMed] [Google Scholar]
- 47.Southern, J. A., D. F. Young, F. Heaney, W. K. Baumgartner, and R. E. Randall. 1991. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J. Gen. Virol. 72:1551-1557. [DOI] [PubMed] [Google Scholar]
- 48.Sun, M., T. A. Rothermel, L. Shuman, J. A. Aligo, S. Xu, Y. Lin, R. A. Lamb, and B. He. 2004. Conserved cysteine-rich domain of paramyxovirus simian virus 5 V protein plays an important role in blocking apoptosis. J. Virol. 78:5068-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 50.tenOever, B. R., M. J. Servant, N. Grandvaux, R. Lin, and J. Hiscott. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 76:3659-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas, S. M., R. A. Lamb, and R. G. Paterson. 1988. Two mRNAs that differ by two nontemplated nucleotides encode the amino co-terminal proteins P and V of paramyxovirus SV5. Cell 54:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulane, C. M., A. Kentsis, C. D. Cruz, J. P. Parisien, K. L. Schneider, and C. M. Horvath. 2005. Composition and assembly of STAT-targeting ubiquitin ligase complexes: paramyxovirus V protein carboxyl terminus is an oligomerization domain. J. Virol. 79:10180-10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Unterstab, G., S. Ludwig, A. Anton, O. Planz, B. Dauber, D Krappmann, G. Heins, C. Ehrhardt, and T. Wolff. 2005. Viral targeting of the interferon-beta inducing TRAF family member-associated NF-κB activator (TANK)-binding kinase-1. Proc. Natl. Acad. Sci. USA 102:13640-13645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wansley, E. K., and G. D. Parks. 2002. Naturally occurring substitutions in the P/V gene convert the noncytopathic paramyxovirus simian virus 5 into a virus that induces alpha/beta interferon synthesis and cell death. J. Virol. 76:10109-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wansley, E. K., J. M. Grayson, and G. D. Parks. 2003. Apoptosis induction and interferon signaling but not IFN-beta promoter induction by an SV5 P/V mutant are rescued by coinfection with wild-type SV5. Virology 316:41-54. [DOI] [PubMed] [Google Scholar]
- 56.Wansley, E. K., P. J. Dillon, M. D. Gainey, J. Tam, S. D. Cramer, and G. D. Parks. 2005. Growth sensitivity of a recombinant simian virus 5 P/V mutant to type I interferon differs between tumor cell lines and normal primary cells. Virology 335:131-144. [DOI] [PubMed] [Google Scholar]
- 57.Weber, F., V. Wagner, S. B. Rasmussen, R. Hartmann, and S. R. Paludan. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young, V. A., P. J. Dillon, and G. D. Parks. 2006. Variants of the paramyxovirus simian virus 5 with accelerated or delayed viral gene expression activate proinflammatory cytokine synthesis. Virology 350:90-102. [DOI] [PubMed] [Google Scholar]