Abstract
Herpes simplex virus type 1 (HSV-1) genomes become associated with structures related to cellular nuclear substructures known as ND10 or promyelocytic leukemia nuclear bodies during the early stages of lytic infection. This paper describes the relationship between HSV-1 genomes and ND10 in human fibroblasts that maintain the viral genomes in a quiescent state. We report that quiescent HSV-1 genomes detected by fluorescence in situ hybridization (FISH) are associated with enlarged ND10-like structures, frequently such that the FISH-defined viral foci are apparently enveloped within a sphere of PML and other ND10 proteins. The number of FISH viral foci in each quiescently infected cell is concordant with the input multiplicity of infection, with each structure containing no more than a small number of viral genomes. A proportion of the enlarged ND10-like foci in quiescently infected cells contain accumulations of the heterochromatin protein HP1 but not other common markers of heterochromatin such as histone H3 di- or trimethylated on lysine residue 9. Many of the virally induced enlarged ND10-like structures also contain concentrations of conjugated ubiquitin. Quiescent infections can be established in cells that are highly depleted for PML. However, during the initial stages of establishment of a quiescent infection in such cells, other ND10 proteins (Sp100, hDaxx, and ATRX) are recruited into virally induced foci that are likely to be associated with HSV-1 genomes. These observations illustrate that the intimate connections between HSV-1 genomes and ND10 that occur during lytic infection also extend to quiescent infections.
Herpes simplex virus type 1 (HSV-1) is an important human pathogen that establishes a latent state in neuronal cells after a primary infection in epithelial tissues (for a general review, see reference 44). During latency, transcription of the great majority of the viral genome is repressed and only a family of related latency-associated transcripts is detectable (39, 58). This contrasts with lytic infection, during which the whole viral genome is transcriptionally active, expressing a large number of genes that can be grouped into immediate-early (IE), early, and late classes according to the time course of their expression. The mechanisms that regulate the balance between lytic and latent infection are of intense interest because latency ensures that infected individuals retain the virus for life, allowing periodic episodes of reactivation and the ensuing clinical symptoms.
Transcription of the HSV-1 genome is controlled primarily by three viral proteins. VP16 is a virion component that activates the expression of IE genes, IE protein ICP4 is essential for the transcription of E and L genes, and IE protein ICP0 is very important in certain cell types for initiation of the viral transcription program. Viruses with mutations that inactivate VP16 and/or ICP0 do not enter productive replication after low-multiplicity infection of human fibroblasts and certain other cultured cells but instead are retained for many days in a repressed, quiescent state (18, 19, 34, 35, 41, 46, 51). In some respects, the quiescent state resembles latency (39). Human fibroblasts are suitable for the study of quiescent infection because they are particularly nonpermissive for HSV-1 mutants lacking VP16 and ICP0 function, and they can be maintained for relatively long periods in the laboratory. The quiescent genome is not detectably expressed and appears to be unresponsive to stimulation by a range of transcription factors, suggesting that it is organized into an inactive chromatin configuration (34, 41, 46). Exogenous provision of ICP0, or the human cytomegalovirus (HCMV) tegument protein pp71, results in disruption of the quiescent state and resumption of viral gene expression (19, 34, 40, 41, 46).
The status and location of HSV-1 genomes in latently or quiescently infected cells are long-standing questions of considerable interest. Maul and colleagues were the first to observe that the parental genomes of HSV-1, HCMV, and adenovirus are commonly closely associated with small nuclear substructures known as ND10 or promyelocytic leukemia (PML) nuclear bodies (called ND10 below) during the early stages of lytic virus infection (20, 30, 31). It is at these sites that IE gene transcription occurs, and viral replication compartments develop from these sites (reviewed in references 8 and 30). Although the role of ND10 in herpesvirus infections remains incompletely understood, it has been firmly established that regulatory proteins ICP0 of HSV-1 and ie72 (IE1) of HCMV localize to these structures during the earliest stages of infection and then bring about their disruption and that these activities correlate with the efficiency at which the viruses enter the lytic cycle (reviewed in references 8, 10, and 30). These and other observations have prompted the hypothesis that ND10 have a repressive effect on viral gene expression. This concept has been confirmed by recent studies demonstrating that depletion of PML increases both viral expression and plaque formation of ICP0-null mutant HSV-1 and ie72-deficient HCMV, although not to wild-type levels (13, 55). Furthermore, depletion of hDaxx (a prominent component of ND10 and a PML interaction partner) overcomes the repression of HCMV IE gene expression that occurs in the absence of the viral tegument activator protein pp71 (4, 42, 45, 60), and depletion of Sp100 (another core component of ND10) decreases the sensitivity of HSV-1 gene expression to interferon (36) and increases the replication of ICP0-null HSV-1 (C. Parada, C. M. Preston, A. Orr, and R. D. Everett, unpublished data).
It has been established that the association of HSV-1 genomes with ND10 occurs not via migration of the genomes through the nucleoplasm to preexisting ND10 but rather because ND10 proteins accumulate at novel sites that are closely associated with the viral genomes (12, 14). These events are readily detectable in the early stages of lytic infection of human fibroblasts, and they imply that the association of viral genomes with ND10 occurs as a consequence of a cellular response to the entry of the genomes into the nucleus. In contrast to the extensive information on the location of herpesvirus genomes during lytic infection, the location of HSV-1 genomes in quiescently infected cells has not been described previously.
In this study we made use of a system based on the HSV-1 mutant in1312, which has lesions that inactivate VP16, ICP0, and ICP4 (43). Infection of human fibroblasts with in1312-based mutants enables the establishment of cultures in which most cells contain at least one quiescent viral genome. By combining fluorescence in situ hybridization (FISH) with immunocytochemistry to detect PML and other protein components of ND10, we observed that HSV-1 genomes are frequently associated with enlarged ND10 structures. In addition to the presence of other common ND10 components (Sp100, hDaxx, and ATRX), we also found that these structures characteristically contained readily detectable accumulations of conjugated ubiquitin. We found that neither abundant viral transcription nor de novo viral protein synthesis is required for viral genome association with ND10, implying that chromatin assembly and/or other events occurring on the incoming viral genome are sufficient to trigger the accumulation of ND10 proteins at the sites of the viral genomes. Extensive depletion of PML from human fibroblasts increased (albeit to a limited degree) the number of cells in which HSV-1 genomes escaped initial repression. We conclude that repressed HSV-1 genomes in quiescently infected human fibroblasts are stably and intimately associated with ND10-like structures.
MATERIALS AND METHODS
Cells and viruses.
Baby hamster kidney (BHK) cells were grown in Glasgow modified Eagle medium supplemented with 10% newborn calf serum and 10% tryptose phosphate broth. Human fetal foreskin fibroblast cells (HFFF-2; European Collection of Cell Cultures), primary human fibroblasts isolated from human foreskin tissue (HFS cells) (Department of Urology, University of Erlangen), and U2OS, Vero, and 293T cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum (FCS). All cell growth media were supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin. Lentivirus-transduced cells were maintained in the appropriate medium with puromycin selection (2 μg/ml).
HSV-1 strain 17 syn+ (17+) was the wild-type (wt) strain used, from which all other HSV-1 strains were derived. HSV-1 mutant tsK expresses a temperature-sensitive variant of the major viral transcriptional activator ICP4 (5, 38). vEYFP-ICP4 is an otherwise wt HSV-1 strain that expresses ICP4 linked to enhanced yellow fluorescent protein (EYFP) (14). The following viruses were used in quiescent-infection experiments: in1312, a virus derived from the VP16 insertion mutant in1814 (1) that also carries the tsK lesion and a deletion/frameshift mutation in the ICP0 open reading frame (43); viruses in1374 and in1382, which contain insertions of an HCMV-lacZ reporter cassette into the UL43 and TK genes of in1312, respectively (40); virus in1330, a VP16 rescuant of in1312 (C. M. Preston, unpublished data); in1388, which is in1312 with an internal ribosome entry site element and lacZ coding sequences in the latency-associated transcript (LAT) region (29); and viruses in1365 and in1368, ICP0 and ICP4 rescuants of in1388, respectively (29). All HSV-1 strains were grown in BHK cells and titrated in U2OS cells. Virus tsK was grown and titrated at 31°C. Viruses derived from in1312 were grown and titrated at 31°C in the presence of 3 mM hexamethylene bisacetamide (32). Baculovirus Ac.CMV.ECFP-PML expresses PML isoform IV linked to enhanced cyan fluorescent protein (ECFP) from the HCMV enhancer/promoter (49).
Infections.
To establish quiescently infected cell populations, HFFF-2 cells were infected at a multiplicity of infection (MOI) of 3 PFU per cell and were maintained at a restrictive temperature (38.5°C) in a medium containing reduced levels of FCS (2%) for 6 to 8 days before analysis as described below.
Superinfection with tsK to determine proportions of quiescently infected cells.
Monolayers were infected with 2 PFU of tsK per cell, maintained overnight at 38.5°C, and stained for expression of β-galactosidase followed by treatment with Carmalum stain, as described previously (21). Digital images of stained monolayers were obtained by use of a Nikon TS100 microscope and a SPOT INSIGHT camera and software. The proportion of cells that expressed β-galactosidase was determined by counting cells on prints of images.
Isolation of PML-depleted human fibroblasts.
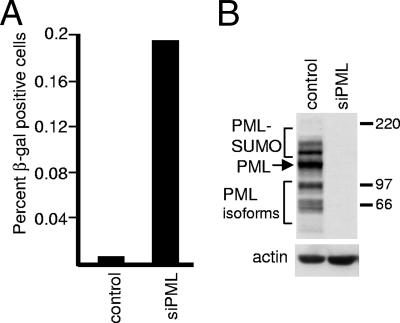
HFS cells were transduced with lentivirus vectors expressing either a control anti-luciferase short hairpin RNA (shRNA) or an anti-PML shRNA to produce the HFS/shLuci and HFS/shPML1 cell lines, respectively. The anti-luciferase and anti-PML shRNA sequences were the same as those utilized to prepare the siLuci and siPML2 cells described previously (13). Note that the siPML2 sequence of the earlier study is the same as that named shPML1 in this paper. The lentivirus vector was based on pLKO.1puro, and the methods used to isolate lentivirus stocks have been described previously (13). Transduced cells were selected with puromycin (2 μg/ml) and maintained in a medium containing puromycin. More than 95% of HFS/shPML1 cells were negative for PML in immunofluorescence experiments, and PML was undetectable by Western blotting of extracts of these cells.
FISH.
HFFF-2 cells on 13-mm-diameter glass coverslips were washed twice with phosphate-buffered saline (PBS) containing 1% FCS and then fixed by incubation for 5 min at −20°C with precooled 95% ethanol-5% acetic acid. The cells were then washed three times with PBS-1% FCS and were stored at 4°C for further use. The probe for FISH was cosmid 56, containing a 39.7-kb segment of the HSV-1 genome between coordinates 24699 and 64405 (33). After DNase I treatment, the probe was labeled by nick translation using Cy3-dCTP (PA 53021; Amersham) according to the manufacturer's protocol. The cells were prehybridized by incubation for 30 min at 37°C with 20 μl hybridization buffer (50% formamide, 10% dextran sulfate, 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) in a humidified microarray hybridization chamber (Camlab). The coverslips were removed from the chamber and blotted dry. The probe was added to the hybridization buffer to a concentration of 1 ng/μl; then each coverslip was incubated at 95°C for 2 min to denature the probe and the sample. The coverslips and hybridization mixture were placed in the humidified chamber, and the hybridization was continued overnight at 37°C. The cells were washed for 5 min at 60°C with 2× SSC and once with 2× SSC at room temperature. After a further wash with PBS-1% FCS, the coverslips were incubated with the appropriate primary and secondary antibodies as for a normal immunofluorescence experiment (see below). For most FISH experiments, the antibodies used were anti-PML rabbit serum r8 (3) and fluorescein isothiocyanate-conjugated sheep anti-mouse immunoglobulin G (Sigma). The cells were washed several times, air dried, and then mounted on glass slides using a glycerol-PBS mounting solution (CitiFluor AF1). The samples were examined using a Zeiss LSM 510 confocal microscope, with 488-nm and 543-nm laser lines, scanning each channel separately under image capture conditions that eliminated channel overlap. The images were exported as tagged-image format files and then processed using Photoshop. For detailed analysis, Z-stacks of typical FISH foci were captured (30 to 50 slices of 0.1-μm depth covering a total depth of 2 to 3 μm, with cubic voxel dimensions and gain settings so that the channels had similar gray scale values at a maximum of about 80% saturation). The stacks were imported into Autodeblur and Autovisualize software (Media Cybernetics) for 3-dimensional (3-D) reconstruction and production of rendered images.
Immunofluorescence.
When FISH was combined with immunofluorescence, hybridization was performed first as described above, and then the coverslips were incubated with the relevant primary and secondary antibodies as described previously (12). In simple immunofluorescence experiments, the cells were fixed with formaldehyde and incubated with primary antibodies and the relevant secondary antibodies as described previously (12). The antibodies used to detect other proteins were as follows: mouse anti-ICP4 monoclonal antibody (MAb) 58S (48) and rabbit anti-ICP4 polyclonal antibody r74 (7); anti-PML MAb 5E10 (52) and anti-PML rabbit serum r8 (3); anti-hDaxx rabbit serum r1866 (37); anti-Sp100 rabbit serum SpGH and anti-Sp100 rat serum sp26 (50); anti-ATRX rabbit serum sc-15408 (Santa Cruz Biotechnology); anti-conjugated ubiquitin MAb FK2 (BioMol); anti-HP1 mouse serum (11); and anti-dimethyl and anti-trimethyl lysine 9 histone H3 (Abcam).
qPCR.
HFFF-2 cells were either mock infected or infected with in1374 at an MOI of 3 to produce quiescently infected cultures as described above; then, 7 days postinfection, the cells were reseeded into 24-well dishes. Parallel samples were analyzed for the proportions of cells containing quiescent genomes that could be reactivated by superinfection with virus tsK (as described above), while DNA was prepared for quantitative PCR (qPCR) from others, as follows. The cells were harvested into 250 μl of 10 mM Tris-HCl (pH 8.0)-50 mM EDTA-1% sodium dodecyl sulfate and then incubated with 100 μg/ml proteinase K overnight at 37°C. The mixture was sonicated in a sonicator water bath twice for 1 min; then the DNA was purified using Qiaquick PCR purification columns (QIAGEN) according to the supplier's recommendations. PCR mixtures were prepared using Taqman universal PCR master mix (Applied Biosystems) with the recommended amounts of primer oligonucleotides (Sigma Genosys) and qPCR probes (Applied Biosystems). The primer and probe sequences were as follows: ICP0 forward primer, 5′-GGAAAGGCGTGGGGTATAA; ICP0 reverse primer, 5′-AACGTAGGCGGGGCTTC; ICP0 probe, 6-carboxyfluorescein (6FAM)-TCGCATTTGCACCTCGGCAC-6-carboxytetramethylrhodamine (TAMRA); glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward primer, 5′-CGGCTACTAGCGGTTTTACG; GAPDH reverse primer, 5′-AAGAAGATGCGGCTGACTGT; GAPDH probe, 6FAM-CACGTAGCTCAGGCCTCAAGACCT-TAMRA. Each experiment included a 10-fold dilution series of ICP0 and GAPDH plasmid DNAs of known concentrations, and 5-μl samples plus two further 10-fold dilutions of infected and uninfected DNA samples. All reactions were performed in triplicate using the standard recommended conditions in a 96-well format using an Applied Biosystems 7500 fast real-time PCR system. Using the software provided with the system, the data were analyzed to calculate the numbers of cellular and viral genome equivalents in each experimental sample by comparison with the known copy numbers in the control plasmid samples.
RESULTS
Detection of HSV-1 genomes in association with PML in quiescently infected human fibroblasts.
During the early stages of lytic HSV-1 infection, parental HSV-1 genomes are closely associated with ND10 (20, 31) through a mechanism that results in the accumulation of ND10 proteins at sites juxtaposed to the incoming viral DNA molecules (12). We set out to determine whether HSV-1 genomes were similarly associated with ND10 in cells that had established a quiescent infection. HFFF-2 human fibroblasts were infected with HSV-1 mutant in1374, which carries lesions in ICP0, ICP4, and VP16 and is therefore highly defective in HSV-1 gene expression (40, 43). This virus also carries a lacZ gene linked to the HCMV promoter-enhancer, whose activity is also repressed in in1374-infected HFFF-2 cells (40, 41). Human fibroblasts can be infected with in1374 at a multiplicity that results in most cells maintaining a viral genome(s) whose presence in the overwhelming majority of cases is undetectable by immunofluorescence staining for viral proteins or by β-galactosidase activity by 24 h after infection (40). At later times after infection, the number of β-galactosidase-positive cells declines to zero, although the viral genomes are stably retained for extended periods and can be reactivated to express β-galactosidase by later superinfection with tsK, a virus that expresses ICP0 (40).
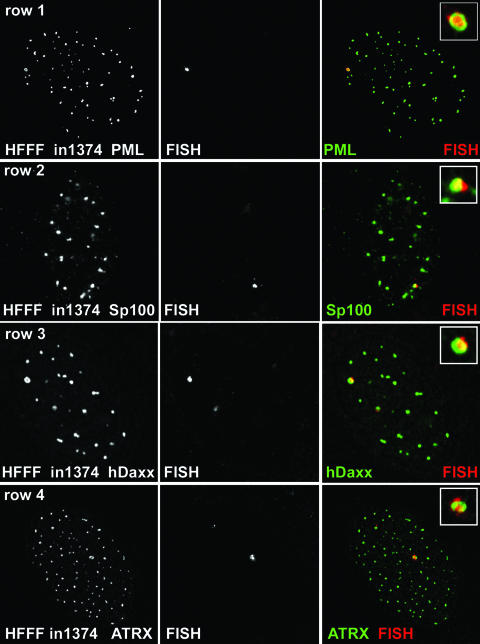
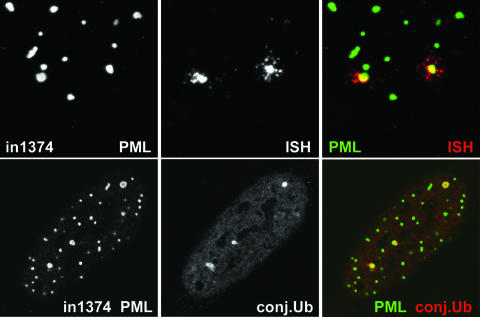
HFFF-2 cells were infected with in1374 (MOI, 3 PFU per cell); then, 7 or 8 days later, the samples were prepared for FISH and immunocytochemistry to detect viral genomes and PML, respectively. At this stage, no cells expressed detectable levels of β-galactosidase (40). For a high proportion of the cells, we observed distinct sites of hybridization that were invariably associated with PML protein, frequently in the form of an apparent ring-like structure of PML surrounding the FISH signal, suggesting sequestration of the viral genome within a modified form of ND10 (Fig. 1, row 1). Such FISH signals were never observed in mock-infected cells (data not shown). Analysis of ND10 appearance and composition in mock-infected cells that had been maintained under the same culture conditions as the infected samples and subjected to the FISH procedure indicated that the enlarged ring-like ND10 were not an artifact of the experimental methods (data not shown). The enlarged ring-like ND10 were defined for the purposes of this study as apparently annular structures with a clear central core whose diameter approximates half of the external diameter. When similar samples were costained for Sp100 and hDaxx (two other major ND10 proteins), we also observed association of these proteins with the viral FISH signal (Fig. 1, rows 2 and 3). ATRX was also a component of these structures (Fig. 1, row 4). In the latter three cases, the FISH procedure resulted in loss of sensitivity to the antibodies to a greater extent than for the detection of PML by antibody r8, resulting in a decrease in image quality. Accordingly, we were unable to assign significance to any apparent differences in detail in the relative locations of the PML, Sp100, and hDaxx signals in the cells in Fig. 1.
FIG. 1.
Detection of HSV-1 genomes in quiescently infected cells. HFFF cells were infected with virus in1374 (MOI, 3 PFU per cell) and then maintained at 38.5°C for 8 days before being processed for FISH detection of HSV-1 genomes (center panel of each row) and staining for ND10 components by immunofluorescence (left panels). (Row 1) PML; (row 2) Sp100; (row 3) hDaxx; (row 4) ATRX. The merged signals are shown in color on the right (proteins in green; FISH signal in red), and the insets show the FISH foci at a higher magnification.
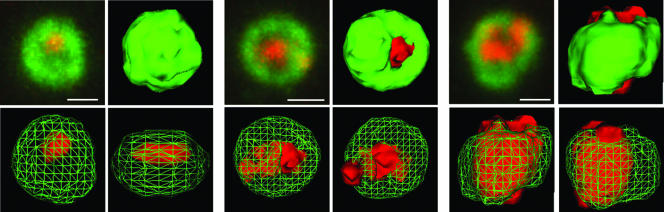
Z-stack analysis indicated that the ring-like structures were in fact spherical, with PML forming a surface layer and the FISH signal in the interior (Fig. 2). In some cases, more than one FISH signal was present within a PML structure, while in others the FISH signal broke beyond the confines of the PML signal (Fig. 2). Note that ring-like ND10 structures can be observed in many other situations and cell types, particularly if PML proteins have been exogenously expressed, so they are not specific to cells quiescently infected with HSV-1. However, because they were observed at a low frequency in mock-infected HFFF-2 cells, the presence of two or more of these structures in a particular cell could be used as a reliable indicator of the presence of quiescent in1374 genomes.
FIG. 2.
Z-stack and 3-D reconstruction analysis of PML foci containing viral DNA. Image stacks of typical examples of viral FISH foci, as in Fig. 1, row 1, were imported into Autodeblur and Autovisualize software (Media Cybernetics) for 3-D reconstruction and production of rendered images. Three examples are shown. In each case, the top left image of the set presents a projection of the complete Z-stack onto a single plane (bar, 0.5 μm); the top right shows a 3-D reconstruction of the outer rendered surface; and the lower two images show alternative views with the PML signal (green) represented as a cage to reveal the FISH signal (red) in the interior.
As controls for the authenticity of the FISH signal, we prepared probes from cosmids covering different segments of the entire HSV-1 genome. All gave results similar to those obtained with the cosmid 56 probe normally employed. In contrast, probes derived from vector or HCMV sequences gave negative results (data not shown). Taking these together with the negative results obtained with mock-infected cells, we conclude that the FISH signals that we observed are derived from HSV-1 genomes in the quiescently infected cells.
We considered the possibility that viral RNA transcripts might contribute to the FISH signal. The signal was eliminated if the sample was not denatured prior to hybridization to a denatured probe (indicating that the probe was hybridizing to a double-stranded template), while preincubation of the sample with a variety of different RNase enzymes had no discernible effect on the quantity or quality of the FISH signal, despite eliminating the enhanced staining of nucleoli with propidium iodide that arises from rRNA transcripts (data not shown). While these observations appear to rule out single-stranded RNA as being the source of the FISH signal, we cannot exclude the possibility that part of the FISH signal is derived from double-stranded or small RNAs that are RNase resistant in fixed cell preparations.
Similar FISH results were observed with a panel of viruses related to in1374. For example, the FISH signal was present in cells infected with virus in1312, which does not have an HCMV IE promoter or lacZ reporter gene, thereby excluding any effect of the insertion of a strong heterologous promoter. Furthermore, UV-inactivated in1312 also produced FISH-positive cells, indicating that residual viral protein expression from the mutant HSV-1 was not required for the appearance of the FISH signal or its association with PML (data not shown). Infection with virus mutant derivatives of in1312 in which the lesions in the genes expressing ICP0, VP16, and ICP4 had been repaired individually also resulted in the formation of PML-associated viral FISH foci in quiescently infected cells (data not shown). Therefore, the appearance of the viral FISH signal was not attributable to any of the individual lesions in the viral genes in the in1374 genome or to the presence of either the HCMV IE promoter or the lacZ gene.
Viral gene expression can be reactivated from cells containing viral FISH foci by superinfection with a virus that expresses ICP0.
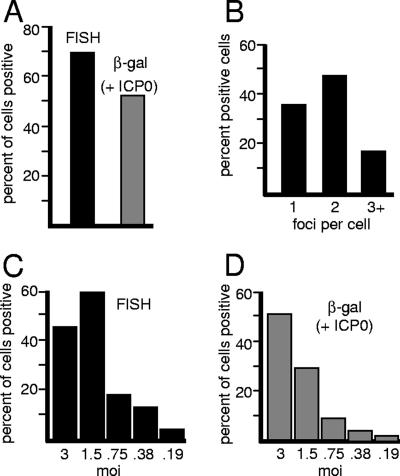
To determine if cells containing positive FISH foci could be induced to reactivate β-galactosidase expression, HFFF-2 cells quiescently infected with in1374 were established as described above. At 9 days postinfection, samples were examined by FISH, while parallel samples were superinfected with virus tsK (which expresses ICP0) to reactivate transcription of the reporter gene in in1374. A total of 143 cells in four different fields of view were scored in the FISH sample, and 70% of these gave positive FISH signals. This compares with approximately 52% of cells that showed detectable β-galactosidase expression after tsK superinfection (Fig. 3A). The number of cells that expressed β-galactosidase in the absence of tsK superinfection at this time after in1374 infection was zero (data not shown, but see reference 40). The slight difference between the number of FISH-positive and reactivated cells is not considered significant because of potential inaccuracies in the determination of precise cell counts on stained monolayers and the possibility that the tsK infection had not been 100% efficient. We conclude that all or most of the cells that harbor a detectable FISH signal can be induced to reactivate expression from the quiescent viral genome by superinfection with a virus that expresses ICP0.
FIG. 3.
Quantification of quiescently infected cells exhibiting the HSV-1-specific FISH signal. (A) HFFF cells were infected with in1374 (MOI, 3) as for Fig. 1; then, at 9 days postinfection, the samples were processed for FISH and immunofluorescence detection of PML. Four random fields of view were analyzed by counting the proportion of cells exhibiting FISH foci in association with PML. A total of 143 cells were analyzed (filled bar). At the same time, parallel samples were infected with 2 PFU of the ICP0-expressing virus tsK per cell to induce reactivation of β-galactosidase expression; then the samples were stained for β-galactosidase activity and counterstained with Carmalum. The proportion of β-galactosidase-positive cells was calculated (shaded bar). (B) The number of positive FISH foci was determined in each positive cell counted for panel A, and the proportion of positive cells exhibiting 1, 2, or 3 or more foci is shown. (C and D) The numbers of cells that exhibit FISH foci and that can be reactivated to express β-galactosidase decrease in parallel with decreasing MOIs. (C) HFFF cells were infected with in1374 at decreasing MOIs as shown, and the proportions of cells with PML-associated FISH signals were determined at 9 days postinfection as for panel A. (D) At the same time, parallel samples were infected with tsK to determine the proportions of cells from which β-galactosidase expression could be reactivated.
Determination of the numbers of FISH-positive foci and quiescent viral genomes in quiescently infected cells.
The number of FISH foci within positive cells varied over a small range: 36% of cells had only one FISH dot, 47% had two, and 17% had three or more (Fig. 3B). The percentages of FISH-positive cells were proportional to the input MOI (Fig. 3C), and these percentages were also similar to those of β-galactosidase-positive cells produced by coinfection of parallel samples with tsK (Fig. 3D). Again, the minor differences between the percentages of FISH- and β-galactosidase-positive cells at a given multiplicity were not considered significant.
The presence of multiple FISH foci in the positive cells was not unexpected. The particle-to-PFU ratio of the in1374 stock used was 50 (as determined by electron microscopy), and therefore the initial load of viral genomes exceeded the nominal input MOI in terms of PFU. Time course experiments demonstrated that HFFF-2 cells infected with in1374 exhibited large numbers of viral particles on their surfaces during the early stages of infection, and only a minority of these particles appeared to gain entry into the nucleus (data not shown). This property is also exhibited by wt HSV-1 (data not shown) and may in part explain why the particle-to-PFU ratios of even wt HSV-1 stocks are usually in the range of 20 to 50. To eliminate any problems of misinterpretation that could be caused by viral particles remaining adhered to the outside of the cell, rather than genomes being within the nucleus, the cells were routinely cultured for several days after infection and then trypsinized and reseeded the day prior to being fixed for FISH. In addition, Z-stack analysis confirmed that the FISH signal was in the same focal plane as that of PML within the nucleus (Fig. 2).
The numbers of viral genomes in the quiescently infected cells were determined by qPCR. By comparison with qPCR results for samples of plasmids including ICP0 and GAPDH coding sequences of known concentrations, we determined that quiescently infected cell populations in which 50% to 80% of cells contained quiescent virus, as judged by reactivation by infection with tsK, contained an average of 4.3 viral genome equivalents per cell (range from four independent experiments, 1.3 to 10.0 genome equivalents per cell). The accuracy of the qPCR procedure was assessed by reconstitution experiments, which demonstrated that the expected number of viral gene copies was detected when a mock DNA sample was supplemented with an amount of ICP0 gene plasmid equivalent to 10 genomes per cell. Given the data on the numbers of FISH foci in each positive cell (Fig. 3B), it follows that each FISH-defined structure contains only a small number of viral genomes. Note that high-resolution analysis indicated that certain FISH foci contained more than one distinct FISH signal (Fig. 2).
Further characterization of the enlarged PML foci that contain quiescent viral genomes.
Detailed characterization of the viral genome FISH foci in cells quiescently infected with in1374 was technically difficult because many antibodies to ND10 proteins gave poor signals after the FISH procedure. Although such structures could also be observed in a minority of mock-infected HFFF-2 cells (1% of cells had one such structure, and 2.2% had more than one; 488 cells were counted), in1374-infected monolayers contained a far greater proportion of such cells (24% of cells with one PML ring structure and 27% with more than one; 196 cells counted). The percentage of in1374-infected cells that contained an enlarged ring-like PML structure(s), detected by PML staining alone, is consistent with the proportion that were positive for HSV-1 genomes (Fig. 3A). Therefore, in comparison with mock-infected parallel samples, it was possible to analyze the components of these structures in cells quiescently infected with in1374 by simple immunofluorescence.
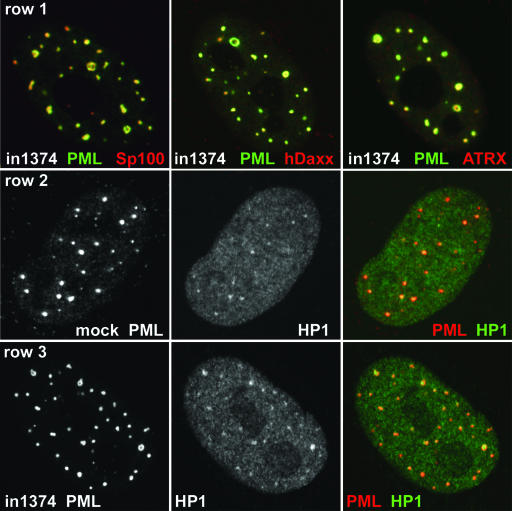
We found that the major ND10 proteins Sp100, hDaxx, and ATRX all colocalized with PML in the ring-like structures, albeit with some minor differences in apparent local concentrations (Fig. 4, row 1). As in previous studies (11, 28, 47), we found that HP1 was present in ND10 in the mock-infected cells to an extent that varied both between cells and between individual ND10 within a cell (Fig. 4, row 2). Distinct accumulations of HP1 within the PML ring-like structures in in1374-infected cells were observed in more than 85% of the foci examined (n = 38) (Fig. 4, row 3). However, the presence of HP1 is not a specific characteristic of the enlarged ND10 foci in quiescently infected cells, because it is also present in a proportion of both normal ND10 and the rare ring-like PML structures in mock-infected cells (data not shown). Nonetheless, the intensity of the HP1 staining in foci in in1374-infected cells could be used as a reliable predictor of the presence of a PML ring-like structure before the PML signal was examined.
FIG. 4.
Recruitment of cellular proteins to PML ring structures in cells quiescently infected with in1374. HFFF cells were infected with in1374 to establish quiescently infected cells. Then, 8 days later, the samples were processed for immunofluorescence. Cells harboring quiescent viral genomes could be identified by the characteristic ring-like structures of PML. (Row 1) Cells were stained for PML (green) and Sp100, hDaxx, or ATRX (red) as indicated. (Row 2) Mock-infected HFFF cells incubated in parallel were costained for PML (red) and HP1 (green), illustrating the variable presence of HP1 within ND10 in human fibroblasts. (Row 3) HFFF cells quiescently infected with in1374 were costained for PML (red) and HP1 (green). Many of the ring-like PML structures contained prominent HP1 staining.
The presence of HP1 did not extend to other prominent markers of condensed heterochromatin. Neither di- nor trimethyl lysine 9 histone H3 was enriched within the PML ring-like structures in quiescently infected cells (data not shown). We also investigated the presence of other ND10 proteins, such as the histone chaperone HIRA (62), the DNA helicase BLM, and the DNA repair factor NBS1, in the ring-like PML structures in quiescently infected cells. All of these proteins could be detected in the annuli of such structures, but not in any way that distinguished them from their colocalization with PML in normal ND10 in uninfected cells (data not shown).
The viral FISH foci are frequently complex in nature, and frequently contain accumulations of conjugated ubiquitin.
Detailed examination revealed that many of the larger viral FISH foci were more complex than a single discrete locus and frequently larger than those derived from virus particles adsorbed to the cell surface during the initial stages of the infection. In some instances, small “satellite” foci could be seen surrounding the parent body (Fig. 5, top panels). These observations are difficult to analyze, because they depend in part on the efficiency of the FISH procedure and the sensitivity of the probe, and perhaps also on the degree of compaction or accessibility of the viral genomes. These factors may be subject to considerable variation. We considered the following explanations for the nature of these complex signals: (i) that they are derived from viral transcripts whose levels and products are undetectable by other methods; (ii) that they are derived from multiple viral genomes; and (iii) that they reflect an uncompacted status of the viral genome. To date, we have been unable to resolve these issues. The control experiments to test the possibility of viral RNAs contributing to the FISH signal (described above) appear to discount the presence of major viral RNA transcripts, but a role for small or double-stranded RNAs could not be discounted. The qPCR experiments eliminate the possibility that large numbers of viral genomes are present in these foci, and given that the viral genomes are functionally repressed, it appears unlikely that they are in a loose or extended configuration. Further work will be required to determine the basis of these intriguing complex FISH signals.
FIG. 5.
The HSV-1 genome foci in quiescently infected HFFF cells can have a complex nature, and the associated PML ring structures invariably contain local accumulations of conjugated ubiquitin. (Top) HFFF cells were infected with virus in1374 (MOI, 3) and then maintained at 38.5°C for 8 days before being processed for FISH detection of HSV-1 genome foci (red) and immunofluorescence staining for PML (green). Images show details of a cell with prominent FISH foci that exhibit a complex nature, with smaller foci surrounding the main central portion. (Bottom) HFFF cells quiescently infected with in1374 were costained for PML (green) and conjugated ubiquitin (conj.Ub) (red). As in Fig. 4, the quiescently infected cells could be identified by the presence of two or more ring-like PML structures. Such rings invariably contained localized concentrations of conjugated ubiquitin.
Finally, we found that the PML ring-like structures that characterize cells quiescently infected with in1374 frequently contained accumulations of conjugated ubiquitin (Fig. 5, bottom panels). Although conjugated ubiquitin has been observed to be present to a variable extent in normal ND10 (2, 9), the situation in the quiescently infected cells was different, in that instead of colocalizing with PML, the ubiquitin signal was clearly in the interior of the PML sphere. The proportions of the PML ring-like foci that contained distinct accumulations of conjugated ubiquitin ranged from 49% to 75% (average, 63%) in five independent samples (a total of 347 foci were examined). Less than 10% of any similar ring-like PML structures in parallel mock-infected samples contained conjugated ubiquitin within their interiors.
Factors affecting the recruitment of ND10 proteins to viral genomes.
The results described above demonstrate that HSV-1 genomes in quiescently infected human fibroblasts are associated with ND10-like structures that contain PML, Sp100, hDaxx, and ATRX. This finding raises questions about how the association occurs and its functional consequences. Since the initial observation of viral genomes in association with ND10 (20, 31), there have been several studies on the factors that underlie this phenomenon. In the case of transfected simian virus 40 replicon plasmids, association with ND10 occurred only if large T antigen and a simian virus 40 replication origin were present (53). In studies using HSV-1 amplicons, expression of ICP4 and ICP27 was required for ND10 association (54), and the degree of association was increased if an early gene transcription cassette and ICP4 binding sites were included in the amplicon genome (49). Although differing in detail, these observations suggest that transcription from or viral regulatory factor binding to the viral genome increases the frequency of ND10 association, an implication in line with the recent demonstration that the association is due to ND10 protein deposition at the sites of incoming HSV-1 genomes (12, 14). Accordingly, ND10 association is a cellular response to the presence of the viral genome, and this response is perhaps easier to explain if the viral genomes reveal their presence through transcription and/or the binding of viral factors. Why then does this association still occur in cells infected with in1374 in which gene expression from the viral genomes is undetectable by Western blotting and immunofluorescence?
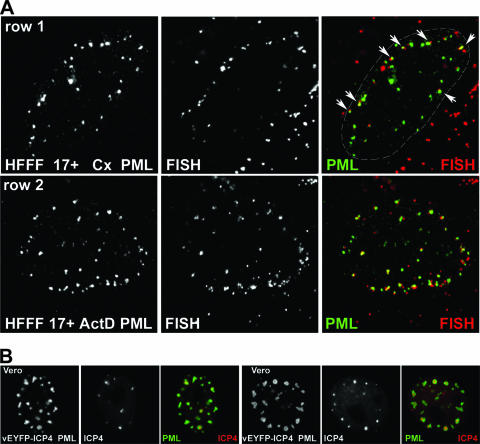
We note that in the original studies, neither actinomycin D nor cycloheximide eliminated the association of viral genomes with ND10 (20, 31). Since these observations are relevant to our current work and are in apparent conflict with the later studies cited above, we have repeated these experiments in a manner that allows visualization of the recruitment of ND10 proteins to the sites of parental viral genomes. As described previously (12, 14), infection of HFFF-2 cells leads to an asymmetric distribution of viral genomes within the nucleus, thereby allowing clear conclusions on the deposition of ND10 proteins at the sites of the viral genomes, rather than the converse. While this effect is most pronounced in cells at the edges of developing plaques, it is also detectable in cells infected at high multiplicity (14). Therefore, HFFF-2 cells were first infected with wt HSV-1 at an MOI of 10 in the presence of either actinomycin D or cycloheximide and then analyzed by FISH and staining for PML at 4 h after infection. In both instances we observed cells with viral genomes located near the nuclear periphery and in association with deposits of PML in similar asymmetrical distributions (Fig. 6A, rows 1 and 2). At this multiplicity and at this time after infection, many virus particles remain adsorbed to the surface of the cell or the genomes have yet to gain entry into the nucleus, causing the detection of nonnuclear genomes, which are particularly prominent in the cycloheximide example presented (Fig. 6A, row 1). These results confirm the original conclusions of Maul and colleagues that neither de novo viral protein expression nor extensive viral transcription is required for genome association with ND10 (20, 31). However, abundant IE gene transcription takes place in the presence of protein synthesis inhibitors in HSV-1-infected cells, and actinomycin D will not inhibit the assembly of transcription complexes containing the viral particle component VP16 on wt viral genomes. In both situations, the viral genomes will interact with a variety of host chromatin and other DNA binding proteins, so the genomes are not completely inert.
FIG. 6.
(A) HSV-1 gene expression is not required for the recruitment of ND10 proteins to the sites of incoming viral genomes. HFFF cells were infected with wt HSV-1 17+ (MOI, 10). At 4 h after infection, the cells were processed for FISH detection of viral genomes (red) and immunofluorescence detection of PML (green) as indicated. For rows 1 and 2, the cells had been pretreated for 15 min with cycloheximide (Cx) (100 μg/ml) or actinomycin D (ActD) (2.5 μg/ml), respectively, before the addition of virus; then infections were conducted, and the cells were maintained in the presence of the drugs throughout. At the early times of a high-MOI infection, many viral particles remain bound to the cell surface or are present in the cytoplasm, giving the extranuclear punctate red staining seen in these images. Prominent examples of genomes associated with PML are arrowed in the merged image of row 1, and the nuclear periphery is delineated by a dashed line. The genome foci at the lower right of this image are from particles that have remained adsorbed to the cell surface or in the cytoplasm. (B) Recruitment of exogenously expressed PML to ring-like foci that surround accumulations of ICP4 during the early times of a low-multiplicity infection. Vero cells were infected with a baculovirus that expresses ECFP-linked PML from the HCMV promoter. On the following day, the cells were infected with HSV-1 vEYFP-ICP4 at an MOI of 0.01. The cells were monitored by live-cell microscopy. Two cells exhibiting ICP4 foci enveloped by PML are shown. The set of images on the left shows a cell at 150 min postinfection, and that on the right shows a cell after 4 h, in both cases prior to the onset of DNA replication and before ICP0 had been able to degrade the highly overexpressed PML (approximately 100-fold greater than the level of endogenous PML [16]).
Although the mutant VP16 contained within in1374 particles is unable to form a complex with host cell factors and viral DNA (1), it is reasonable to assume that many host chromatin and regulatory proteins interact with the mutant viral genomes. Despite the multiple mutations, some cells infected with this virus exhibit transient expression of β-galactosidase (and in some cells, ICP4) and the viral genomes are therefore partially responsive to cellular transcription factors for the first few hours after infection (41). We suggest that this process and/or viral chromatin assembly with host cell proteins, and/or a cellular DNA damage response, instigates the process that leads to the deposition of ND10 proteins in close association with the viral genomes. In the case of the repressed in1374 genomes, the process appears to proceed until the genomes become surrounded by a shell-like structure containing PML.
In addition to the envelopment of viral genome foci by PML in quiescently infected cells, we have observed that a similar phenomenon can occur during the early stages of a wt HSV-1 infection. Vero cells expressing high levels of exogenous ECFP-linked PML (isoform IV) were infected at a low multiplicity with virus vEYFP-ICP4 (an essentially wt HSV-1 strain that expresses ICP4 linked to EYFP), and the relative locations of PML and ICP4 were studied by live cell microscopy. Foci of ICP4 (representing the recruitment of the protein to viral genomes [14]) were commonly enveloped by ring-like structures of exogenously expressed PML (Fig. 6B). In this experiment, the rate of ICP0-induced PML degradation has been minimized by the low MOI and the excessive expression of exogenous PML (on the order of 100-fold more than the endogenous level [16]). It is reasonable to suggest that the formation of annular PML structures surrounding the ICP4 foci in this situation reflects processes that are initiated during a normal infection but that are strongly inhibited by the activity of ICP0.
Initial repression of in1374 is decreased in PML-depleted cells.
Given recent data demonstrating that depletion of PML from human fibroblasts increases the gene expression and plaque formation efficiencies of ICP0-null mutant HSV-1 (13), it was of interest to determine whether the degree of repression of in1374 genomes is reduced in cells lacking PML. Human fibroblasts can be extensively depleted of PML by retrovirus- or lentivirus-transduced expression of an anti-PML shRNA (13, 55). HFS cells were infected with a lentivirus expressing either an anti-PML or an anti-luciferase control shRNA to create cell line HFS-shPML1 or HFS-shLuci, respectively. PML was undetectable by Western blotting in HFS-shPML1 cells (Fig. 7B), and >95% of the cells contained levels of PML that were undetectable by immunofluorescence staining (data not shown; but see reference 13). The two cell lines were infected in parallel with in1374 and then stained for β-galactosidase expression the following day. Only a tiny proportion of in1374-infected HFS-shLuci cells expressed the marker gene, despite the fact that most of the cells contained the viral genome (as detected by its activation by infection with the ICP0-expressing virus tsK) (Fig. 7A). In contrast, a higher proportion of HFS-shPML1 cells expressed β-galactosidase at the same time point (Fig. 7A). These results indicate that repression of the in1374 genome is less efficient in PML-depleted cells, although, considering that only a minority of cells containing the viral genome express β-galactosidase at this time point, it is clear that repression is still occurring in the majority of cells.
FIG. 7.
A proportion of PML-depleted human fibroblasts have a reduced ability to repress β-galactosidase expression from in1374. (A) HFS cells transduced with lentiviruses expressing an anti-luciferase control shRNA or an anti-PML shRNA were infected with in1374 (MOI, 3). On the following day, the cells were stained for β-galactosidase expression, and the number of blue cells in wells containing approximately 1 × 105 cells in total was determined. A parallel control coinfection of samples of the same cells with in1374 and tsK indicated that in both cases more than 50% of the cells had received in1374 genomes (data not shown). The increases in blue cell numbers in PML-depleted cells over those in control cells in a series of repeated experiments were as follows: 17.0-, 31.0-, 32.5-, 22.7-, 12.8-, 30.1-, 19.3-, and 19.2-fold. (B) Western blot illustrating PML expression in typical isolates of fibroblasts transduced with a lentivirus expressing a control anti-luciferase shRNA or an anti-PML shRNA.
We investigated whether the quiescent in1374 genomes in PML-depleted cells were enveloped by the ND10 proteins Sp100, hDaxx, and ATRX even in the absence of PML. This was difficult to do by FISH due to the detrimental effect of the procedure on antibody detection of these proteins. However, depletion of PML causes the almost complete dispersal of hDaxx and ATRX into diffuse nuclear localizations, while Sp100 forms small punctate foci on a background of increased nuclear diffuse staining (13). Therefore, relocation of these proteins, particularly hDaxx and ATRX, to the sites of quiescent in1374 genomes would be expected to provide a means of distinguishing infected from uninfected cells, especially if these proteins form characteristic ring-like structures that are stable and are maintained for extended periods. However, we observed no differences between infected and uninfected PML-depleted cells in these experiments at 8 days postinfection (data not shown). We conclude that the presence of Sp100, hDaxx, and ATRX in ring-like structures that are stably maintained at the sites of quiescent in1374 genomes (Fig. 1 and 4) is dependent on PML. Since repression of in1374 gene expression still occurs in the majority of infected PML-depleted cells, it appears that maintenance of visible ring-like structures containing these particular ND10 proteins is not an obligatory part of continued repression of the viral genome.
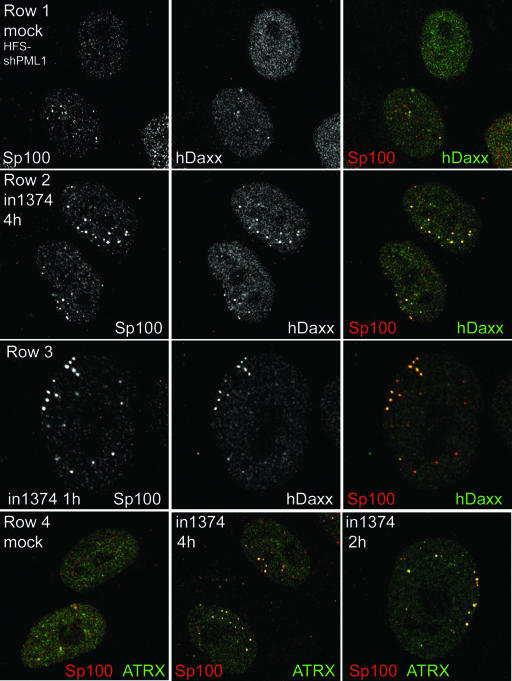
This result is in some ways surprising, since even in the absence of PML, the ND10 proteins Sp100 and hDaxx are recruited to the sites of HSV-1 genomes at early times of infection (13), and this recruitment in PML-positive cells does not require viral gene expression (Fig. 6A). Therefore we investigated whether these other ND10 proteins might be relocated to the sites of in1374 genomes even in the absence of PML during the early stages of infection. HFS-shPML1 cells were infected with in1374 at an MOI of 10 and were then fixed for immunofluorescence analysis at 1, 2, 4, and 6 h after virus adsorption. Consistent with previous observations (13), mock-infected PML-depleted cells exhibited mainly diffuse distributions of hDaxx and ATRX, with Sp100 forming variable small foci in a proportion of the cells. There was little evidence of colocalization of the proteins (Fig. 8, rows 1 and 4, left). The small number of PML-positive cells in the population were readily distinguishable from the PML-depleted cells in both mock-infected and infected samples because the presence of PML resulted in intensely stained ND10 foci that were spread evenly through the nucleoplasm. Such cells were not considered further in this analysis. After in1374 infection, in contrast to the mock-infected cells, distinct foci of colocalizing Sp100 and hDaxx were readily detectable in a high proportion of in1374-infected cells at all time points examined (Fig. 8, row 2; also data not shown). These colocalizing foci were observed as early as 1 h after virus adsorption (Fig. 8, row 3). Similarly, foci of colocalizing Sp100 and ATRX were also induced after in1374 infection (Fig. 8, row 4, center and right). Significantly, such foci were routinely observed in asymmetric patterns close to the nuclear periphery (see Fig. 8, rows 3 and 4, right panels, for clear examples). This pattern is highly reminiscent of normal and PML-depleted human fibroblasts infected with the ICP0-null mutant virus dl1403; in those instances, the ND10-like foci were demonstrated to form adjacent to viral genomes that had entered the nucleus (12, 13). We were unable to test this inference directly in the current experiment, because in1374 does not express DNA binding-competent ICP4 (which can be used as a marker for viral genomes [12, 14, 15]) and because the FISH procedure is too detrimental for the relatively weak immunofluorescence detection of Sp100, hDaxx, and ATRX in PML-depleted cells. Nonetheless, the precedents cited above and the asymmetric distribution of the foci of colocalizing Sp100, hDaxx, and ATRX near the nuclear periphery in in1374-infected cells suggest that it is highly probable that these foci form in association with the viral genomes. It is striking that this occurs within 1 h of infection and in the absence of significant viral gene expression, illustrating the rapidity of this cellular response to the presence of the viral genome. The hypothesis that such recruitment is in some way connected to the initial repression of the in1374 genome is compelling. However, as noted above, we did not observe stable, long-term association of Sp100, hDaxx, and ATRX in ring-like foci in HFS-shPML1 cells quiescently infected with in1374, implying that the presence of these proteins (and/or their associates) may not be required for maintenance of the repressed state.
FIG. 8.
Initial recruitment of ND10 proteins Sp100, hDaxx, and ATRX into in1374-induced foci in PML-depleted cells. Human fibroblasts transduced with a lentivirus expressing an anti-PML shRNA (HFS-shPML1 cells) were infected with in1374 (MOI, 10) and then fixed at 1, 2, 4, and 6 h after infection. Mock-infected cells were fixed in parallel. Coverslips were stained with combinations of anti-Sp100 (rat sp26) and rabbit anti-hDaxx or rabbit anti-ATRX antibodies as indicated. (Row 1) Mock-infected HFS-shPML1 cells exhibit variable foci of Sp100 and mainly diffuse hDaxx staining, with little colocalization. (Row 2) HFS-shPML1 cells infected with in1374 contain pronounced foci of colocalizing Sp100 and hDaxx. (Row 3) This effect was visible in some cells as early as 1 h after virus adsorption. (Row 4) There is little colocalization between Sp100 and ATRX in mock-infected HFS-shPML1 cells (left), but pronounced colocalizing foci form at early times of in1374 infection (center and right).
DISCUSSION
The organization and location of HSV-1 genomes in the nuclei of quiescently infected cells are important parameters that may have considerable influence on the regulation of viral gene expression by cellular mechanisms. Our observations can be considered at several levels: first, how the structures associated with quiescent viral genomes relate to the enlarged PML foci that have been described previously; second, the potential relationship between our observations for quiescently infected fibroblasts and the situation of latently infected neurons; and third, the relationship of our findings to the repression of HSV-1 gene expression that occurs in human fibroblasts in the absence of viral activator proteins such as ICP0.
The moderately enlarged PML structures that we observed in quiescently infected cells, and which are associated with viral genomes, appear as ring-like structures in confocal microscopy sections, but Z-stack analysis indicates that they are actually spherical in nature. Apparently similar enlarged spherical ND10 structures can be observed in many transformed cell lines, especially in cells expressing exogenous PML at high levels, but in uninfected HFFF-2 fibroblasts such structures are relatively rare. There are a number of cellular conditions under which large ring-like PML-defined structures are common, including cells induced to enter senescence (23). However, the PML foci associated with viral genomes in in1374-infected cells do not resemble senescence-associated heterochromatin foci, because the latter are not enveloped by PML and they contain readily detectable levels of histone H3 that has been di- or trimethylated on lysine residue 9 (62). Enlarged ring-like PML structures have also been observed in cells from patients with ICF (immunodeficiency, centromeric instability, and facial dysmorphy) syndrome (28). The ICF-associated PML structures also contain DNA (in this case, juxtacentromeric satellite DNA) and other major ND10 proteins (including HP1), and therefore they resemble the quiescent in1374 genome foci morphologically. There are also parallels between the in1374-associated foci and the enlarged PML-defined structures containing telomeric DNA sequences that are found in some ALT (alternative lengthening of telomere) tumor cell lines (61). Therefore, the concept that enlarged spherical ND10-like structures can form and contain particular DNA sequences is not restricted to cells containing quiescent HSV-1 genomes; rather, it represents a phenomenon that cells have an intrinsic ability to invoke.
How the status of quiescent HSV-1 genomes in fibroblasts compares to truly latent viruses in neurons is an important and intriguing question. At the level of epigenetic modifications, quiescent viral genomes in fibroblasts associate with HP1 and bind to modified histones similar to those found in viral chromatin in latently infected mouse neurons (24, 25, 59; S. Efstathiou, personal communication). Despite the readily detectable levels of LATs in latently infected mouse neurons, as yet there are no reports of successful detection of the viral genomes themselves by FISH in explanted ganglia. However, PML can be detected in human neurons analyzed after explantation from autopsy samples (56). By electron microscopy, the PML bodies in stressed neurons consist of concentric layers containing the PML protein that surround an inner core (56). Therefore, PML structures that morphologically resemble the viral genome foci in quiescently infected fibroblasts have the potential to form in human neurons.
The presence of conjugated ubiquitin in a substantial proportion of the PML ring-like structures associated with viral genomes presents intriguing possibilities. If the production of the conjugated ubiquitin is related to the presence of the viral genomes, it implies that repression may be an ongoing process and that the repressed viral genomes are not completely inert, since the cell continues to mark their locations for extended periods. One class of enlarged PML-defined structures, named clastosomes, contains proteasomes, ubiquitin conjugates, and aggregated proteins (6, 17, 22, 26, 27). Similar structures have been described in human neurons (57). These findings provide a link between PML structures and protein degradation, and it is possible that similarities exist between this class of PML body and those at the sites of quiescent HSV-1 genomes.
Taken together with earlier studies (12), our observations suggest that an early cellular response to the entry of HSV-1 genomes into the nucleus is the accumulation of several cellular proteins in close association with the viral DNA. This process does not require viral gene expression but may depend on cellular responses including chromatin assembly and the binding of other factors to the viral DNA. It is reasonable to assume that among these factors are proteins that repress viral gene expression, whose activity is compromised if ICP0 is expressed. When ICP0-null mutant in1374 genomes become repressed in quiescently infected fibroblasts, PML and other ND10 proteins are recruited to the sites of the viral genomes. The initial recruitment of certain ND10 proteins seems not to require PML (Fig. 8), whereas their stable maintenance in ring-like structures is PML dependent. Although PML depletion increases the proportion of cells in which in1374 gene expression escapes initial repression, the recruitment of PML itself cannot be an essential step in the repression pathway, since most PML-depleted cells remain able to repress in1374 gene expression. Therefore, other factors, which may or may not be related to ND10, must also be involved in the repression of HSV-1 gene expression in quiescently infected cells. As discussed in relation to Fig. 8, the PML-independent recruitment of Sp100, hDaxx, and ATRX to the likely sites of in1374 genomes is extremely rapid, perhaps one of the earliest observable consequences of viral genome entry into the nucleus, and it is possible that recruitment of these proteins and/or their associated factors is in some way involved in the rapid repression of the in1374 genome.
Acknowledgments
This work was supported by the Medical Research Council.
We thank Duncan McGeoch for comments on the manuscript, members of the R.D.E. and C.M.P. laboratories for helpful discussions, Thomas Stamminger and colleagues for their initial work on PML depletion, Heather Coleman and Stacey Efstathiou for advice and materials concerning qPCR, Richard Adair for help with the qPCR experiments, Jim Aitken for particle counts, Peter Adams for HIRA and Asf1 antibodies, Hans Will for anti-Sp100 rabbit serum SpGH, and Roel van Driel for anti-PML MAb 5E10.
Footnotes
Published ahead of print on 1 August 2007.
REFERENCES
- 1.Ace, C. I., T. A. McKee, J. M. Ryan, J. M. Cameron, and C. M. Preston. 1989. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J. Virol. 63:2260-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antón, L. C., U. Schubert, I. Bacik, M. F. Princiotta, P. A. Wearsch, J. Gibbs, P. M. Day, C. Realini, M. C. Rechsteiner, J. R. Bennink, and J. W. Yewdell. 1999. Intracellular localization of proteasomal degradation of a viral antigen. J. Cell Biol. 146:113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boddy, M. N., K. Howe, L. D. Etkin, E. Solomon, and P. S. Freemont. 1996. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971-982. [PubMed] [Google Scholar]
- 4.Cantrell, S. R., and W. A. Bresnahan. 2006. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J. Virol. 80:6188-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davison, M. J., V. G. Preston, and D. J. McGeoch. 1984. Determination of the sequence alteration in the DNA of the herpes simplex virus type 1 temperature-sensitive mutant ts K. J. Gen. Virol. 65:859-863. [DOI] [PubMed] [Google Scholar]
- 6.Dovey, C. L., A. Varadaraj, A. H. Wyllie, and T. Rich. 2004. Stress responses of PML nuclear domains are ablated by ataxin-1 and other nucleoprotein inclusions. J. Pathol. 203:877-883. [DOI] [PubMed] [Google Scholar]
- 7.Everett, R., A. Cross, J. Tyler, and A. Orr. 1993. An epitope within the DNA-binding domain of the herpes simplex virus immediate early protein Vmw175 is conserved in the varicella-zoster virus gene 62 protein. J. Gen. Virol. 74:1955-1958. [DOI] [PubMed] [Google Scholar]
- 8.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 9.Everett, R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74:9994-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett, R. D. 2006. Interactions between DNA viruses, ND10 and the DNA damage response. Cell. Microbiol. 8:365-374. [DOI] [PubMed] [Google Scholar]
- 11.Everett, R. D., W. C. Earnshaw, A. F. Pluta, T. Sternsdorf, A. M. Ainsztein, M. Carmena, S. Ruchaud, W. L. Hsu, and A. Orr. 1999. A dynamic connection between centromeres and ND10 proteins. J. Cell Sci. 112:3443-3454. [DOI] [PubMed] [Google Scholar]
- 12.Everett, R. D., and J. Murray. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D., S. Rechter, P. Papior, N. Tavalai, T. Stamminger, and A. Orr. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, R. D., G. Sourvinos, C. Leiper, J. B. Clements, and A. Orr. 2004. Formation of nuclear foci of the herpes simplex virus type 1 regulatory protein ICP4 at early times of infection: localization, dynamics, recruitment of ICP27, and evidence for the de novo induction of ND10-like complexes. J. Virol. 78:1903-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, R. D., G. Sourvinos, and A. Orr. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 77:3680-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, R. D., and A. Zafiropoulos. 2004. Visualization by live-cell microscopy of disruption of ND10 during herpes simplex virus type 1 infection. J. Virol. 78:11411-11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu, L., Y. S. Gao, A. Tousson, A. Shah, T. L. Chen, B. M. Vertel, and E. Sztul. 2005. Nuclear aggresomes form by fusion of PML-associated aggregates. Mol. Biol. Cell 16:4905-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris, R. A., and C. M. Preston. 1991. Establishment of latency in vitro by the herpes simplex virus type 1 mutant in1814. J. Gen. Virol. 72:907-913. [DOI] [PubMed] [Google Scholar]
- 19.Hobbs, W. E., D. E. Brough, I. Kovesdi, and N. A. DeLuca. 2001. Efficient activation of viral genomes by levels of herpes simplex virus ICP0 insufficient to affect cellular gene expression or cell survival. J. Virol. 75:3391-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamieson, D. R. S., L. H. Robinson, J. I. Daksis, M. J. Nicholl, and C. M. Preston. 1995. Quiescent viral genomes in human fibroblasts after infection with herpes simplex virus Vmw65 mutants. J. Gen. Virol. 76:1417-1431. [DOI] [PubMed] [Google Scholar]
- 22.Janer, A., E. Martin, M. P. Muriel, M. Latouche, H. Fujigasaki, M. Ruberg, A. Brice, Y. Trottier, and A. Sittler. 2006. PML clastosomes prevent nuclear accumulation of mutant ataxin-7 and other polyglutamine proteins. J. Cell Biol. 174:65-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, W. Q., and N. Ringertz. 1997. Altered distribution of the promyelocytic leukemia-associated protein is associated with cellular senescence. Cell Growth Differ. 8:513-522. [PubMed] [Google Scholar]
- 24.Kubat, N. J., A. L. Amelio, N. V. Giordani, and D. C. Bloom. 2004. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J. Virol. 78:12508-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubat, N. J., R. K. Tran, P. McAnany, and D. C. Bloom. 2004. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J. Virol. 78:1139-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumada, S., T. Uchihara, M. Hayashi, A. Nakamura, E. Kikuchi, T. Mizutani, and M. Oda. 2002. Promyelocytic leukemia protein is redistributed during the formation of intranuclear inclusions independent of polyglutamine expansion: an immunohistochemical study on Marinesco bodies. J. Neuropathol. Exp. Neurol. 61:984-991. [DOI] [PubMed] [Google Scholar]
- 27.Lafarga, M., M. T. Berciano, E. Pena, I. Mayo, J. G. Castano, D. Bohmann, J. P. Rodrigues, J. P. Tavanez, and M. Carmo-Fonseca. 2002. Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol. Biol. Cell 13:2771-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luciani, J. J., D. Depetris, Y. Usson, C. Metzler-Guillemain, C. Mignon-Ravix, M. J. Mitchell, A. Megarbane, P. Sarda, H. Sirma, A. Moncla, J. Feunteun, and M. G. Mattei. 2006. PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. J. Cell Sci. 119:2518-2531. [DOI] [PubMed] [Google Scholar]
- 29.Marshall, K. R., R. H. Lachmann, S. Efstathiou, A. Rinaldi, and C. M. Preston. 2000. Long-term transgene expression in mice infected with a herpes simplex virus type 1 mutant severely impaired for immediate-early gene expression. J. Virol. 74:956-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660-667. [DOI] [PubMed] [Google Scholar]
- 31.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67-75. [DOI] [PubMed] [Google Scholar]
- 32.McFarlane, M., J. I. Daksis, and C. M. Preston. 1992. Hexamethylene bisacetamide stimulates herpes simplex virus immediate early gene expression in the absence of trans-induction by Vmw65. J. Gen. Virol. 73:285-292. [DOI] [PubMed] [Google Scholar]
- 33.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 34.Minaker, R. L., K. L. Mossman, and J. R. Smiley. 2005. Functional inaccessibility of quiescent herpes simplex virus genomes. Virol. J. 2:85-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mossman, K. L., and J. R. Smiley. 1999. Truncation of the C-terminal acidic activation domain of herpes simplex virus VP16 renders expression of the immediate-early genes almost entirely dependent on ICP0. J. Virol. 73:9726-9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Negorev, D. G., O. V. Vladimirova, A. Ivanov, F. Rauscher III, and G. G. Maul. 2006. Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. J. Virol. 80:8019-8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pluta, A. F., W. C. Earnshaw, and I. G. Goldberg. 1998. Interphase-specific association of intrinsic centromere protein CENP-C with HDaxx, a death domain-binding protein implicated in Fas-mediated cell death. J. Cell Sci. 111:2029-2041. [DOI] [PubMed] [Google Scholar]
- 38.Preston, C. M. 1979. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J. Virol. 29:275-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preston, C. M. 2000. Repression of viral transcription during herpes simplex virus latency. J. Gen. Virol. 81:1-19. [DOI] [PubMed] [Google Scholar]
- 40.Preston, C. M., and M. J. Nicholl. 2005. Human cytomegalovirus tegument protein pp71 directs long-term gene expression from quiescent herpes simplex virus genomes. J. Virol. 79:525-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preston, C. M., and M. J. Nicholl. 1997. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J. Virol. 71:7807-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preston, C. M., and M. J. Nicholl. 2006. Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 87:1113-1121. [DOI] [PubMed] [Google Scholar]
- 43.Preston, C. M., A. Rinaldi, and M. J. Nicholl. 1998. Herpes simplex virus type 1 immediate early gene expression is stimulated by inhibition of protein synthesis. J. Gen. Virol. 79:117-124. [DOI] [PubMed] [Google Scholar]
- 44.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed.,vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 45.Saffert, R. T., and R. F. Kalejta. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samaniego, L. A., L. Neiderhiser, and N. A. DeLuca. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seeler, J. S., A. Marchio, D. Sitterlin, C. Transy, and A. Dejean. 1998. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl. Acad. Sci. USA 95:7316-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sourvinos, G., and R. D. Everett. 2002. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. EMBO J. 21:4989-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sternsdorf, T., H. H. Guldner, C. Szostecki, T. Grotzinger, and H. Will. 1995. Two nuclear dot-associated proteins, PML and Sp100, are often co-autoimmunogenic in patients with primary biliary cirrhosis. Scand. J. Immunol. 42:257-268. [DOI] [PubMed] [Google Scholar]
- 51.Stow, E. C., and N. D. Stow. 1989. Complementation of a herpes simplex virus type 1 Vmw110 deletion mutant by human cytomegalovirus. J. Gen. Virol. 70:695-704. [DOI] [PubMed] [Google Scholar]
- 52.Stuurman, N., A. de Graaf, A. Floore, A. Josso, B. Humbel, L. de Jong, and R. van Driel. 1992. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101:773-784. [DOI] [PubMed] [Google Scholar]
- 53.Tang, Q., P. Bell, P. Tegtmeyer, and G. G. Maul. 2000. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J. Virol. 74:9694-9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang, Q., L. Li, A. M. Ishov, V. Revol, A. L. Epstein, and G. G. Maul. 2003. Determination of minimum herpes simplex virus type 1 components necessary to localize transcriptionally active DNA to ND10. J. Virol. 77:5821-5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tavalai, N., P. Papior, S. Rechter, M. Leis, and T. Stamminger. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villagra, N. T., J. Berciano, M. Altable, J. Navascues, I. Casafont, M. Lafarga, and M. T. Berciano. 2004. PML bodies in reactive sensory ganglion neurons of the Guillain-Barre syndrome. Neurobiol. Dis. 16:158-168. [DOI] [PubMed] [Google Scholar]
- 57.Villagra, N. T., J. Navascues, I. Casafont, J. F. Val-Bernal, M. Lafarga, and M. T. Berciano. 2006. The PML-nuclear inclusion of human supraoptic neurons: a new compartment with SUMO-1- and ubiquitin-proteasome-associated domains. Neurobiol. Dis. 21:181-193. [DOI] [PubMed] [Google Scholar]
- 58.Wagner, E. K., and D. C. Bloom. 1997. Experimental investigation of herpes simplex virus latency. Clin. Microbiol. Rev. 10:419-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, Q. Y., C. Zhou, K. E. Johnson, R. C. Colgrove, D. M. Coen, and D. M. Knipe. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. USA 102:16055-16059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woodhall, D. L., I. J. Groves, M. B. Reeves, G. Wilkinson, and J. H. Sinclair. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 281:37652-37660. [DOI] [PubMed] [Google Scholar]
- 61.Yeager, T. R., A. A. Neumann, A. Englezou, L. I. Huschtscha, J. R. Noble, and R. R. Reddel. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59:4175-4179. [PubMed] [Google Scholar]
- 62.Zhang, R., M. V. Poustovoitov, X. Ye, H. A. Santos, W. Chen, S. M. Daganzo, J. P. Erzberger, I. G. Serebriiskii, A. A. Canutescu, R. L. Dunbrack, J. R. Pehrson, J. M. Berger, P. D. Kaufman, and P. D. Adams. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8:19-30. [DOI] [PubMed] [Google Scholar]