Abstract
Digital transcript subtraction (DTS) was developed to subtract in silico known human sequences from expression library data sets, leaving candidate nonhuman sequences for further analysis. This approach requires precise discrimination between human and nonhuman cDNA sequences. Database comparisons show high likelihood that small viral sequences can be successfully distinguished from human sequences. DTS analysis of 9,026 20-bp tags from an expression library of BCBL-1 cells infected with Kaposi's sarcoma-associated herpesvirus (KSHV) resolved all but three candidate sequences. Two of these sequences belonged to KSHV transcripts, and the third belonged to an unannotated human expression sequence tag. Overall, 0.24% of transcripts from this cell line were of viral origin. DTS analysis of 241,122 expression tags from three squamous cell conjunctival carcinomas revealed that only 21 sequences did not align with sequences from human databases. All 21 candidates amplify human transcripts and have secondary evidence for being of human origin. This analysis shows that it is unlikely that distinguishable viral transcripts are present in conjunctival carcinomas at 20 transcripts per million or higher, which is the equivalent of approximately 4 transcripts per cell. DTS is a simple screening method to discover novel viral nucleic acids. It provides, for the first time, quantitative evidence against some classes of viral etiology when no viral transcripts are found, thereby reducing the uncertainty involved in new pathogen discovery.
Although infection contributes to over 20% of human cancers worldwide, the list of confirmed carcinogenic infectious agents is surprisingly short (26). Epidemiologic studies strongly suggest that novel infectious agents remain to be discovered and contribute to a broad range of diseases, including cancers, autoimmune disorders, and degenerative diseases. A major problem for new pathogen discovery, however, is that when a search fails to identify an infection, it is not possible to know whether the search failed because of technical reasons or because the disease is actually noninfectious and no unique agent is present. Complicating this issue, infection may be transient (hit and run) or, in the case of autoimmune diseases, distant from the site of disease.
This problem is illustrated by the search for human tumor viruses. Tumorigenic infectious agents have been classified into two broad categories (27): indirect carcinogens, which lead to tumorigenesis through chronic infection and nonspecific inflammation, and direct carcinogens, agents that express intracellular oncogenes that directly contribute to cell transformation. Indirect agents, such as Helicobacter pylori and possibly the hepatitis viruses, can ultimately be lost from the tumor mass. Direct carcinogens, such as papillomaviruses, Epstein-Barr virus, and Kaposi's sarcoma-associated virus (KSHV), are present in the tumor mass with at least one genome copy per cell and express a foreign oncogene transcript.
Indirect and direct infectious carcinogens have distinct epidemiologic properties. For example, immunosuppression may lessen the risk of indirect carcinogenesis by reducing chronic inflammation. In contrast, immunosuppression dramatically increases the risk of direct carcinogenic tumors by reducing host immune surveillance. Thus, patterns of disease occurrence may give clues as to which types of tumors are most likely to have a direct or indirect infectious carcinogenic trigger.
A tumor that has long been suspected of having a direct infectious origin is squamous cell conjunctival carcinoma (SCCC). SCCC is typically a low-grade malignancy with little potential for invasion and metastasis (34). This cancer is uncommon in the United States and is primarily a disease of the elderly (20). There has been, however, a sharp increase in the incidence of SCCC among patients infected with human immunodeficiency virus (HIV) (4, 14, 28, 40), as well as others with immunodeficiency, such as transplant recipients (17, 33). SCCC has emerged along with the AIDS epidemic as a very common cancer in Uganda and parts of sub-Saharan Africa over the past two decades (18, 21, 41, 44). The striking association of SCCC with HIV infection and its geographic localization in regions of Africa are consistent with a direct carcinogenic infectious etiology. Papillomaviruses are a leading candidate for causing this tumor, but current evidence for their role in this disease is contradictory. Some investigators have found an excess occurrence of cutaneous papillomavirus types in SCCCs by PCR (3, 35). Others have failed to find consistent evidence of papillomavirus infection by serology (24, 42) and by papillomavirus consensus PCR (Y. Chang, unpublished results) using primers HVP2/B5, CN1F/R, CP62/69, and EN1F/R (16) and GP+b-Gp5/6 (19). We also failed to find evidence for the presence of adenovirus, herpesvirus, or polyomavirus DNA in SCCCs by consensus PCR.
Molecular methods to discover new pathogens can also be divided into two broad categories (13, 29). One approach uses sequence information from known pathogens to identify related but undiscovered agents through cross amplification (consensus PCR) (25, 37) or cross hybridization (microarray hybridization) (36). Other approaches do not make assumptions about sequence homology for the agent. These techniques include cDNA library panning, used previously to discover hepatitis C virus (8), and representational difference analysis, used to discover KSHV (7).
Although each of these methods has been successful in identifying human viruses, current pathogen discovery approaches suffer from several shortcomings. None are quantitative, so if no candidate sequence is found, it is not possible to estimate how likely it is that an agent is present but missed in the search. Some approaches, such as representational difference analysis, rely on isogenic control samples that differ only in the presence or absence of agent nucleic acids. Since tumors may arise from rare cell populations, identifying an appropriate comparison control tissue may be difficult. Finally, there are no simple ways to scale up pathogen discovery with these methods short of analyzing additional samples.
It is unlikely that any technique can universally identify all human viral pathogens. But with the near completion of the human genome project, high-throughput sequencing may be exploited to examine specific classes of disease, such as direct infectious tumors. In theory, it should be possible to distinguish viral from human transcripts if sufficient sequence length is available. Longer sequences will increase the specificity of screening but at the cost of reduced transcriptome sampling for any given sequencing project. Further, sequences should be vetted for accuracy since sequencing errors will prevent alignment with deposited human sequences and a missequenced human nucleic acid may be wrongly described as nonhuman.
We have investigated long serial analysis of gene expression (L-SAGE) as a means to sample cellular transcriptomes for the presence of viral transcripts. L-SAGE quantitatively concatenates ∼21-bp cDNA tags from the 3′ ends of mRNA transcripts, allowing the measurement of gene expression via high-throughput sequencing (32). If sufficiently stringent conditions are placed on L-SAGE sequencing accuracy, this should be a valuable method for rapidly searching for exogenous viral mRNAs. For a typical L-SAGE tag, there is a high level of confidence for the first 20 of 21 bp. Variations at the 21st bp position result from lax type II enzyme site cutting, leading to uncertainty in base pair assignment during ditag generation (47).
We have evaluated a novel approach to search for exogenous viral transcripts by using L-SAGE libraries, digital transcript subtraction (DTS). Physical transcriptome subtraction to identify differential or novel transcripts has been described previously (23). In addition, the direct examination of expressed-sequence-tag (EST) libraries has been proposed as a means to identify human pathogens (45, 46). Our approach differs in that we performed in silico subtraction by the alignment of short sequence tags with deposited human sequence data. Allowing a 1-in-20-base misalignment, we achieved complete in silico human transcriptome subtraction from a KSHV-infected cell line library, leaving candidate sequences belonging to KSHV. Performing the same analysis on a large L-SAGE library from three SCCCs, we identified 21 candidate sequences, all of which have experimental evidence for being polymorphic human sequences. These results show that DTS can screen human expression sequence data to identify sequences most likely to be of viral origin. DTS shows promise for rapidly identifying some types of new human tumor viruses and, when no viral transcripts are found, can establish quantitative limits on the presence or absence of exogenous mRNA.
MATERIALS AND METHODS
Cell culture.
Cells of the BCBL-1 line (primary effusion lymphoma [PEL]-derived B-cell line infected with KSHV) (30) were maintained in RPMI 1640 medium (Cellgro, Herndon, VA) supplemented with 10% fetal bovine serum (GIBCO, Grand Island, NY) containing penicillin-streptomycin (100 U/ml) and l-glutamine (2 mM) at 37°C in the presence of 5% CO2.
Clinical samples and RNA preparation.
SCCC tissues were obtained from a large case-control study (involving more than 800 cases) by the Uganda Eye Project (43). Tissues were made anonymous with respect to patient identifiers and assigned accession numbers prior to analysis. After surgical extraction, specimens were stored under liquid nitrogen or in RNAlater solution (Ambion Inc., Austin, TX) until processed for RNA extraction. Prior to L-SAGE, cryostat-processed frozen sections of tissues were examined to identify uniform tumor tissues to be used for RNA extraction. Three SCCCs were included in this study: (i) tumor tissue from an HIV-negative female (patient 448), (ii) tumor tissue from an HIV-positive male (patient 1811), and (iii) tumor tissue from an HIV-positive female (patient 1795). The tumor from patient 448 (tumor 448) was snap-frozen after surgery, whereas tumors 1811 and 1795 were placed in RNAlater (Ambion) immediately after surgery. These patients were negative for human papillomavirus types 16 and 18 in previous serological studies (24).
Total RNA from unstimulated BCBL-1 cells was extracted by TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Total RNA from SCCC tissues was extracted after homogenization with the RNeasy midi kit (QIAGEN, Alameda, CA). RNAs were assayed for integrity using the Agilent 2100 bioanalyzer (Quantum Analytics, Foster City, CA) with the RNA 6000 Nano reagent kit. Tumor 448 showed marked degradation (an RNA integrity number of 2), while tumors 1811 and 1795 had high integrity scores (RNA integrity numbers of 8.2 and 7.5, respectively).
L-SAGE.
Four L-SAGE libraries were generated from RNAs from BCBL-1 cells and from the three SCCCs by using the I-SAGE kit according to the instructions of the manufacturer (Invitrogen, Carlsbad, CA). Library 1811 was spiked with BCBL-1 RNA prior to L-SAGE to serve as an internal control for our ability to detect viral sequences in the tumor library. In brief, poly(A) RNA was captured onto magnetic beads and converted into double-stranded cDNA. The cDNA was cleaved with NlaIII and then ligated to oligonucleotide adapters containing recognition sites for MmeI. The linked cDNA was then released from magnetic beads by digestion with MmeI. Released tags were ligated to each other to create ditags and amplified by PCR. Amplified ditags were subsequently concatenated and cloned into the SphI site of pZero-1. Agar lawns were sent for sequencing of the inserts with the M13 forward primer to either the University of Pittsburgh Core Sequencing Facility (Pittsburgh, PA) or Agencourt Biosciences Corporation (Beverly, MA).
DTS.
All sequences were trimmed at a phred score of 20 (12), a stringent measure of base-calling accuracy for automated sequencer traces. Tags were extracted from 32- to ∼38-bp ditags using the SAGE2000 4.5 analysis software (Invitrogen) (39) and analyzed with UNIX Perl scripts (available on request) on a Mac Pro desktop computer. For SCCC DTS analysis, L-SAGE data from the three separate tumors were combined into a single data set. SAGE tag data files used in this analysis are available at http://www.kshv.pitt.edu/.
To reduce sequencing errors, the following criteria were used to generate high-fidelity (HiFi) sequences: tags with ambiguous base calls were eliminated, and only tags identified independently two or more times were included. After performing DTS, tags that were not subtracted were examined for linker sequences and missequencing errors on electropherograms. These tags were eliminated post hoc from the candidate sequences after initial alignment with sequences from reference databases. To generate the HiFi sequences, each unique sequence from HiFi tags was placed into a text file regardless of the frequency of occurrence. Since DTS does not rely on gene expression levels once a tag has been verified, highly abundant and infrequent tags have equal weights in the analysis.
HiFi sequences were examined using stand-alone BLAST (BLAST-2.2.14-univeral-macosx at ftp://ftp.ncbi.nih.gov/BLAST/excutables) against multiple nucleotide databases. The detailed information for BLAST parameters can be viewed at the NCBI website (http://www.ncbi.nlm.nih.gov/staff/tao/URLAPI/blastall/blastall_node23.html). The following parameter set was used to identify exact matches among sequences from human RefSeq RNA and genome databases: r = 1 (matched reward), q = −3 (mismatch penalty), F = F (no filtering of input sequence), W = 20 (word size), and e = 10,000 (expectation cutoff). For the lower-level-homology alignment, the same databases were searched for short, near-exact matches with the following parameters for cross-species sequence exploration (22): (r = 1, q = −1, G = 1 [gap opening cost], E = 2 [gap extension cost], F = F, W = 7, and e = 10,000) and (r = 5, q = −4, G = 25, E = 10, F = F, W = 7, and e = 10,000). Comparison of candidate sequences to sequences in viral nonredundant (NR), human NR, and human EST databases was performed manually using the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST/) and the following parameter set for short, near-exact matches: r = 5, q = −4, G = 25, E = 10, F = F, W = 7, and e = 10,000. To ensure that NR and EST sequences aligning to candidate tags were not nonhuman sequences inadvertently deposited into the databases, MegaBLAST was used to track aligning sequences back to the human genome (45).
Sequential database comparisons of the HiFi sequences were performed using the human RefSeq RNA database first and then the human RefSeq genome database (http://www.ncbi.nlm.nih.gov/RefSeq/). The latter database includes human mitochondrial sequences. Further analysis was performed by comparing sequences to viral NR, human NR, and human EST database entries. All databases were downloaded between September and November 2006 from NIH NCBI (ftp://ftp.ncbi.nih.gov/BLAST/db).
Virtual herpesvirus expression sequence tag analysis.
Genomic sequences of the human herpesviruses (HHV), including HHV-1 (NC_001806), HHV-2 (NC_001798), HHV-3 (NC_001348), HHV-4 (NC_007605), HHV-5 (NC_001347), HHV-6 (NC_001664), HHV-6B (NC_000898), HHV-7 (NC_001716), and HHV-8 (NC_003409), were downloaded from GenBank (NCBI). Virtual SAGE tags with the NlaIII CATG recognition sequence were extracted with Perl scripts. The pool of virtual herpesvirus tags included all sites with CATG sequences in both coding and noncoding regions. To examine KSHV gene expression in BCBL-1 experiments, 80 open reading frames (ORFs) from the deposited KSHV sequence (KSU75698) (31) were analyzed for NlaIII sites by using MacVector (Accelrys Inc.) to retrieve the 20-mer tag most proximal to the 3′ end of the transcript.
Reverse transcription-PCR.
Candidate sequences from BCBL-1 and SCCC SAGE libraries were used with rapid amplification of cDNA ends (RACE) or reverse SAGE (rSAGE) to obtain 3′ sequences. Conventional RACE was performed with BCBL-1 cells and two SCCCs, those from patients 1811 and 1795, by using the 3′ RACE system according to the instructions of the manufacturer (Invitrogen). rSAGE followed the protocol of Jian Yu (University of Pittsburgh; http://www.sagenet.org/protocol) to isolate cDNA fragments corresponding to novel SAGE tags. Briefly, poly(A) RNA from an SCCC from patient 1795 was used to synthesize double-stranded cDNA with a specifically designed biotin-labeled primer, BRS1, containing an AscI restriction enzyme site. The cDNA was digested with NlaIII and then bound to streptavidin beads. This complex was then bound to linker 2A/B and digested with AscI to release the 3′ cDNA fragments from the beads. These 3′ cDNA fragments were then enriched by PCRs to obtain a sufficient quantity of DNA (amplified rSAGE library). A second PCR amplification was performed with L-SAGE tag-specific primers and a common M13 forward primer to obtain specific products. PCR products from both RACE and rSAGE were TA cloned into pCR2.1 vector (Invitrogen), and the sequencing of the PCR products (at least two colonies) was used to verify the authenticity of a given L-SAGE tag and to obtain additional nucleotide sequences that could be used in database searches.
RESULTS
Virtual analysis of herpesvirus L-SAGE tags.
To empirically determine if L-SAGE tags have sufficient sequence diversity to distinguish viral from human sequences, we generated a virtual pool of L-SAGE tags from known HHV genomes (Table 1). The 9,484 virtual NlaIII site tags from nine HHV genomes, representing nearly 1.4 million bp, were aligned with sequences in the NCBI human RefSeq RNA and RefSeq genome databases. For this analysis, we included all NlaIII sites, not just 3′ coding sites, as would occur in experimental SAGE. Only 27 (0.28%) 21-bp and 94 (1.0%) 20-bp virtual herpesvirus SAGE tags align perfectly with the human genome. Of 80 virtual ORF tags from the 3′ ends of KSHV transcripts, none was found to match human sequences. Since herpesviruses extensively pirate host regulatory and DNA synthesis genes (10), this is likely to be a conservative approximation of sequence similarity for other viruses. Standard L-SAGE analyses assume a 21-bp tag length, but the accuracy of only the first 20 bp can be assured with a high level of confidence due to variability in MmeI cutting. As expected, a modest increase in the tag length from 20 to 21 bases markedly increased alignment specificity.
TABLE 1.
Comparison of virtual HHV tags with human RefSeq database entries to assess the degree of divergence between human and viral sequencesa
| Virus | Size (bp) of whole genome | No. of virtual tags | No. of tags matching RefSeq sequence over:
|
No. (%) of remaining 19-of-20-base tags | ||
|---|---|---|---|---|---|---|
| 21 of 21 bases | 20 of 20 bases | 19 of 20 bases | ||||
| HHV-1 | 152,261 | 842 | 1 | 4 | 112 | 730 (86.70) |
| HHV-2 | 154,746 | 786 | 2 | 4 | 84 | 702 (89.31) |
| HHV-3 | 124,884 | 891 | 0 | 5 | 255 | 636 (71.38) |
| HHV-4 | 171,823 | 1,242 | 4 | 15 | 356 | 886 (71.34) |
| HHV-5 | 230,287 | 1,624 | 1 | 15 | 286 | 1,338 (82.39) |
| HHV-6 | 159,321 | 1,027 | 4 | 11 | 320 | 707 (68.84) |
| HHV-6B | 162,114 | 1,076 | 6 | 16 | 338 | 738 (68.59) |
| HHV-7 | 153,080 | 941 | 6 | 17 | 425 | 516 (54.83) |
| HHV-8/KSHV | 137,508 | 1,055 | 3 | 7 | 243 | 812 (76.97) |
| Total | 1,446,024 | 9,484 | 27 | 94 | 2,419 | 7,065 (74.49) |
| KSHVb | 80 | 0 | 0 | 16 | 64 (80) | |
Virtual tags were extracted from nine herpesviruses and compared by BLAST search against human RefSeq sequences by using perfect-match (21-of-21- and 20-of-20-base) and one-mismatch (19-of-20-base) criteria. Virtual tags represent all NlaIII sites rather than actual 3′ cDNA SAGE tag sites to increase viral genome sampling in this comparison. The numbers and percentages of remaining tags were calculated with the cutoff of one mismatch in 20-bp tags.
Actual L-SAGE tag sequences in KSHV coding regions.
To identify viral candidates, polymorphic human sequences with near-exact alignment with reference database sequences must be excluded. Allowing for mismatches, however, increases the likelihood that true viral sequences will be mistaken for human. Allowing for a 1-in-20-base mismatch results in alignments for 2,419, or 25.5%, of 20-bp herpesvirus tags with human sequences. If three viral transcripts are present, the likelihood that all three tags will be misidentified as human drops to 1.66% ([0.255]3). This calculation suggests that it is practical to use a 1-in-20 mismatch alignment to screen for viral sequences in human expression libraries.
DTS analysis of PEL.
We next sought to determine if a known tumor virus can be identified de novo using in silico subtraction and to determine the abundance of viral transcripts. KSHV was discovered in 1993 (7) using a physical subtraction process, representational difference analysis, that first isolated two small viral DNA fragments but allowed subsequent full characterization of the virus (31). To determine if DTS can achieve similar results, we performed L-SAGE with the PEL cell line BCBL-1 (30), which is latently infected with approximately 30 viral genome copies per cell. A total of 9,026 tags from the BCBL-1 L-SAGE library were sequenced (Table 2).
TABLE 2.
Frequencies of tags in the L-SAGE librariesa
| No. of occurrences and/or description | No. of tags with unique sequences (total no. of tags) in library generated from:
|
||
|---|---|---|---|
| BCBL-1 cells | Tumor 1811 spiked with BCBL-1 RNA | Combined 1811, 1795 and 448 SCCC data sets | |
| >74 | 2 (198) | 18 (4,124) | 268 (73,198) |
| 25-74 | 31 (1,259) | 59 (2,473) | 648 (25,517) |
| 5-24 | 204 (1,828) | 527 (4,794) | 4,945 (46,089) |
| 2-4 | 757 (1,851) | 1,984 (4,925) | 12,343 (30,926) |
| ≥2 (HiFi) | 994 (5,136) | 2,588 (16,316) | 18,204 (175,730) |
| 1 | 3,688 (3,688) | 7,351 (7,351) | 61,878 (61,878) |
| Unambiguous | 4,682 (8,824) | 9,939 (23,667) | 80,082 (237,608) |
| Total | (9,026) | (23,700) | (241,122) |
The total pool of tags was analyzed for tag frequency. Data in italics correspond to the tags identified more than once in the SAGE libraries, which were used as HiFi sequences for DTS analysis.
Since missequenced or misextracted tags may be falsely identified as nonhuman, we reduced the SAGE data set to a smaller, HiFi data set composed of unique sequences. This reduction was achieved by extracting sequences with a phred score of 20, eliminating tags with one or more ambiguous base calls. We then compiled unique sequences from tags independently sequenced two or more times. To account for variation in MmeI cutting (47), tag lengths were initially restricted to 20 bp, although longer tag lengths can be confirmed by manually inspecting smaller groups of ditags. This approach generated a HiFi data set of 994 sequences representing 5,136, or 56.90% (5,136 of 9,026), of the original BCBL-1 L-SAGE tags.
Sequential DTS database comparisons (Fig. 1) of the 994 HiFi sequences eliminated all but 34 sequences as having exact 20-of-20-bp alignment with deposited human sequences. An additional 31 sequences diverged from the human database sequences at 1 of 20 bp, leaving 3 candidate sequences represented by 19 tags (0.21%) for analysis. Two of these three sequences exactly map to transcripts of the KSHV genome (gi:2065526) T1.1 locus (nucleotides 29640 to 29659; 11 tags) and K12 locus (nucleotides 117460 to 117441; 6 tags), previously described as being present in BCBL-1 cells (48). These viral transcripts are highly abundant, accounting for 1,219 and 665 transcripts per million (TPM), respectively, in the BCBL-1 library. The viral origin of these sequences was confirmed by experimental 3′ RACE with product sequencing (data not shown). The third candidate sequence precisely matches an unannotated human EST (gi:78486988; two tags). Reanalysis of the entire 8,824-tag library for KSHV-specific transcript tags identified five additional KSHV tags sequenced once, and therefore not included in our HiFi data set, for KSHV genes ORFK2 (encoding viral interleukin-6), ORFK13 (encoding v-FLIP), ORF58, ORF64, and ORF38. Taken together, KSHV transcripts constituted 0.24% [(11 + 6 + 1 + 1 + 1 + 1 + 1)/9,026] of the total transcriptome from this uniformly virus-infected cancer cell line. These results indicate that complete in silico subtraction of HiFi human L-SAGE tags using DTS is feasible.
FIG. 1.
Schematic diagram for in silico subtraction of BCBL-1 L-SAGE tags. L-SAGE was performed on BCBL-1 cell line RNA to generate 9,026 20-bp tags. HiFi sequences from two or more tags were chosen after the exclusion of ambiguous tags and single-reading tags from the total group of tags. HiFi sequences were then compared against human RefSeq database (RNA and genomic) entries downloaded from the NCBI website for exact and near-exact (19-in-20-base) matches. The identities of KSHV T1.1 and K12 sequences were confirmed by 3′ RACE. Hum, human.
DTS analysis of SCCC.
We next extracted mRNA from three SCCCs, tumors 1811 (AIDS associated), 1795 (AIDS associated), and 448 (non-AIDS associated), for L-SAGE library generation. Tumor tissues were dissected prior to extraction to reduce the contamination of healthy-tissue RNA. L-SAGE libraries were separately generated and sequenced, and then tag data from the three libraries were combined for DTS analysis. To validate DTS, BCBL-1 RNA was spiked (1:20) into one tumor (1811) prior to L-SAGE. KSHV RNA was absent from this tumor prior to spiking by PCR (data not shown). BCBL-1 RNA spiking prevents accurate gene expression determination for this library but should not affect DTS, which relies only on sequence occurrence, not tag frequency. L-SAGE data from tumors 1811 (23,700 tags), 1795 (137,224 tags), and 448 (80,198 tags) were combined to generate an SCCC library pool containing 241,122 L-SAGE tags from the three tumors (Table 2). Of these 241,122 SAGE tags, 175,730 (72.88%) met our criteria for HiFi, forming a HiFi data set of 18,204 different 20-bp sequences to be used in DTS.
The SCCC HiFi sequences were subtracted as shown in Fig. 2. Exact alignments to human RefSeq RNA and RefSeq genome sequences were found for all but 1,120 SCCC HiFi sequences. By allowing one mismatch, an additional 974 HiFi sequences aligned with RefSeq database entries, leaving 146 candidate sequences for analysis. To reduce the candidate pool to a high-likelihood data set, the 146 candidate sequences were compared to the sequences in the human NR and EST databases. This step subtracts unannotated and polymorphic tag sequences but also may subtract viral sequences that have been inadvertently deposited into these databases. To avoid this problem, all NR and EST sequences matching the candidate sequences were compared with sequences in the RefSeq databases by using MegaBLAST to allow mapping back to human chromosomes. Of the 146 candidate sequences, 96 aligned with NR and EST sequences, and the majority of these contained 3′ polyadenylation tracts that were not included in RefSeq database sequences. The remaining group of 50 20-base sequences did not align with human sequences at 19 or more bases. Of these 50 sequences, manual analysis revealed that 24 arose artifactually from duplicated ditags and 2 contained sequencing errors found by the inspection of electropherograms. These 26 sequences were eliminated from the HiFi data set, leaving 24 candidate sequences.
FIG. 2.
In silico subtraction analysis of SCCC transcriptome. Three separate L-SAGE libraries were generated from SCCCs (two AIDS associated and one non-AIDS associated), resulting in a combined 241,122-SAGE-tag pool and 18,204 unique HiFi sequences. Sequential subtraction generated 146 sequences that were compared to EST and NR database entries to generate a candidate list of 21 HiFi sequences. These 21 sequences were examined by rSAGE and 3′ RACE for evidence of human origin. Hum, human.
Of the 24 remaining sequences, 2 were SAGE linker sequences and 1 sequence, represented by three SAGE tags found exclusively in the spiked 1811 library, corresponded to the KSHV T1.1 sequence previously identified in the BCBL-1 experiments. This sequence was again confirmed by experimental 3′ RACE (data not shown). The abundance of this tag was 127 TPM (3 of 23,700 tags in the 1811 library), which is not significantly higher than predicted (61 TPM; chi-square value = 0.58). Thus, 21 sequences were identified as candidate viral transcript sequences by DTS.
Analysis of SCCC DTS candidate sequences.
The 21 nonaligning sequences (Table 3) remaining after DTS screening were used for reverse transcription. rSAGE and 3′ RACE products from all 21 sequences were cloned and sequenced, and all products corresponded to human transcript sequences, evidence that no distinguishable viral transcript sequences were present in our HiFi data set. Due to potential mispriming with these short sequences, however, each sequence was examined in greater detail. Four of the 21 sequences aligned with immunoglobulin variable-region gene sequences, and two aligned with highly polymorphic mitochondrial DNA sequences. Two additional sequences were deposited from multiple libraries into the NIH SAGEmap database (http://www.ncbi.nlm.nih.gov/projects/SAGE/). These eight sequences were considered likely to be human and excluded from further analysis (Table 4).
TABLE 3.
Representative results of DNA sequencing of RACE and rSAGE products of 24 HiFi sequences
| Tag sequence | No. of occurrences | GenBank accession no. of corresponding human sequence | Description |
|---|---|---|---|
| CATGTCGGACGTACATCGTT | 81 | Adapter in L-SAGE | |
| CATGTCGGATATTAAGCCTA | 22 | Adapter in L-SAGE | |
| CATGTTATACTTTTGACAAT | 3 | U66521 | KSHV T1.1 mRNA transcript |
| CATGCGCTGGGCATCTACCC | 7 | AB064111 | Homo sapiens mRNA for immunoglobulin κ light-chain VL-J region |
| CATGGTAAGTGCACTTGGAC | 6 | DQ523681 | H. sapiens isolate 2798T mitochondrion gene |
| CATGGTATTACCAACTGGTT | 4 | BC073764 | H. sapiens immunoglobulin κ constant gene |
| CATGCAAAGTTTACAACTTC | 2 | BC063599 | H. sapiens immunoglobulin κ variable-region gene sequence |
| CATGGCCACCTATCACACCC | 2 | EF060266 | H. sapiens isolate Paudibhuiya_84 mitochondrion gene |
| CATGCAGACTACACAGTGGC | 2 | BC073791 | H. sapiens immunoglobulin κ constant gene |
| CATGGTAGGGGGCTTGAGGA | 2 | NM_003564 | H. sapiens transgelin 2 gene |
| CATGGATAGCACGCCGCATA | 2 | NM_002107 | H. sapiens H3 histone gene, family 3A |
| CATGCAGCTGGCAATCAATA | 4 | NM_000526 | H. sapiens keratin 14 gene |
| CATGGCCCACATTAGAATAA | 3 | NM_001025158 | H. sapiens CD74 molecule gene, major histocompatibility complex |
| CATGGGGTGTCAATAAAGCT | 2 | NM_002178 | H. sapiens insulin-like growth factor binding protein 6 gene |
| CATGGGATAGGTGGGGCGGG | 2 | NM_032818 | H. sapiens chromosome 9 ORF100 |
| CATGCAGGTTTCACATTCTA | 2 | NM_004887 | H. sapiens chemokine (C-X-C motif) ligand 14 gene |
| CATGGACCTGAATTGCCTGG | 2 | AK123483 | H. sapiens cDNA FLJ41489 fis |
| CATGTAATATTATCCTGTCC | 2 | NM_144691 | H. sapiens calpain 12 gene |
| CATGGAGATTGCCCCTGCTG | 2 | NM_006142 | H. sapiens stratifin gene |
| CATGTATGCTGGTTCCACCA | 2 | AL161669 | Human chromosome 14 DNA sequence BAC R-736N17 of library RPCI-11 from chromosome 14 |
| CATGTTGGACGTACATCGTT | 2 | NM_002213 | H. sapiens β5 integrin gene |
| CATGCAGCTGGACCCTGCTT | 3 | NM_000526 | H. sapiens keratin 14 gene |
| CATGTCGGACGTACACGTTA | 3 | AL034344 | Human DNA sequence from clone RP1-118B18 on chromosome 6p24.1-25.3 |
| CATGCCGACGATGCCCAGAA | 2 | NM_022873 | H. sapiens alpha interferon-inducible protein 6 gene |
TABLE 4.
Virtual analysis of 21 candidates with one extra base
| Tag sequence | Sequence with extra basea | Aligns with immunoglobulin or mitochondrion sequence | Present in SAGEmap database (no. of libraries) | No. of matching bases/no. of bases analyzed relative to:
|
Accession no. and description of best-match virus or viral gene or region | |
|---|---|---|---|---|---|---|
| Best-match human NR sequence | Best-match viral NR sequence | |||||
| CATGCGCTGGGCATCTACCC | CATGCGCTGGGCATCTACCCg | Yes | 19/21 | 18/21 | AB154196.1, hepatitis C virus type 1b polyprotein gene | |
| CATGGTAAGTGCACTTGGAC | CATGGTAAGTGCACTTGGACg | Yes | Yes (13) | 18/21 | 17/21 | DQ279927.1, human herpesvirus 4 strain AG876 |
| CATGGTATTACCAACTGGTT | CATGGTATTACCAACTGGTTa | Yes | 19/21 | 17/21 | AF039490.1, gp41 (env) gene of HIV-2 isolate PO1 clone C1.6 from Portugal | |
| CATGCAAAGTTTACAACTTC | CATGCAAAGTTTACAACTTCc | Yes | 19/21 | 18/21 | U37488.1, human papillomavirus type 54 | |
| CATGGCCACCTATCACACCC | CATGGCCACCTATCACACCCc | Yes | 19/21 | 18/21 | AF478169.1, porcine lymphotropic herpesvirus long unique region | |
| CATGCAGACTACACAGTGGC | CATGCAGACTACACAGTGGCc | Yes | 18/21 | 17/21 | AF331718.1, Alkhurma virus strain 1176 polyprotein gene | |
| CATGGTAGGGGGCTTGAGGA | CATGGTAGGGGGCTTGAGGAa | Yes (10) | 19/21 | 18/21 | AY309060.1, mumps virus isolate Dg1062/Korea/98 | |
| CATGGATAGCACGCCGCATA | CATGGATAGCACGCCGCATAc | Yes (2) | 18/21 | 18/21 | AY864330.1, Chrysodeixis chalcites nucleopolyhedrovirus | |
| CATGCAGCTGGCAATCAATA | CATGCAGCTGGCAATCAATAc | Yes(1) | 19/21 | 18/21 | AF512031.3, Choristoneura fumiferana multicapsid nucleopolyhedrovirus polyhedrin gene | |
| CATGGCCCACATTAGAATAA | CATGGCCCACATTAGAATAAa | Yes (1) | 19/21 | 18/21 | AY355259.1, human rhinovirus 78 VP1 capsid protein gene | |
| CATGGGGTGTCAATAAAGCT | CATGGGGTGTCAATAAAGCTg | Yes (1) | 19/21 | 17/21 | DQ127618.1, Emiliania huxleyi virus 163 clone | |
| CATGGGATAGGTGGGGCGGG | CATGGGATAGGTGGGGCGGGa | Yes (1) | 19/21 | 18/21 | AY564767.1, hepatitis C virus isolate BO-124 polyprotein gene | |
| CATGCAGGTTTCACATTCTA | CATGCAGGTTTCACATTCTAa | 19/21 | 19/21 | AF160139.1, coxsackievirus B4 isolate 8033 polyprotein gene | ||
| CATGGACCTGAATTGCCTGG | CATGGACCTGAATTGCCTGGc | 19/21 | 18/21 | AY729654.1, Sudan Ebola virus strain Gulu | ||
| CATGTAATATTATCCTGTCC | CATGTAATATTATCCTGTCCc | 19/21 | 18/21 | AY552664.1, feline immunodeficiency virus isolate Ple-349 subtype B polyprotein (pol) gene | ||
| CATGGAGATTGCCCCTGCTG | CATGGAGATTGCCCCTGCTGa | 19/21 | 19/21 | AY618418.1, maize mosaic virus | ||
| CATGTATGCTGGTTCCACCA | CATGTATGCTGGTTCCACCAg | 19/21 | 18/21 | DQ116961.1, West Nile virus isolate goshawk-Hungary/04 | ||
| CATGTTGGACGTACATCGTT | CATGTTGGACGTACATCGTTa | Yes (1) | 18/21 | 18/21 | AJ781163.1, Amasya cherry disease-associated chrysovirus ORF4 | |
| CATGCAGCTGGACCCTGCTT | CATGCAGCTGGACCCTGCTT | 18/20 | 17/20 | AF333347.1, cetacean morbillivirus phosphoprotein (P) gene | ||
| CATGTCGGACGTACACGTTA | CATGTCGGACGTACACGTTAg | 17/21 | 18/21 | AF325155.1, Spodoptera litura nucleopolyhedrovirus strain G2 | ||
| CATGCCGACGATGCCCAGAA | CATGCCGACGATGCCCAGAAt | 18/21 | 18/21 | DQ504428.1, Clanis bilineata nucleopolyhedrosis virus isolate DZ1 | ||
Extra bases are in lowercase.
Direct inspection of ditags allowed high-level-confidence elongation of 12 of the 13 remaining candidate sequences to 21 bases (Table 4). Short-sequence, exact BLAST alignment with human NR sequences revealed nine of these sequences to match human sequences at 19 of 21 base pairs, providing additional evidence for their human origin. For the remaining four sequences, comparison to virus NR database entries revealed that all but one sequence had lower degrees of homology to viral than to human sequences. A transcript belonging to a virus causing SCCC would be expected to be found independently in more than one SCCC library, but tags from all four remaining sequences were found in only one library (1795).
Although these 21 DTS candidates should be considered for future studies, present evidence suggests that all 21 sequences are of human origin. If none of the candidate tags belong to viral transcripts, and assuming a Poisson distribution for sequencing a viral transcript, the upper 95% confidence limit that a viral transcript exceeding our matching criteria is present in SCCC is 20 TPM. It is estimated that there are approximately 200,000 mRNA transcripts in a cell, corresponding to 5 TPM for a single mRNA copy per cell (5, 38). Using this value as a comparison, we sequenced to a level of two to three transcripts per cell (11 TPM) and can conclude with a 95% confidence level that DTS-identifiable viral transcripts are not present in conjunctival carcinoma at four transcripts per cell or higher (20 TPM).
DISCUSSION
Short human expression tags can be reliably subtracted to screen for exogenous viral transcript sequences, opening the possibility of using high-throughput sequencing to discover new viral pathogens. L-SAGE is attractive for DTS since it uses polyadenylated mRNA and will not include most bacterial RNAs found in nonsterile-site tissues such as conjunctival tumors. DTS can be applied to other expression sampling techniques, such as massively parallel sequencing, and will improve with tags longer than those in this study. DTS differs from standard SAGE in that only HiFi sequence data are used and comparison can be made without control tissue samples. Once a tag sequence is accepted as valid, tag frequency is not taken into account.
Our analysis suggests that 20 bases is the minimum useful length for DTS. Early attempts to perform DTS with 14-base SAGE tags failed to discriminate between human and viral sequences (Y. Chang and P. S. Moore, unpublished results). At shallow sequencing depths as used for the BCBL-1 library, 20-bp DTS readily identified viral sequences and allowed complete transcriptome subtraction. When more extensive sequencing is performed, as seen with our SCCC library, the numbers of ambiguous and rare tag sequences increase, generating a large list of candidates (146) that require further subtraction using NR and EST library databases and manual inspection. We anticipate that even small increases in tag lengths should be able to markedly reduce the candidate pool size, as well as increase the discrimination between viral and human sequences.
Another important advantage in using SAGE or other quantitative expression profiling techniques for DTS is that a direct measurement of transcript levels can also be performed. This capacity is shown by our analysis of viral transcripts in a tumor virus-infected cell line (BCBL-1). KSHV transcripts constitute a high percentage (0.24%) of the total mRNA from BCBL-1 cells. Transcripts identified in PEL cells are identical to those previously found by Cornelissen and colleagues using short SAGE of Kaposi's sarcoma (KS) tumors (9). T1.1 is commonly referred to as a lytic gene product, but multiple studies have shown that it is also readily detected in resting PEL cells. Whether this is due to transcriptional control outside of the latent-lytic replication cycle or to a fraction of cells in culture undergoing lytic replication cannot be addressed by our study. An examination of unpublished 293 cell line SAGE data from the Cancer Genome Anatomy Project (293 human embryonic kidney cells) (6) for 41,955 tags shows similar levels of viral transcript abundance (0.31%). In contrast to BCBL-1 cells, 293 cells are artificially transformed with adenovirus DNA rather than naturally infected with a virus (15).
Tumor tissues are likely to have a far lower level of viral gene expression than cell lines. This is due to lower viral copy numbers and infiltration by uninfected, nontumor cells. Cornelissen et al. examined two KS tumors from HIV-infected patients by SAGE and found 24 of 45,913 tags (522 TPM, or 104 transcripts per cell) and 3 of 47,316 tags (63 TPM, or 12 transcripts per cell) belonging to KSHV (9). These levels are comparable to the transcript abundance found for KSHV (127 TPM) in our library 1811 spiking experiments. We did not identify a viral transcript after sequencing to a depth of two to three transcripts per cell (11 TPM), and we can exclude with 95% confidence the possibility that distinguishable viral transcripts are present in SCCC at approximately four transcripts per cell (20 TPM) or higher—a level far below that found empirically for KSHV in KS tumors. To confirm these results, we performed an entirely new long-EST analysis of library 1811, analyzing an additional 150,000 50- to 200-bp ESTs, but did not identify a candidate viral sequence (H. Feng, Y. Chang, and P. S. Moore, unpublished results). For technical reasons, these data cannot be directly added to our current DTS analysis, but they provide additional confidence in the validity of the analysis and substantially extend the lower limit for the detection of a unique viral transcript in SCCCs.
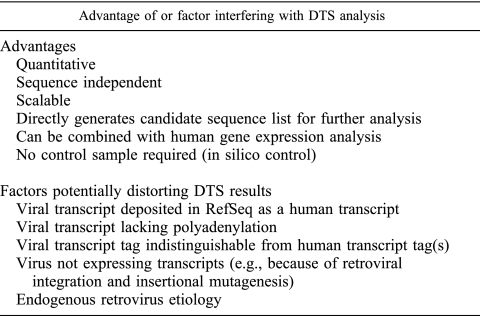
Despite DTS evidence against a direct viral carcinogen's being present in SCCC, there are important caveats to concluding that SCCC does not have an infectious etiology. Viral NlaIII SAGE tags indistinguishable from human transcripts will be missed by DTS. Our virtual herpesvirus tag analysis suggests that this would occur for approximately 25% of viral tags. This uncertainty is reduced if a virus expresses multiple transcripts. Similarly, if a viral transcript does not contain a suitable NlaIII site, it will not be processed into a SAGE tag. Subtraction using unannotated EST transcripts also increases the probability that a viral sequence previously identified during EST screening will falsely be subtracted. We have minimized this risk by mapping all unannotated EST sequences back to the human genome, but it remains a possible pitfall. While most RNA viruses and all DNA viruses express mRNA transcripts, those viruses that do not have polyadenylated transcripts will be missed. Finally, DTS is unlikely to identify endogenous retroviral sequences that have been annotated as human or retroviruses integrating into the host genome and acting through insertional mutagenesis (11). These and other caveats to DTS analysis are listed in Fig. 3 and should be kept in mind when interpreting DTS results. For these reasons, it remains possible that SCCC has an infectious etiology, but it is highly unlikely that this tumor is caused by a direct viral carcinogen (e.g., human papillomavirus, Epstein-Barr virus, or KSHV).
FIG. 3.
Advantages that distinguish DTS as a valuable tool for identifying conditions with viral etiologies are listed, along with factors to consider in interpreting DTS results.
DTS has marked advantages over other existing methods for new viral pathogen discovery. Physical enrichment of encapsidated viral nucleic acids has been successfully used to identify new human parvoviruses (2) and polyomaviruses (1) from respiratory secretions. This is unlikely to be useful for tumor viruses that initiate tumorigenesis during latent infection, in which the viral episome is not encapsidated. In silico subtraction, however, shows promise for directly identifying viral sequences within the context of the host cell transcriptome. If DTS is used with a quantitative expression technology like L-SAGE, it provides estimates of the presence or absence of a distinguishable viral transcript. For diseases in which the epidemiology and biology implicate a detectable exogenous virus, DTS may prove to be a powerful tool for either identifying or eliminating a viral etiology.
Acknowledgments
We thank Giovanna Bestetti and Eian Murphy for preliminary SAGE analyses, Harold Jaffe for epidemiologic insights on SCCC, Paul Woods for help with SAGE sequencing, Tao Tao from NCBI for valuable suggestions on BLAST alignment, Jian Yu for the helpful suggestion on rSAGE technique, and Richard Wood for comments on the manuscript. We also thank Cuauhtemoc Gomez, Kelly Bogda, Elizabeth Weigand, and Beth Schnieder for technical assistance. This study was made possible through the essential help of Robert Downing, CDC, and other members of the CDC-Uganda team who collected and characterized patient specimens.
This project was supported in part by funds from the Public Health Service, CDC (EIN 1250965591), and a National Cancer Institute, NIH, grant (R01 CA67391) and in part under a grant from the Pennsylvania Department of Health.
The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Footnotes
Published ahead of print on 8 August 2007.
REFERENCES
- 1.Allander, T., K. Andreasson, S. Gupta, A. Bjerkner, G. Bogdanovic, M. A. A. Persson, T. Dalianis, T. Ramqvist, and B. Andersson. 2007. Identification of a third human polyomavirus. J. Virol. 81:4130-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 102:12891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ateenyi-Agaba, C., E. Weiderpass, A. Smet, W. Dong, M. Dai, B. Kahwa, H. Wabinga, E. Katongole-Mbidde, S. Franceschi, and M. Tommasino. 2004. Epidermodysplasia verruciformis human papillomavirus types and carcinoma of the conjunctiva: a pilot study. Br. J. Cancer 90:1777-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beral, V., and R. Newton. 1998. Overview of the epidemiology of immunodeficiency-associated cancers. J. Natl. Cancer Inst. Monogr. 1998:1-6. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, J. O., J. G. Morton, M. Rosbash, and M. Richardson. 1974. Three abundance classes in HeLa cell messenger RNA. Nature 250:199-204. [DOI] [PubMed] [Google Scholar]
- 6.Boon, K., E. C. Osorio, S. F. Greenhut, C. F. Schaefer, J. Shoemaker, K. Polyak, P. J. Morin, K. H. Buetow, R. L. Strausberg, S. J. De Souza, and G. J. Riggins. 2002. An anatomy of normal and malignant gene expression. Proc. Natl. Acad. Sci. USA 99:11287-11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 265:1865-1869. [DOI] [PubMed] [Google Scholar]
- 8.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 9.Cornelissen, M., A. C. van der Kuyl, R. van den Burg, F. Zorgdrager, C. J. van Noesel, and J. Goudsmit. 2003. Gene expression profile of AIDS-related Kaposi's sarcoma. BMC Cancer 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davison, A. 2007. Comparative analysis of the genomes, p. 10-26. In A. Arvin, G. Campadelli-Fiume, E. Mocarski, P. S. Moore, B. Roizman, R. Whitley, and K. Yamanishi (ed.), Human herpesvirus: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom.
- 11.Derse, D., B. Crise, Y. Li, G. Princler, N. Lum, C. Stewart, C. F. McGrath, S. H. Hughes, D. J. Munroe, and X. Wu. 2007. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J. Virol. 81:6731-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 13.Gao, S.-J., and P. S. Moore. 1996. Molecular approaches to the identification of unculturable infectious agents. Emerg. Infect. Dis. 2:159-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goedert, J. J., and T. R. Cote. 1995. Conjunctival malignant disease with AIDS in USA. Lancet 346:257-258. [DOI] [PubMed] [Google Scholar]
- 15.Graham, F. L., and A. J. Van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 16.Harwood, C. A., P. J. Spink, T. Surentheran, I. M. Leigh, E. M. de Villiers, J. M. McGregor, C. M. Proby, and J. Breuer. 1999. Degenerate and nested PCR: a highly sensitive and specific method for detection of human papillomavirus infection in cutaneous warts. J. Clin. Microbiol. 37:3545-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hon, C., W. Y. Au, and R. H. Liang. 2004. Conjunctival carcinoma as a novel post-stem cell transplantation malignancy. Bone Marrow Transplant. 34:181-182. [DOI] [PubMed] [Google Scholar]
- 18.Kaimbo Wa Kaimbo, D., R. Parys-Van Ginderdeuren, and L. Missotten. 1998. Conjunctival squamous cell carcinoma and intraepithelial neoplasia in AIDS patients in Congo Kinshasa. Bull. Soc. Belge Ophtalmol. 268:135-141. [PubMed] [Google Scholar]
- 19.Karlsen, F., M. Kalantari, A. Jenkins, E. Pettersen, G. Kristensen, R. Holm, B. Johansson, and B. Hagmar. 1996. Use of multiple PCR primer sets for optimal detection of human papillomavirus. J. Clin. Microbiol. 34:2095-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karp, C. L., I. U. Scott, T. S. Chang, and S. C. Pflugfelder. 1996. Conjunctival intraepithelial neoplasia. A possible marker for human immunodeficiency virus infection? Arch. Ophthalmol. 114:257-261. [DOI] [PubMed] [Google Scholar]
- 21.Kestelyn, P., A. M. Stevens, A. Ndayambaje, M. Hanssens, and P. van de Perre. 1990. HIV and conjunctival malignancies. Lancet 336:51-52. [DOI] [PubMed] [Google Scholar]
- 22.Korf, I., M. Yandell, and J. Bedell. 2003. Blast, 1st ed. O'Reilly & Associates, Sebastopol, CA.
- 23.Li, L., D. Techel, N. Gretz, and A. Hildebrandt. 2005. A novel transcriptome subtraction method for the detection of differentially expressed genes in highly complex eukaryotes. Nucleic Acids Res. 33:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newton, R., J. Ziegler, C. Ateenyi-Agaba, L. Bousarghin, D. Casabonne, V. Beral, E. Mbidde, L. Carpenter, G. Reeves, D. M. Parkin, H. Wabinga, S. Mbulaiteye, H. Jaffe, D. Bourboulia, C. Boshoff, A. Touze, and P. Coursaget. 2002. The epidemiology of conjunctival squamous cell carcinoma in Uganda. Br. J. Cancer 87:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichol, S. T., C. F. Spiropoulou, S. Morzunov, P. E. Rollin, T. G. Ksiazek, H. Feldmann, A. Sanchez, J. Childs, S. Zaki, and C. J. Peters. 1993. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 262:914-917. [DOI] [PubMed] [Google Scholar]
- 26.Parkin, D. M. 2006. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer 118:3030-3044. [DOI] [PubMed] [Google Scholar]
- 27.Parsonnet, J. (ed.). 1999. Microbes and malignancy, p. 3-24. Oxford University Press, New York, NY.
- 28.Rabkin, C. S. 1998. Association of non-acquired immunodeficiency syndrome-defining cancers with human immunodeficiency virus infection. J. Natl. Cancer Inst. Monogr. 1998:23-25. [DOI] [PubMed] [Google Scholar]
- 29.Relman, D. A. 1999. The search for unrecognized pathogens. Science 284:1308-1310. [DOI] [PubMed] [Google Scholar]
- 30.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 31.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saha, S., A. B. Sparks, C. Rago, V. Akmaev, C. J. Wang, B. Vogelstein, K. W. Kinzler, and V. E. Velculescu. 2002. Using the transcriptome to annotate the genome. Nat. Biotechnol. 20:508-512. [DOI] [PubMed] [Google Scholar]
- 33.Shelil, A. E., C. L. Shields, J. A. Shields, and R. C. Eagle, Jr. 2003. Aggressive conjunctival squamous cell carcinoma in a patient following liver transplantation. Arch. Ophthalmol. 121:280-282. [DOI] [PubMed] [Google Scholar]
- 34.Spraul, C. W., and H. E. Grossniklaus. 1996. Tumors of the cornea and conjunctiva. Curr. Opin. Ophthalmol. 7:28-34. [DOI] [PubMed] [Google Scholar]
- 35.Tornesello, M. L., M. L. Duraturo, K. M. Waddell, B. Biryahwaho, R. Downing, S. Balinandi, S. B. Lucas, L. Buonaguro, and F. M. Buonaguro. 2006. Evaluating the role of human papillomaviruses in conjunctival neoplasia. Br. J. Cancer 94:446-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urisman, A., R. J. Molinaro, N. Fischer, S. J. Plummer, G. Casey, E. A. Klein, K. Malathi, C. Magi-Galluzzi, R. R. Tubbs, D. Ganem, R. H. Silverman, and J. L. Derisi. 2006. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.VanDevanter, D. R., P. Warrener, L. Bennett, E. R. Schultz, S. Coulter, R. L. Garber, and T. M. Rose. 1996. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 34:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velculescu, V. E., S. L. Madden, L. Zhang, A. E. Lash, J. Yu, C. Rago, A. Lal, C. J. Wang, G. A. Beaudry, K. M. Ciriello, B. P. Cook, M. R. Dufault, A. T. Ferguson, Y. Gao, T. C. He, H. Hermeking, S. K. Hiraldo, P. M. Hwang, M. A. Lopez, H. F. Luderer, B. Mathews, J. M. Petroziello, K. Polyak, L. Zawel, K. W. Kinzler, et al. 1999. Analysis of human transcriptomes. Nat. Genet. 23:387-388. [DOI] [PubMed] [Google Scholar]
- 39.Velculescu, V. E., L. Zhang, B. Vogelstein, and K. W. Kinzler. 1995. Serial analysis of gene expression. Science 270:484-487. [DOI] [PubMed] [Google Scholar]
- 40.Verma, N., and J. Kearney. 1996. Ocular manifestations of AIDS. P. N. G. Med. J. 39:196-199. [PubMed] [Google Scholar]
- 41.Wabinga, H. R., D. M. Parkin, F. Wabwire-Mangen, and S. Nambooze. 2000. Trends in cancer incidence in Kyadondo County, Uganda, 1960-1997. Br. J. Cancer 82:1585-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waddell, K., J. Magyezi, L. Bousarghin, P. Coursaget, S. Lucas, R. Downing, D. Casabonne, and R. Newton. 2003. Antibodies against human papillomavirus type 16 (HPV-16) and conjunctival squamous cell neoplasia in Uganda. Br. J. Cancer 88:2002-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waddell, K. M., R. G. Downing, S. B. Lucas, and R. Newton. 2006. Corneo-conjunctival carcinoma in Uganda. Eye 20:893-899. [DOI] [PubMed] [Google Scholar]
- 44.Waddell, K. M., S. Lewallen, S. B. Lucas, C. Atenyi-Agaba, C. S. Herrington, and G. Liomba. 1996. Carcinoma of the conjunctiva and HIV infection in Uganda and Malawi. Br. J. Ophthalmol. 80:503-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weber, G., J. Shendure, D. M. Tanenbaum, G. M. Church, and M. Meyerson. 2002. Identification of foreign gene sequences by transcript filtering against the human genome. Nat. Genet. 30:141-142. [DOI] [PubMed] [Google Scholar]
- 46.Xu, Y., N. Stange-Thomann, G. Weber, R. Bo, S. Dodge, R. G. David, K. Foley, J. Beheshti, N. L. Harris, B. Birren, E. S. Lander, and M. Meyerson. 2003. Pathogen discovery from human tissue by sequence-based computational subtraction. Genomics 81:329-335. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto, M., T. Wakatsuki, A. Hada, and A. Ryo. 2001. Use of serial analysis of gene expression (SAGE) technology. J. Immunol. Methods 250:45-66. [DOI] [PubMed] [Google Scholar]
- 48.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 93:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]