Abstract
Turnip yellow mosaic virus (TYMV), a positive-strand RNA virus belonging to the alphavirus-like supergroup, encodes its nonstructural replication proteins as a 206K precursor with domains indicative of methyltransferase (MT), proteinase (PRO), NTPase/helicase (HEL), and polymerase (POL) activities. Subsequent processing of 206K generates a 66K protein encompassing the POL domain and uncharacterized 115K and 85K proteins. Here, we demonstrate that TYMV proteinase mediates an additional cleavage between the PRO and HEL domains of the polyprotein, generating the 115K protein and a 42K protein encompassing the HEL domain that can be detected in plant cells using a specific antiserum. Deletion and substitution mutagenesis experiments and sequence comparisons indicate that the scissile bond is located between residues Ser879 and Gln880. The 85K protein is generated by a host proteinase and is likely to result from nonspecific proteolytic degradation occurring during protein sample extraction or analysis. We also report that TYMV proteinase has the ability to process substrates in trans in vivo. Finally, we examined the processing of the 206K protein containing native, mutated, or shuffled cleavage sites and analyzed the effects of cleavage mutations on viral infectivity and RNA synthesis by performing reverse-genetics experiments. We present evidence that PRO/HEL cleavage is critical for productive virus infection and that the impaired infectivity of PRO/HEL cleavage mutants is due mainly to defective synthesis of positive-strand RNA.
Many positive-strand RNA viruses produce their replication proteins as polyprotein precursors that are subsequently cleaved to generate functional viral gene products. Such proteolytic-processing events allow the expression of multiple intermediate products that may perform various functions in viral replication, possibly distinct from those performed by mature products, thus providing additional ways of regulating the viral multiplication cycle (37, 45). Understanding the proteolytic-processing pathways of viral polyprotein precursors can thus help decipher the molecular processes directing the assembly, function, and regulation of viral replication machineries.
Here, we address this question by studying the proteolytic processing of the replication polyprotein of Turnip yellow mosaic virus (TYMV), the type member of the genus Tymovirus. TYMV is a spherical plant virus that shares replication features with other positive-strand RNA viruses in the alphavirus-like supergroup (14, 21) and has proven useful in the study of fundamental aspects of viral multiplication (10).
The two extensively overlapping open reading frames (ORFs) encoded by the 6.3-kb genomic RNA (Fig. 1) produce a 69K protein that serves as the viral movement protein and RNA interference suppressor and a 206-kDa precursor protein (206K) that is the only viral protein necessary for replication (10). Viral replication is initiated by the synthesis of a negative-strand RNA complementary to the genomic positive-strand RNA, which in turn serves as a template for the synthesis of new positive-strand genomic RNA and of a subgenomic RNA that allows expression of the 20-kDa viral coat protein (CP). Compared with alphaviruses, little is known about the processing of the TYMV 206K protein and the role of such processing, if any, in the regulation of replication.
FIG. 1.
Schematic representation of the genomic organization of TYMV RNA. The open bars denote viral ORFs. The encoded 206K protein is proteolytically processed at a peptide bond (indicated by a filled triangle). Protein domains are indicated. The locations of epitopes recognized by the anti-PRR and anti-HEL antibodies used in this study are indicated by filled squares.
The 206K protein contains sequence domains indicative of methyltransferase (MT), proteinase (PRO), NTPase/helicase (HEL), and RNA-dependent RNA polymerase (POL) activities, as well as an ∼200-amino-acid (aa)-long proline-rich region (PRR) that may constitute a hinge between the MT and PRO domains (Fig. 1). The PRO domain, which is characterized by a Cys783-His869 catalytic dyad, belongs to the subgroup of viral papain-like proteases (2, 40, 41). Previous in vitro studies have demonstrated its involvement in the cleavage of 206K between residues 1259 and 1260, leading to the synthesis of an N-terminal product of 140 kDa (140K) containing the MT, PRR, PRO, and HEL domains and a C-terminal 66-kDa protein (66K) encompassing the POL domain (4, 20, 34) (Fig. 1).
This HEL/POL cleavage has been demonstrated to be functional in vivo (39) and appears to be essential for viral replication (2). However, while 66K is readily detected in infected samples using specific antibodies (39), only trace amounts of 140K are detected in infected cells using an antibody raised against the PRR domain (hereafter called anti-PRR) (18) (Fig. 1). Instead, two shorter products of 115 and 85 kDa, referred to as 115K and 85K, respectively, are detected in infected cells (18), suggesting that the 140K protein may be further processed in vivo. The 115K protein is of particular interest because it has long been known to be a major component of the purified TYMV replicase (6, 36).
The goal of the present work was to study the remaining cleavages occurring in vivo in the TYMV nonstructural polyprotein using a transient expression system in plant cells and specific antisera and to determine whether these cleavage events are essential for viral infectivity. Here, we demonstrate that the TYMV proteinase mediates one additional cleavage within the 206K polyprotein, generating 115K and a product encompassing the HEL domain. Using reverse-genetics experiments, we also report that processing at the novel cleavage site is critical for viral infectivity.
MATERIALS AND METHODS
Plasmid construction.
All DNA manipulations were performed using standard techniques (1, 42). The full-length TYMV cDNA clone E17, which produces infectious transcripts, and its derivative E17-stopΔ, in which the 206K is truncated at aa 1259, have been described previously (11, 38). Plant expression vectors were derived from pΩ-206K, pΩ-140K, or pΩ-66K (18, 38). Mutations were introduced by PCR-mediated site-directed mutagenesis or by subcloning of restriction fragments. The overall structures of all plasmids were confirmed by restriction analysis, and the sequences of PCR-generated DNA fragments were confirmed by DNA sequencing. When proteins are truncated, the encoded amino acids are indicated within parentheses in the plasmid name. Primer sequences and cloning details will be made available on request.
Preparation and transfection of Arabidopsis protoplasts.
Protoplasts of Arabidopsis thaliana were prepared as described previously (18, 19) and transfected with 5 μg expression vector or capped in vitro transcripts generated from linearized DNA templates as described previously (11).
Protein extraction, SDS-PAGE, antibodies, and immunodetection analyses.
Total-protein extraction from protoplasts, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and immunodetection using anti-66K, anti-CP, or anti-140K (anti-PRR) antibodies were performed as described previously (18, 19, 39).
The polyclonal anti-HEL antiserum raised against the TYMV HEL domain was obtained by injecting rabbits with a mixture of two synthetic peptides (aa 1079 to 1093 and aa 1245 to 1259) conjugated to keyhole limpet hemocyanin. Peptide synthesis, coupling, immunization, and affinity purification of antibodies were performed by Eurogentec (Belgium) according to standard double-XP protocols. For detection of the TYMV HEL domain, proteins were separated by 10% SDS-PAGE and subjected to immunodetection with antibodies purified against peptide 1245-1259 (hereafter called anti-HEL), used at a 1/200 dilution.
RNA extraction and Northern blot hybridization.
Total RNA extraction from protoplasts, agarose-formaldehyde electrophoresis, blotting, hybridization with TYMV strand-specific riboprobes, and signal quantitation were performed as previously described (5, 19).
Fluorescence microscopy.
Fluorescence microscopy of transfected protoplasts and image acquisition were performed as previously described (18, 38).
Sequence analysis.
Alignment of primary sequences for replication proteins of tymoviruses was performed using ClustalW (47) and shaded with Multiple Align Show (http://bioinformatics.org/sms/multi_align.html). The accession numbers of the viral replication proteins of tymoviruses are as follows: TYMV, NP_663297; Eggplant mosaic virus, NP_040968; Kennedya yellow mosaic virus, NP_044328; Ononis yellow mosaic virus, NP_041257; Erysimum latent virus, NP_047920; Physalis mottle virus, NP_619756; Chayote mosaic virus, NP_067737; Dulcamara mottle virus, YP_406375; Plantago mottle virus, AAW88526; Scrophularia mottle virus, AAW88520; and Anagyris vein yellowing virus, AAW88529.
RESULTS
The viral proteinase generates 115K, but not 85K.
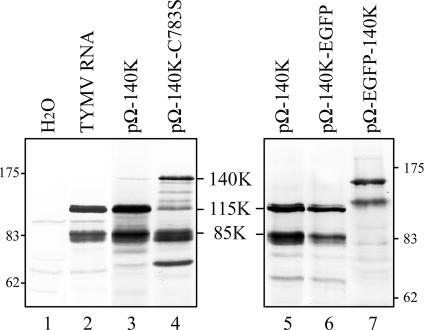
Immunodetection using anti-PRR antiserum has previously shown that 115K and 85K proteins are readily detected in infected cells (18) (Fig. 2, lane 2) and that these products derive exclusively from the 140K protein, as both could be detected upon transfection of Arabidopsis protoplasts with the pΩ-140K vector, which expresses the TYMV 140K protein (18) (Fig. 2, lane 3). These data suggested the occurrence of at least two additional cleavages within the 140K protein.
FIG. 2.
Characterization of 115K and 85K proteins. Arabidopsis protoplasts were transfected with water (lane 1), TYMV RNA (lane 2), or the expression vectors indicated. The cells were harvested 48 h posttransfection, and total proteins were subjected to 8% SDS-PAGE and immunoblot analysis with anti-PRR polyclonal antibodies. The positions of viral proteins and molecular mass markers (Biolabs) are indicated (in kDa).
To determine whether these cleavage events require an active viral proteinase or whether the responsible proteolytic activity is of cellular origin, the expression vector pΩ-140K-C783S, which encodes a 140K protein debilitated in its proteolytic activity due to a point mutation in the proteinase active site (41), was transfected into Arabidopsis protoplasts. Upon immunodetection of protein extracts with anti-PRR (Fig. 2, lane 4), we observed that such a mutation was detrimental to the accumulation of 115K, with the 140K unprocessed protein being detected instead, while the 85K product was produced in amounts comparable to that expressed by the pΩ-140K expression vector (lane 3). This result indicates that the 115K protein results from the proteolytic activity of the viral proteinase whereas the 85K product does not.
The 115K and 85K proteins correspond to N-terminal cleavage products of 140K.
To determine whether the 115K and 85K proteins correspond to N- or C-terminal cleavage products of 140K, Arabidopsis protoplasts were transfected with the expression vectors pΩ-EGFP-140K and pΩ-140K-EGFP, which encode the 140K protein fused at its N or C terminus, respectively, to enhanced green fluorescent protein (EGFP). Upon immunodetection of protein extracts with anti-PRR, we observed that pΩ-140K-EGFP gave rise to products identical in size to 115K and 85K (Fig. 2, lane 6), whereas pΩ-EGFP-140K led to the expression of slower-migrating products whose estimated molecular masses were consistent with that expected for the fusion of EGFP to 85K and 115K proteins (Fig. 2, lane 7). This result indicates that both the 115K and 85K proteins correspond to N-terminal cleavage products of the 140K protein and that the cleavage sites are located downstream of the PRR.
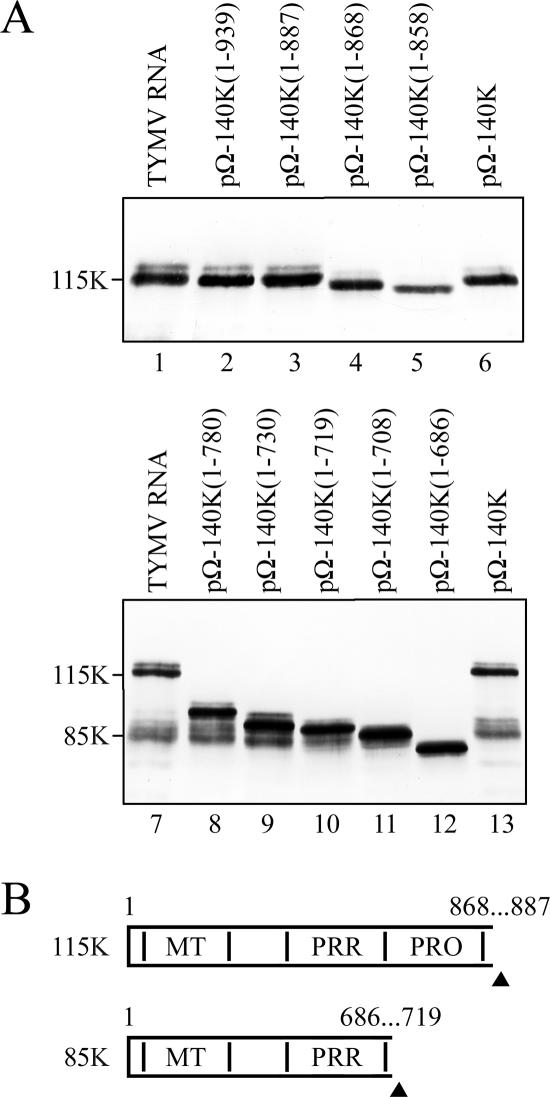
Deletion mapping of the cleavage sites.
To locate the cleavage sites leading to the appearance of 115K and 85K products, a series of constructs encoding C-terminally truncated 140K derivatives were transfected into Arabidopsis protoplasts and the corresponding protein extracts were analyzed by immunoblotting using anti-PRR (Fig. 3A). From the electrophoretic mobilities of the encoded proteins and comparison with the 115K and 85K products derived from the full-length 140K protein (lanes 6 and 13), the cleavage site giving rise to the 115K protein was mapped to between residues 868 and 887, whereas the 85K product, which contains several subspecies with similar mobilities, appeared to be generated by a cleavage(s) located between residues 686 and 719.
FIG. 3.
Deletion mapping of the cleavage sites generating the 115K and 85K proteins. (A) Arabidopsis protoplasts were transfected with TYMV RNA (lanes 1 and 8) or the expression vectors indicated. The cells were harvested 48 h posttransfection, and total proteins were subjected to 6% (top) or 8% (bottom) SDS-PAGE and immunoblot analysis with anti-PRR polyclonal antibodies. The positions of viral proteins are indicated. (B) Schematic representation of the 115K and 85K proteins. Cleavage sites and protein domains are designated as in Fig. 1.
These observations indicate that the 85K protein carries the MT and PRR domains, while the 115K protein results from a cleavage between the PRO and HEL domains and encompasses the MT, PRR, and PRO domains (Fig. 3B).
Fine mapping of the 115K cleavage site.
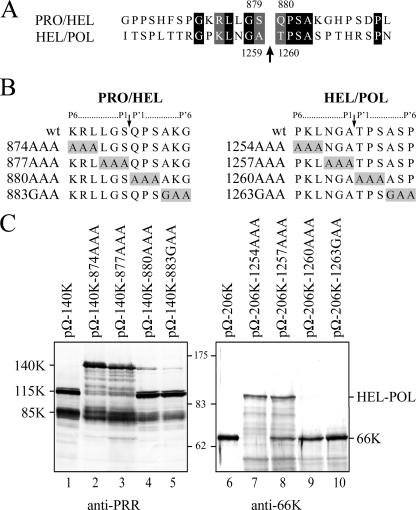
As the 115K protein is generated by a cleavage between the PRO and HEL domains (Fig. 3) mediated by the viral proteinase (Fig. 2), we examined the corresponding region of the 140K protein for the presence of a potential viral papain-like proteinase cleavage site. Based on sequence alignments and similarities with the previously identified HEL/POL cleavage site between aa 1259 and 1260 (4, 20), a serine-glutamine dipeptide corresponding to aa 879 and 880 of the 206K ORF was identified as a potential PRO/HEL cleavage site (Fig. 4A). Processing at this site would result in the production of cleavage products in perfect agreement with those observed (Fig. 3).
FIG. 4.
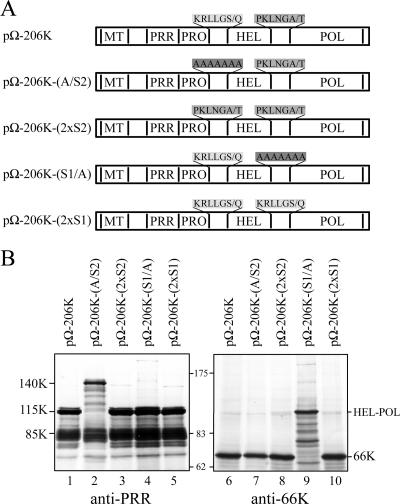
Mutagenesis of the proposed cleavage site generating the 115K protein. (A) Amino acid sequence alignment of the PRO/HEL and HEL/POL cleavage sites. Identical or similar residues are boxed in black or gray, respectively. The arrow indicates the previously characterized HEL/POL and the proposed PRO/HEL cleavage sites. (B) Amino acid sequence of the PRO/HEL (left) and HEL/POL (right) cleavage sequence mutants. The mutated residues are boxed in gray. P6 to P1 and P′1 to P′6 refer to position of the residues relative to the previously characterized or proposed cleavage sites. (C) Arabidopsis protoplasts were transfected with the expression vectors indicated. The cells were harvested 48 h posttransfection, and total protein extracts were subjected to 8% SDS-PAGE and immunoblot analysis with anti-PRR (left) or anti-66K (right). The positions of viral proteins and molecular mass markers are indicated.
To test the above prediction, mutagenesis of the putative PRO/HEL site was conducted. To this end, alanine or glycine substitutions spanning residues 874 to 886 of the 140K protein (positions P6 to P′6 relative to the predicted cleavage site, according to the nomenclature of Schechter and Berger [43]) were introduced into the expression vector pΩ-140K (Fig. 4B, left). The corresponding mutant proteins were transiently expressed in Arabidopsis protoplasts, and cleavage of the 140K protein was assessed by immunodetection with anti-PRR (Fig. 4C, lanes 1 to 5). As a control, identical substitutions within the previously characterized HEL/POL cleavage site were introduced into the expression vector pΩ-206K (Fig. 4B, right), and their effects on the generation of the 66K protein were followed by Western blotting experiments using anti-66K (Fig. 4C, lanes 6 to 10).
Substitutions of aa 874 to 879 (P6 to P1) strongly inhibited PRO/HEL processing, as the 140K protein was detected in place of the 115K protein (Fig. 4C, lanes 2 and 3). In contrast, substitutions at positions 880 to 885 (P′1 to P′6) had less influence on the PRO/HEL cleavage, with the 115K protein being the major product detected (Fig. 4C, lanes 4 and 5). These results are strikingly similar to those observed for the corresponding HEL/POL mutants affecting P6 to P1 and P′1 to P′6 (Fig. 4C, lanes 7 to 10). Taken together, the sequence analyses and experimental data strongly support the identification of Ser-879/Gln-880 as the probable PRO/HEL cleavage site in the TYMV 140K.
In vivo analyses of the cleavages generating 85K and 115K.
To determine whether the cleavages giving rise to the 85K and 115K proteins occur in vivo in intact cells or whether the appearance of these proteins is the result of nonspecific degradation processes during extraction or analysis, we took advantage of the fact that the 140K protein is targeted to chloroplasts in vivo (38) and that determinants of chloroplast targeting are located within its N-terminal region (18; I. Jupin, unpublished data). We therefore constructed the expression vectors pΩ-140K(1-719)-EGFP and pΩ-140K(1-886)-EGFP, which encode fusion proteins in which the green fluorescent protein (GFP) moiety is located downstream of the mapped cleavage sequences (Fig. 5A). We reasoned that cleavage at the predicted sites would release the GFP moiety, which would thus become cytoplasmic, whereas in the absence of processing, it would remain fused to the N terminus of the 140K protein and would retain its chloroplastic distribution.
FIG. 5.
In vivo analyses in intact cells of the cleavage site generating the 115K and 85K proteins. (A) Schematic representation of EGFP fusion proteins. Cleavage sites and protein domains are designated as in Fig. 1, while the mutated cleavage site is represented by ×. (B) Arabidopsis protoplasts were transfected with plasmids pΩ-140K(1-719)-EGFP (i to iii), pΩ-140K(1-886)-EGFP (iv to vi), and pΩ-140K(1-886)-874AAA-EGFP (vii to ix). Single protoplasts were observed by epifluorescence microscopy 25 h posttransfection, and EGFP localization (green) was observed (i, iv, and vii). To visualize the locations of chloroplasts, the chlorophyll autofluorescence (red) was acquired (ii, v, and viii) and superimposed onto the EGFP fluorescence (iii, vi, and ix). Scale bars, 10 μm.
Upon transfection of Arabidopsis protoplasts with the corresponding expression vectors, the green fluorescence of the GFP moiety allows monitoring of its subcellular distribution by fluorescence microscopy in living cells, while the simultaneous observation of chlorophyll red autofluorescence permits detection of the chloroplasts. As shown in Fig. 5B (i to iii), protoplasts expressing the 140K (1 to 719)-EGFP fusion displayed a bright fluorescent staining in the shape of rings around the chloroplasts, indicating that the fusion protein is targeted to the chloroplasts. This result therefore demonstrates that the cleavage between aa 686 and 719 does not occur in vivo and that the 85K protein is likely to result from nonspecific proteolytic degradation occurring during protein sample extraction or analysis.
In contrast, when the EGFP moiety was placed downstream of residue 886 of the 140K protein, a diffuse fluorescence characteristic of cytosoluble GFP was observed throughout the cells (Fig. 5B, iv to vi), demonstrating that the cleavage event leading to the 115K occurred in vivo. This processing was inhibited when the 874AAA mutation, shown above to inhibit the PRO/HEL cleavage, was introduced in the pΩ-140K(1-886)-874AAA-EGFP construct, as the corresponding protein then retained its chloroplastic distribution (Fig. 5B, vii to ix). These findings therefore demonstrate that the PRO/HEL cleavage that generates the 115K protein indeed takes place in vivo at the predicted site.
Detection of the viral helicase in infected cells.
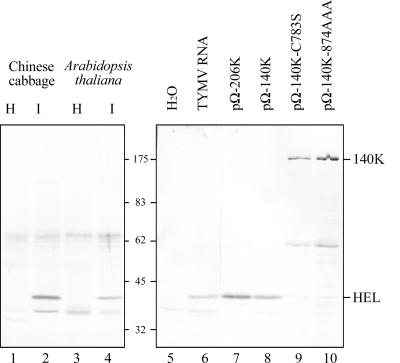
To detect the C-terminal cleavage product, i.e., the viral polypeptide encompassing the HEL domain, polyclonal antibodies were raised against synthetic peptides derived from the helicase amino acid sequence. The resulting antibodies (anti-HEL) were affinity purified and then used in immunoblotting experiments. As shown in Fig. 6, an immunoreactive protein with an apparent molecular mass of ∼42 kDa (42K) was specifically detected in protein extracts of TYMV-infected plant and protoplast samples (lanes 1 to 6), providing the first evidence for the existence of a mature product encompassing the HEL domain in TYMV-infected samples. Its observed molecular mass is consistent with that expected to result from processing at residue 880 (42,145 Da). As expected, this 42K product was detected upon expression of the 206K precursor polyprotein (lane 7) and was found to derive exclusively from the 140K protein, as it was also detected upon transfection of Arabidopsis protoplasts with the pΩ-140K vector (lane 8). Transfection of the expression vectors pΩ-140K-C783S and pΩ-140K-874AAA, in which the PRO/HEL cleavage is abolished either by mutation of the proteinase or by mutation of the cleavage site, respectively, led to immunodetection of the 140K protein in place of the 42K protein (lanes 9 and 10), providing further confirmation that PRO/HEL processing takes place in vivo at the predicted site.
FIG. 6.
Detection of the viral helicase in infected cells. Protein extracts from healthy (H) (lane 1) and TYMV-infected (I) (lane 2) Chinese cabbage leaves and from healthy (lane 3) or TYMV-infected (lane 4) Arabidopsis leaves were subjected to SDS-10% PAGE and immunoblot analysis with anti-HEL antiserum. Arabidopsis protoplasts were transfected with water (lane 5), TYMV RNA (lane 6), or the expression vectors indicated. The cells were harvested 48 h posttransfection, and total proteins were subjected to 10% SDS-PAGE and immunoblot analysis with anti-HEL polyclonal antibodies. The positions of viral proteins and molecular mass markers are indicated.
PRO/HEL and HEL/POL cleavage sequences can be processed in trans in plant cells.
Based on previous in vitro cotranslational assays, the processing of 206K was suggested to occur only in cis (2, 3, 41). To evaluate the ability of the TYMV proteinase produced in vivo to function in trans, the expression plasmids pΩ-206K-C783S (encoding the 206K protein lacking proteinase activity but retaining the cleavage sites to serve as a substrate) and pΩ-140K(1-879) (encoding the viral 115K protein to serve as a protease) were transfected into Arabidopsis protoplasts. Processing at the PRO/HEL and HEL/POL cleavage sites of the 206K substrate was assayed by immunoblotting of the corresponding protein samples using anti-PRR and anti-66K antibodies, respectively (Fig. 7). As shown in lanes 3 and 7, the 115K protein encoded by pΩ-140K(1-879) was capable of processing in trans both the PRO/HEL and HEL/POL cleavage sites, as evidenced by the disappearance of the 206K precursor and the immunodetection of the mature 115K and 66K products. As expected, trans cleavage of the substrate was inhibited upon mutation of the catalytic C783 residue of the proteinase encoded by pΩ-140K(1-879)-C783S (lanes 4 and 8). These experiments therefore demonstrate that the TYMV proteinase has the ability to process substrates in trans when expressed in plant cells.
FIG. 7.
PRO/HEL and HEL/POL cleavage sequences can be processed in trans in plant cells. Arabidopsis protoplasts were transfected with the expression vectors indicated, either alone or in combination. The cells were harvested 48 h posttransfection, and total protein extracts were subjected to 8% SDS-PAGE and immunoblot analysis with anti-PRR (left) or anti-66K (right) antisera. The positions of viral proteins and molecular mass markers are indicated.
Effects of cleavage site mutations on proteolytic processing of 206K.
To uncouple the effects of position and amino acid sequence on 206K processing, we tested the ability of the TYMV proteinase to recognize cleavage sites located at different positions within the 206K protein. To this end, expression vectors were constructed in which the PRO/HEL and HEL/POL cleavage sites were mutated or substituted for each other. Upon transfection into Arabidopsis protoplasts, the effects of the introduced mutations on processing of the 206K protein were assayed by immunoblotting of the corresponding protein extracts using anti-PRR or anti-66K antisera (Fig. 8).
FIG. 8.
Effects of cleavage site mutations on 206K proteolytic processing. (A) Schematic representation of the 206K protein and mutated derivatives. Residues at the PRO/HEL and HEL/POL cleavage sites are indicated. (B) Arabidopsis protoplasts were transfected with the expression vectors indicated. The cells were harvested 48 h posttransfection, and total protein extracts were subjected to 8% SDS-PAGE and immunoblot analysis with anti-PRR (left) or anti-66K (right) antisera. The positions of viral proteins and molecular mass markers are indicated.
First, pΩ-206K-(A/S2), a derivative of the pΩ-206K expression vector, was constructed, in which the P6-P′1 residues of the PRO/HEL cleavage site (KRLLGS/Q) were replaced by seven alanine residues (Fig. 8A). As expected, this substitution inhibited PRO/HEL processing, as evidenced by immunodetection of the 140K protein instead of the 115K protein (Fig. 8B, lane 2). Processing at the HEL/POL cleavage site was unaffected, as evidenced by immunodetection of the 66K protein (lane 7), demonstrating that HEL/POL processing can occur independently of cleavage at the upstream site. When the alanine residues at the PRO/HEL junction were replaced by aa P6 to P′1 of the HEL/POL cleavage site (PKLNGA/T) in the pΩ-206K-(2xS2) expression vector, efficient processing was restored (lane 3), demonstrating that the two cleavage sequences are interchangeable at the PRO/HEL junction.
Conversely, pΩ-206K-(S1/A) was constructed, in which the P6-P′1 residues of the HEL/POL cleavage site were replaced by seven alanine residues, which resulted, as expected, in the inhibition of processing at the HEL/POL site (lane 9). Cleavage at the PRO/HEL site was not affected (lane 4), indicating that it can occur independently of processing at the downstream site. Efficient cleavage at the HEL/POL junction was restored when the alanine residues were replaced with aa P6 to P′1 of the PRO/HEL site (KRLLGS/Q) in the pΩ-206K-(2xS1) expression vector (lane 10), demonstrating that the two cleavage sequences are also interchangeable at the HEL/POL junction.
These results, therefore, demonstrate that the PRO/HEL and HEL/POL cleavage sequences can be efficiently processed when exchanged within the 206K precursor.
Effects of cleavage site mutations on viral infectivity.
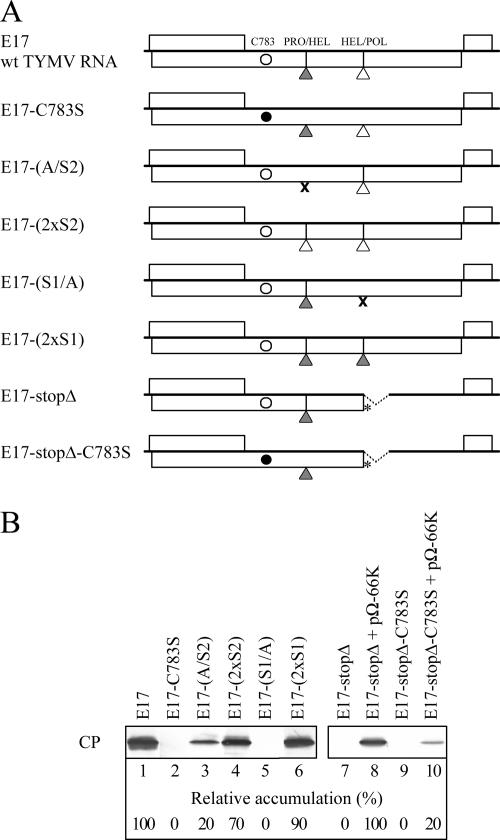
Viral proteinase catalytic activity has previously been reported to be essential for viral replication (2). Thus, we next wanted to determine whether mutations or substitutions of the cleavage sites affected viral infectivity. For this purpose, the mutations shown in Fig. 8 were introduced into plasmid E17 (11), which contains a full-length copy of the TYMV genome and from which infectious viral transcripts can be obtained. Plasmids E17-(A/S2), E17-(2xS2), E17-(S1/A), and E17-(2xS1) were thus obtained, as well as plasmid E17-C783S, which encodes a disabled proteinase (Fig. 9A). Equal amounts of in vitro transcripts were transfected into Arabidopsis protoplasts, and viral infectivity was assessed by detecting accumulation of CP by Western blotting (Fig. 9B).
FIG. 9.
Effects of cleavage site mutations on viral infectivity. (A) Schematic representation of the infectious TYMV in vitro transcript E17 and its derivatives. Residue Cys783 is indicated by an open circle or by a filled circle when mutated to Ser. The PRO/HEL and HEL/POL cleavage sequences are represented by filled and open triangles, respectively. Deletions are indicated by broken lines and introduced stop codons by asterisks. ×, alanine substitutions resulting in impaired cleavage. (B) Arabidopsis protoplasts were transfected with wild-type or mutant in vitro transcripts, alone or in combination with expression vectors as indicated. The cells were harvested 48 h posttransfection, and the abilities of the transcripts to replicate were assessed by immunoblot analysis of equivalent amounts of proteins using anti-CP antibodies. The relative accumulation of CP is indicated below each panel.
Mutation of the viral proteinase in E17-C783S (lane 2) and impairment of the HEL/POL cleavage in E17-(S1/A) (lane 5) both completely abolished accumulation of CP, confirming that proteinase activity and/or HEL/POL processing are essential for viral infectivity, consistent with previous reports (2). Impairment of the PRO/HEL cleavage in E17-(A/S2) resulted in a partial loss of infectivity, as evidenced by a fivefold reduction in the accumulation of viral CP (Fig. 9B, lane 3). These defects were partially restored when cleavage sites were reintroduced into the 206K protein, as in mutants E17-(2xS1) and E17-(2xS2) (Fig. 9B, lanes 4 and 6). These data therefore indicate that the uncleaved 140K protein may be at least partially functional during viral multiplication but that processing at the PRO/HEL cleavage site plays a critical role in viral infectivity.
To confirm this finding, and to ensure that the defect in viral infectivity of the E17-(A/S2) mutant was caused by the lack of 140K processing rather than being due to the amino acid changes at the PRO/HEL junction, a complementary approach, based on the use of proteinase mutants, was undertaken. In the E17-C783S construct, the proteinase is debilitated by a point mutation and is therefore unable to process either the PRO/HEL or HEL/POL junction. As the HEL/POL cleavage event is essential, the putative contribution of the PRO/HEL cleavage to viral infectivity cannot be assessed with this construct. To circumvent this problem, we made use of construct E17-stopΔ (38) (Fig. 9A). Transcripts derived from this construct do not express the 66K protein and cannot replicate, but they can be trans-complemented by the 66K protein expressed from the pΩ-66K expression vector (38) (Fig. 9B, lanes 7 and 8), alleviating the need for the HEL/POL cleavage event. We then constructed the plasmid E17-stopΔ-C783S, with disabled proteinase, and assessed the abilities of the corresponding transcripts to be trans-complemented by pΩ-66K. We reasoned that a decreased efficiency of complementation would reflect a defect in viral infectivity due to the lack of processing at the PRO/HEL site. Indeed, we observed that complementation of E17-stopΔ-C783S transcripts by pΩ-66K was reduced fivefold compared to complementation of E17-stopΔ transcripts (compare lanes 8 and 10), confirming the contribution to viral infectivity of processing at the PRO/HEL site.
Effects of cleavage site mutations on RNA synthesis.
As the reduction in viral infectivity due to defects in 206K cleavages presumably occurs at the level of viral RNA synthesis, we next used strand-specific Northern blotting experiments to examine the abilities of the cleavage mutant viruses to synthesize plus- and minus-strand RNA species (Fig. 10).
FIG. 10.
Effects of cleavage site mutations on RNA synthesis. Arabidopsis protoplasts were transfected with wild-type or mutant in vitro transcripts, alone or in combination with expression vectors as indicated. The cells were harvested 48 h posttransfection, and equivalent RNA amounts were analyzed based on rRNA as a loading control. Plus-strand genomic (g) and subgenomic (sg) TYMV RNAs (top) and minus-strand TYMV RNAs (bottom) were detected by Northern blotting. The relative accumulation of viral RNAs is indicated below each panel.
Consistent with the results described above, duplication of the cleavage sites in the E17(2xS1) and E17(2xS2) viral RNA mutants had only a minor impact on the accumulation of plus- and minus-strand RNAs compared to the levels obtained during a wild-type infection (Fig. 10, lanes 3 and 5). In contrast, E17-(A/S2) (impaired in PRO/HEL cleavage) was severely affected, with plus-strand RNAs accumulating ∼50-fold less than in a wild-type infection. Interestingly, the synthesis of minus-strand RNA was much less affected, with a reduction of only 2.5-fold, indicating that impairment of the PRO/HEL cleavage differentially affects synthesis of plus- and minus-strand RNA species.
This observation was confirmed by results obtained upon trans-complementation of E17-stopΔ-C783S mutant transcripts with pΩ-66K compared to trans-complementation of E17-stopΔ transcripts (Fig. 10, lanes 7 and 9); again, a more severe reduction in plus- than in minus-strand RNA synthesis was observed. Taken together, these results show that inhibition of PRO/HEL processing has more impact on the accumulation of plus-strand RNAs than on minus-strand RNAs, and they support the hypothesis that the PRO/HEL cleavage plays a critical role in the regulation of synthesis of viral RNA during the infectious cycle.
DISCUSSION
Characterization of the 85K and 115K products.
The TYMV 206K replication protein is proteolytically processed in vitro and in vivo to release the C-terminal 66K protein encompassing the POL domain (39). However, its processing pathway remained incompletely understood, as only trace amounts of the N-terminal 140K product were detected in vivo (18). Instead, two products of 115K and 85K were detected, whose origins were unclear. The goal of the present study was to complete the TYMV 206K cleavage map.
Using a viral proteinase mutant, we observed that the occurrence of the 85K protein relied on a host proteinase activity (Fig. 2). However, it is likely that this processing event does not occur in vivo, as evidenced by the absence of cleavage in intact cells (Fig. 5). Given that its C-terminal boundary is located between residues 686 and 719 (Fig. 3), i.e., just downstream of the PRR, which is predicted to be an intrinsically unfolded region (12, 28) and thus is known to display extremely high sensitivity to protease digestion in vitro (13), we conclude that the 85K protein results from a nonspecific degradation process occurring during sample extraction and protein analysis. Consistent with this conclusion was the observation that the amounts and electrophoretic profiles of the 85K product are highly variable from sample to sample (Fig. 4C) (5, 18).
On the other hand, the cleavage event generating the 115K product was demonstrated to occur in vivo (Fig. 5) and to be dependent on TYMV proteinase activity (Fig. 2), indicating that the 206K precursor is processed at an additional cleavage site. Based on the electrophoretic mobilities of deletion derivatives (Fig. 3), sequence comparisons (Fig. 4A), and mutagenesis studies (Fig. 4B and C), the Ser879-Gln880 dipeptide bond was defined as the site most likely cleaved by TYMV proteinase to release the N-terminal 115K protein carrying the MT, PRR, and PRO domains and the C-terminal 42K protein encompassing the HEL domain. This hypothesis was further supported by the detection in infected samples of the C-terminal 42K cleavage product, production of which was prevented by mutagenesis of the proposed cleavage site (Fig. 6). Taken together, our theoretical analysis and experimental data strongly support the identification of Ser879-Gln880 as the probable PRO/HEL cleavage site within the TYMV 206K protein.
The expected molecular masses of the proteins released upon cleavage between Ser879 and Gln880 can be calculated: whereas the C-terminal helicase protein has a calculated molecular mass of 42,145 Da, consistent with that observed experimentally, the N-terminal protein has a calculated molecular mass of 98,404 Da. The reason for the difference between the observed (115-kDa) and the expected (98-kDa) molecular masses is not clear, but it may be related to the presence of the intrinsically unfolded PRR region (17). The reason why the 115K protein was detected as a double band is also not presently known.
We will henceforth refer to the 206K-processing scheme as generating three products: the N-terminal 98K containing the MT, PRR, and PRO domains; the 42K corresponding to the HEL domain; and the 66K encompassing the POL domain (see Fig. 12). We believe that the cleavage map within the TYMV replication protein is now complete, as no additional cleavage products of the EGFP-98K fusion protein could be detected (Fig. 2) and antibodies raised against the MT and the PRO domains also led to detection of the 98K protein (A. Jakubiec and I. Jupin, unpublished data).
FIG. 12.
Comparison of the expression strategies and regulation of replication of different members of the alpha-like supergroup of viruses. Shown is a schematic representation of the nonstructural replication proteins of TYMV, SIN, and RUB. Protein domains are designated as in Fig. 1. Cleavage sites are represented by filled triangles. The number below each cleavage site indicates the sequential order of the cleavage in the precursor protein. Cleavages that are required for minus-strand RNA synthesis are filled in black, whereas those required for switching from minus- to plus-strand synthesis are filled in gray.
Differences between cell-free and in vivo systems.
Previous experiments using various cell-free translation systems programmed with TYMV RNA demonstrated the occurrence of the HEL/POL cleavage in vitro (35, 49). Although a product of 120 kDa has also been detected in some instances (35, 49), it is unlikely to correspond to the 98K protein described here, as the 140K protein was the only product detected with the anti-PRR antibody upon translation of TYMV RNA in reticulocyte lysate (18), thus indicating that processing at the PRO/HEL site does not occur in this in vitro translation assay.
Another unexpected finding was the determination of the ability of TYMV proteinase to cleave in trans when expressed in vivo, as demonstrated in Fig. 7, because such trans-cleavage was not detected in cell-free assays (2, 3, 41) and proteinase activity was thought to be limited to cis processing. Our experiments, however, clearly demonstrated that the TYMV proteinase is able to process substrates in trans, as previously reported for other members of the alphavirus-like supergroup with whom TYMV shares evolutionary relationships, such as the rubivirus rubella virus (RUB) and the alphaviruses Sindbis virus (SIN) and Semliki Forest virus (16, 29, 33). Based on its ability to cleave both in cis and in trans at multiple sites and its location in the central region of the 206K polyprotein, the TYMV PRO domain can be classified as belonging to the main, or M, group of viral papain-like cysteine proteases (15).
The reasons for the observed differences between cell-free translations and in vivo transient-expression assays are unclear, but they may result from differences in active protease concentrations or from the contribution of cellular cofactors and/or the presence of the whole-cell environment. In this respect, it should be pointed out that the TYMV 140K and 98K proteins are membrane-bound proteins that are targeted to the chloroplast envelope in plant cells (18, 38) (Fig. 5), and it is conceivable that membrane association may influence the folding of the viral proteins and hence their cleavage properties, thus contributing to the regulation of 206K processing during viral replication in host cells.
Analysis of the cleavage sites.
Sequence alignments around the TYMV 206K PRO/HEL and HEL/POL cleavage sites revealed strong similarity between the sites, as residues P7, P4, P2, and P2′ to P4′ were identical and residues P5 and P1 were similar, occupied by basic residues and residues with a short side chain, respectively (Fig. 4A). Indeed, analysis of the processing of 206K derivatives demonstrated that the two cleavage sequences can be efficiently processed when substituted for each other within the 206K precursor (Fig. 8B). Whether TYMV proteinase displays a cleavage site preference could not be assessed from analysis of the processing of 206K derivatives, as cleavage at each site could occur independently of the other (Fig. 8B). Further studies are required to establish whether such a preference may exist during viral infection and, if so, what its molecular basis might be.
Mutagenesis experiments revealed that the residues most sensitive to substitution are located upstream of the cleavage sites (positions P6 to P1) (Fig. 4B and C), consistent with previous reports regarding cellular and viral papain-like cysteine proteases, such as cathepsin and foot-and-mouth disease virus or Semliki Forest virus protease (22, 23, 30). The mechanism of cleavage site recognition for TYMV proteinase is unknown, and pursuing the definition of the molecular determinants of its substrate specificity will require further mutagenesis studies.
Alignment of the polyproteins encoded by members of the genus Tymovirus revealed that the junctions between the homologous PRO, HEL, and POL domains are not well conserved. Nevertheless, each tymovirus examined was found to have a potential counterpart for these cleavage sites (Fig. 11), suggesting that processing of the polyprotein precursor into three mature products may constitute a conserved feature among tymoviruses. The most uniform feature of these sites is the presence of amino acids with a short side chain (Gly, Ala, or Ser) in the P2 and P1 positions, consistent with results obtained for rubivirus and alphavirus cleavage sites (7, 30). Positions P5 and P4 were occupied almost exclusively by basic (Lys and Arg) and hydrophobic (Leu, Ile, or Phe) residues, respectively, except in Erysimum latent virus, which appears to be particularly divergent among tymoviruses.
FIG. 11.
Analysis of tymovirus cleavage sites. Protein sequences at the putative PRO/HEL (top) and HEL/POL (bottom) cleavage sites of the replication proteins encoded by members of the genus Tymovirus were aligned using ClustalW. Identical or similar residues are boxed in black or gray, respectively. Amino acid numbers are indicated, with respect to the polyprotein. EMV, Eggplant mosaic virus; KYMV, Kennedya yellow mosaic virus; OYMV, Ononis yellow mosaic virus; ELV, Erysimum latent virus; PhMV, Physalis mottle virus; ChMV, Chayote mosaic virus; DuMV, Dulcamara mottle virus; PlMoV, Plantago mottle virus; SrMV, Scrophularia mottle virus; AVYV, Anagyris vein yellowing virus.
Strikingly, the residues located downstream of the cleavage site appeared rather conserved at the PRO/HEL junction but showed little conservation at the HEL/POL junction, suggesting that conservation of the HEL N-terminal sequences reflects other functional requirements, rather than proteinase substrate specificity determinants. Like alphavirus proteinases, tymovirus proteinase can apparently accommodate a wide variety of residues in the P1′ position, including amino acids with bulky side chains (30, 45).
Related papain-like proteinases are also encoded by maculaviruses and marafiviruses, the other genera constituting the family Tymoviridae, and by some members of the phylogenetically related family Flexiviridae (31, 32, 39). Although it can be speculated that all these tymo-like PRO domains may share common biochemical and biological features, such a comparison awaits further experimental data.
Processing and regulation of viral replication.
Processing of nonstructural proteins allows the precursor protein or partially processed intermediates to perform functions that are distinct from those of the mature cleavage products. This provides a means to temporally regulate the course of viral infection by changing the ratio of polyprotein to mature products and has been reported to be essential for the replication of several RNA viruses, including rubiviruses and alphaviruses (25-27, 44).
To examine the importance of 206K protein processing in TYMV replication, reverse-genetics experiments were performed. Our results (Fig. 9 and 10) confirmed that processing at the HEL/POL junction is absolutely required for TYMV infectivity, as the TYMV HEL/POL mutant was unable to accumulate plus- and minus-strand RNAs. This observation suggests that the HEL/POL cleavage is required to activate one of the catalytic activities carried out by the 206K protein (i.e., polymerase or helicase), which may not be functional when embedded in the precursor protein. Such a cleavage-dependent activation of the polymerase function has been reported for poliovirus (46), and future crystallographic studies may help to determine whether the TYMV polymerase relies on a similar process to adopt an active replication-competent conformation.
On the other hand, processing at the PRO/HEL junction was not essential for viral replication but appeared to contribute to the regulation of viral RNA synthesis, as a TYMV PRO/HEL mutant had more severe defects in plus- rather than minus-strand RNA synthesis (Fig. 9 and 10). This suggests that different cleavage events lead to different patterns of RNA synthesis and emphasizes the importance of an accurate proteolytic-processing scheme in virus replication.
Based on previous studies and our current work, we propose that TYMV RNA replication is regulated as follows (Fig. 12). After its release into the cytoplasm, the genomic RNA is translated into the 206K protein. Cleavage at the HEL/POL junction is believed to occur rapidly in cis, which would lead to the production of a complex consisting of 140K and 66K capable of minus-strand RNA synthesis (Fig. 10). Later in the infection, as the concentration of the 140K increases and/or due to a conformational change induced by its recruitment to chloroplast envelope membranes, the PRO domain becomes capable of trans-cleavage at the PRO/HEL junction, generating the 98K and 42K proteins. The replication complex is then rearranged into a stable form making plus-strand genomic and subgenomic RNAs. Several steps in this process remain to be demonstrated, and further studies addressing these issues are required.
Evolutionary relationship with members of the alphavirus-like supergroup of RNA viruses.
The results obtained in this study allow a comparative analysis of the strategies used for nonstructural-protein expression and regulation of viral replication among togaviruses (comprising alphaviruses and rubiviruses) and tymoviruses, which are animal- or plant-infecting members of the alphavirus-like supergroup, respectively (Fig. 12).
As in tymoviruses, togavirus nonstructural proteins are encoded in the form of a polyprotein containing MT, PRO, HEL, and POL functional domains, as well as an X domain with unknown function. Remarkably, the orders of these functional domains are similar in tymoviruses and rubiviruses but differ from that of alphaviruses, most likely because of genetic rearrangement (9).
In the case of SIN alphavirus, the P1234 precursor contains three cleavage sites and generates four mature products (nsP1 to nsP4) and a number of intermediates (8). Many studies have demonstrated that the temporal regulation of nonstructural-protein processing controls the synthesis of the different RNA species. According to current data (25, 44), uncleaved P1234 is not functional, and a first cleavage at the 3/4 site is required to generate an early replication complex (P123 and nsP4), synthesizing only minus-strand RNA. Upon further cleavage of P123 at the 1/2 site, an intermediate complex (nsP1, P23, and nsP4) is formed, capable of both minus- and plus-strand genomic-RNA synthesis. Upon final cleavage at the 2/3 site, minus-strand synthesis ceases and plus-strand genomic and subgenomic RNAs are generated by a stable complex comprising the four mature products.
In the RUB rubivirus, the situation is much simpler, and only a single cleavage site has been reported (48). The uncleaved P200 precursor is functional and capable of minus-strand synthesis, whereas its cleavage at the PRO/HEL junction plays a critical role in switching the replication complex to synthesis of plus-strand RNA and inhibition of minus-strand synthesis (26, 27).
It thus appears that TYMV occupies an intermediate position, in terms of both genome organization and complexity of the regulation process. A common feature among all three genera is the fact that a cleavage event immediately downstream of the PRO domain is required for switching the replication complex from minus- to plus-strand RNA synthesis. Whether, in the case of TYMV, this cleavage also shuts off synthesis of minus-strand RNA, as reported in togaviruses, is presently unknown. TYMV also shares with alphaviruses the facts that the unprocessed precursor is not functional and that there is an absolute requirement for release of the POL domain to initiate RNA synthesis, in contrast to rubiviruses, whose uncleaved precursor is functional in minus-strand RNA synthesis (24, 26, 27). Finally, one regulatory step that appears specific for alphaviruses relates to the differential synthesis of genomic and subgenomic RNAs promoted by cleavage at the 2/3 site. The mechanisms upon which TYMV and RUB rely for this process are still unknown.
Remarkably, our findings highlight strong similarities between animal- and plant-infecting members of the alphavirus-like supergroup of positive-strand RNA viruses. They also emphasize the importance of the conformational flexibility of viral proteins in the regulation of viral replication, as a change in template specificity or activation of the polymerase is likely to arise from differences in the conformations of the proteins induced by cleavage. Knowledge of the structural characteristics of the various intermediate cleavage products and comparison with those of the final mature products would thus be of great interest, given that those processes are shared by all positive-strand RNA viruses.
Acknowledgments
A.J. was the recipient of a fellowship from the Ministère de l'Education Nationale de la Recherche et de la Technologie (MENRT). This work was supported in part by grants from MENRT and CNRS (Actions Concertées Incitatives “Programme de Microbiologie Fondamentale” and “Biologie Moléculaire, Cellulaire et Structurale”).
We are grateful to L. van Dinten, B. Deiman, and C. W. A. Pleij (Leiden University) for the initial suggestion of the PRO/HEL cleavage site and to S. Pflieger and H. Rothnie for comments on the manuscript. Special thanks are due to C.J. for constant encouragement.
Footnotes
Published ahead of print on 8 August 2007.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Wiley, New York, NY.
- 2.Bransom, K. L., and T. W. Dreher. 1994. Identification of the essential cysteine and histidine residues of the turnip yellow mosaic virus protease. Virology 198:148-154. [DOI] [PubMed] [Google Scholar]
- 3.Bransom, K. L., J. J. Weiland, and T. W. Dreher. 1991. Proteolytic maturation of the 206-kDa nonstructural protein encoded by turnip yellow mosaic virus RNA. Virology 184:351-358. [DOI] [PubMed] [Google Scholar]
- 4.Bransom, K. L., S. E. Wallace, and T. W. Dreher. 1996. Identification of the cleavage site recognized by the turnip yellow mosaic virus protease. Virology 217:404-406. [DOI] [PubMed] [Google Scholar]
- 5.Camborde, L., V. Tournier, M. Noizet, and I. Jupin. 2007. A Turnip yellow mosaic virus infection system in Arabidopsis suspension cell culture. FEBS Lett. 581:337-341. [DOI] [PubMed] [Google Scholar]
- 6.Candresse, T., C. Mouchès, and J. M. Bové. 1986. Characterization of the virus encoded subunit of turnip yellow mosaic virus replicase. Virology 152:322-330. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J.-P., J. H. Strauss, E. G. Strauss, and T. L. K. Frey. 1996. Characterization of the rubella virus nonstructural protease domain and its cleavage site. J. Virol. 70:4707-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Groot, R. J., W. R. Hardy, Y. Shirako, and J. H. Strauss. 1990. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 9:2631-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominguez, G., C. Y. Wang, and T. K. Frey. 1990. Sequence of the genome RNA of rubella virus: evidence for genetic rearrangement during togavirus evolution. Virology 177:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dreher, T. W. 2004. Turnip yellow mosaic virus: transfer RNA mimicry, chloroplasts and a C-rich genome. Mol. Plant Pathol. 5:367-375. [DOI] [PubMed] [Google Scholar]
- 11.Drugeon, G., and I. Jupin. 2002. Stability in vitro of the 69K movement protein of Turnip yellow mosaic virus is regulated by the ubiquitin-mediated proteasome pathway. J. Gen. Virol. 83:3187-3197. [DOI] [PubMed] [Google Scholar]
- 12.Dunker, A. K., C. J. Brown, and Z. Obradovic. 2002. Identification and functions of usefully disordered proteins. Adv. Protein Chem. 62:25-49. [DOI] [PubMed] [Google Scholar]
- 13.Fontana, A., P. Polverino de Laureto, V. De Filippis, E. Scaramella, and M. Zambonin. 1997. Probing the partly folded states of proteins by limited proteolysis. Fold Des. 2:R17-R26. [DOI] [PubMed] [Google Scholar]
- 14.Goldbach, R., and J. Wellink. 1988. Evolution of plus-strand RNA viruses. Intervirology 29:260-267. [DOI] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., E. V. Koonin, and M. M. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy, W. R., and J. H. Strauss. 1989. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol. 63:4653-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iakoucheva, L. M., A. L. Kimzey, C. D. Masselon, R. D. Smith, A. K. Dunker, and E. J. Ackerman. 2001. Aberrant mobility phenomena of the DNA repair protein XPA. Protein Sci. 10:1353-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakubiec, A., J. Notaise, V. Tournier, F. Héricourt, M. A. Block, G. Drugeon, L. van Aelst, and I. Jupin. 2004. Assembly of turnip yellow mosaic virus replication complexes: interaction between the proteinase and polymerase domains of the replication proteins. J. Virol. 78:7945-7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakubiec, A., V. Tournier, G. Drugeon, S. Pflieger, L. Camborde, J. Vinh, F. Héricourt, V. Redeker, and I. Jupin. 2006. Phosphorylation of viral RNA-dependent RNA polymerase and its role in replication of a plus-strand RNA virus. J. Biol. Chem. 281:21236-21249. [DOI] [PubMed] [Google Scholar]
- 20.Kadaré, G., M. Rozanov, and A. L. Haenni. 1995. Expression of the turnip yellow mosaic virus proteinase in Escherichia coli and determination of the cleavage site within the 206 kDa protein. J. Gen. Virol. 76:2853-2857. [DOI] [PubMed] [Google Scholar]
- 21.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 22.Kuehnel, E., R. Cencic, N. Foeger, and T. Skern. 2004. Foot-and-mouth disease virus leader proteinase: specificity at the P2 and P3 positions and comparison with other papain-like enzymes. Biochemistry 43:11482-11490. [DOI] [PubMed] [Google Scholar]
- 23.Lecaille, F., Y. Choe, W. Brandt, Z. Li, C. S. Craik, and D. Bromme. 2002. Selective inhibition of the collagenolytic activity of human cathepsin K by altering its S2 subsite specificity. Biochemistry 41:8447-8454. [DOI] [PubMed] [Google Scholar]
- 24.Lemm, J. A., and C. M. Rice. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 67:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemm, J. A., T. Rumenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang, Y., and S. Gillam. 2000. Mutational analysis of the rubella virus nonstructural polyprotein and its cleavage products in virus replication and RNA synthesis. J. Virol. 74:5133-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang, Y., and S. Gillam. 2001. Rubella virus RNA replication is cis-preferential and synthesis of negative- and positive-strand RNAs is regulated by the processing of nonstructural protein. Virology 282:307-319. [DOI] [PubMed] [Google Scholar]
- 28.Liu, J., and B. Rost. 2003. NORSp: predictions of long regions without regular secondary structure. Nucleic Acids Res. 31:3833-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, X., S. L. Ropp, R. J. Jackson, and T. L. K. Frey. 1998. The rubella virus nonstructural protease requires divalent cations for activity and functions in trans. J. Virol. 72:4463-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lulla, A., V. Lulla, K. Tints, T. Ahola, and A. Merits. 2006. Molecular determinants of substrate specificity for Semliki Forest virus nonstructural protease. J. Virol. 80:5413-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martelli, G. P., S. Sabanadzovic, N. Abou-Ghanem Sabanadzovic, M. C. Edwards, and T. Dreher. 2002. The family Tymoviridae. Arch. Virol. 147:1837-1846. [DOI] [PubMed] [Google Scholar]
- 32.Martelli, G. P., M. J. Adams, J. F. Kreuze, and V. V. Dolja. 2007. Family Flexiviridae: a case study in virion and genome plasticity. Annu. Rev. Phytopathol. 45:4.1-4.28. [DOI] [PubMed] [Google Scholar]
- 33.Merits, A., L. Vasiljeva, T. Ahola, L. Kaariainen, and P. Auvinen. 2001. Proteolytic processing of Semliki Forest virus-specific non-structural polyprotein by nsP2 protease. J. Gen. Virol. 82:765-773. [DOI] [PubMed] [Google Scholar]
- 34.Morch, M. D., G. Drugeon, P. Szafranski, and A. L. Haenni. 1989. Proteolytic origin of the 150-kilodalton protein encoded by turnip yellow mosaic virus genomic RNA. J. Virol. 63:5153-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morch, M. D., W. Zagorski, and A. L. Haenni. 1982. Proteolytic maturation of the turnip-yellow-mosaic-virus polyprotein coded in vitro occurs by internal catalysis. Eur. J. Biochem. 127:259-265. [DOI] [PubMed] [Google Scholar]
- 36.Mouchès, C., T. Candresse, and J. M. Bové. 1984. Turnip yellow mosaic virus RNA-replicase contains host and virus-encoded subunits. Virology 134:78-90. [DOI] [PubMed] [Google Scholar]
- 37.Parsley, T. B., C. T. Cornell, and B. L. Semler. 1999. Modulation of the RNA binding and protein processing activities of poliovirus polypeptide 3CD by the viral RNA polymerase domain. J. Biol. Chem. 274:12867-12876. [DOI] [PubMed] [Google Scholar]
- 38.Prod'homme, D., A. Jakubiec, V. Tournier, G. Drugeon, and I. Jupin. 2003. Targeting of the turnip yellow mosaic virus 66K replication protein to the chloroplast envelope is mediated by the 140K protein. J. Virol. 77:9124-9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prod'homme, D., S. Le Panse, G. Drugeon, and I. Jupin. 2001. Detection and subcellular localization of the turnip yellow mosaic virus 66K replication protein in infected cells. Virology 281:88-101. [DOI] [PubMed] [Google Scholar]
- 40.Rawlings, N. D., F. R. Morton, and A. J. Barrett. 2006. MEROPS: the peptidase database. Nucleic Acids Res. 34:D270-D272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozanov, M. N., G. Drugeon, and A. L. Haenni. 1995. Papain-like proteinase of turnip yellow mosaic virus: a prototype of a new viral proteinase group. Arch. Virol. 140:273-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 43.Schechter, I., and A. Berger. 1967. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27:157-162. [DOI] [PubMed] [Google Scholar]
- 44.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, A. A., and O. B. Peersen. 2004. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 23:3462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao, J., D. Yang, P. Chong, D. Hwang, Y. Liang, and S. Gillam. 1998. Proteolytic processing of rubella virus nonstructural proteins. Virology 246:74-82. [DOI] [PubMed] [Google Scholar]
- 49.Zagorski, W., M. D. Morch, and A. L. Haenni. 1983. Comparison of three different cell-free systems for turnip yellow mosaic virus RNA translation. Biochimie 65:127-133. [DOI] [PubMed] [Google Scholar]