Abstract
Insect BTI-TN-5B1-4 (Tn5) cells have been used extensively with recombinant baculoviruses to express foreign genes. When a recombinant baculovirus containing the hepatitis E virus capsid protein gene was used to infect Tn5 cells, unknown virus particles in addition to the anticipated hepatitis E virus-like particles were produced in the infected cells. The unknown virus particles were 35 nm in diameter and contained RNA that was highly homologous to full-length RNA1 (3,107 bp) and RNA2 (1,383 bp) genomic RNAs of flock house virus. Surprisingly, both RNAs seen in these induced nodavirus particles could be amplified from commercially available Tn5 cells without infection with or induction by a baculovirus. The nucleotide sequences from the purified nodavirus particles and the normal Tn5 cells were identical, demonstrating that the Tn5 cells themselves were latently infected with a nodavirus. However, the generation of nodavirus particles was significantly stimulated by infection with recombinant baculoviruses. Phylogenetic analysis suggested that this new nodavirus belongs to the genus Alphanodavirus in the family Nodaviridae.
The family Nodaviridae comprises small, nonenveloped, and isometric viruses containing positive-sense bipartite RNA genomes (3, 26). Two genera in this family, Alphanodavirus and Betanodavirus, have been distinguished previously (2). Alphanodaviruses infect primarily insects, but their general pathogenicity is not well defined. However, the type virus, Nodamura virus, causes paralysis in infected larvae of the wax moth, Galleria mellonella (1), suckling mice (23, 24), and suckling hamsters (8). In addition to Nodamura virus, the genus Alphanodavirus includes flock house virus (FHV), black beetle virus, Boolarra virus, and Pariacoto virus (5, 22, 23, 28, 32). Viruses belonging to the genus Betanodavirus infect primarily fishes and are the causative agent of viral encephalopathy and retinopathy and viral nervous necrosis, a devastating disease of many species of marine fish cultured worldwide (19). Although a number of betanodavirus strains have been isolated from fish, only five alphanodaviruses have been isolated. Recently, an unclassified nodavirus, Macrobrachium rosenbergii nodavirus, was isolated from the giant freshwater prawn, M. rosenbergii (29).
Nodavirus particles are 32 to 33 nm in diameter, and their buoyant density in CsCl ranges from 1.30 to 1.34 g/cm3 (25). The virion has icosahedral symmetry with T=3 and is composed of 180 copies of a single coat protein of around 40 kDa (12, 30). The genome is composed of two RNA segments, RNA1 (3.1 kb) and RNA2 (1.4 kb). RNA1 carries all the viral information required for RNA transcription and replication and encodes protein A (102 kDa) and protein B (10 kDa). Protein A is a catalytic subunit of the RNA-dependent RNA polymerase that functions in the replication of both RNA segments. RNA2 encodes the coat precursor protein alpha, which is cleaved into beta (38-kDa) and gamma (5-kDa) proteins during the maturation of the particle, a reaction essential for infectivity (27). Both RNA1 and RNA2 are required for infection and are packaged in the same virion (21). Both RNAs have 5′-cap structures, while their 3′ ends lack poly(A) tails (5, 10, 14, 20). During RNA replication, the RNA-dependent RNA polymerase directs the production of the subgenomic RNA3, which encodes nonstructural proteins B1 and B2 in overlapping open reading frames. Nodaviruses provide outstanding model systems for the study of RNA replication, virus assembly, and three-dimensional structure (3, 26).
In our laboratory, the recombinant baculovirus expression system has been a major tool for the generation of recombinant virus proteins. Two insect cell lines, Sf9 and Tn5, were routinely used. During the purification of hepatitis E virus-like particles (HEV-LP) from recombinant baculovirus-infected Tn5 cells, we found a large amount of a 40-kDa protein (p40). This protein appeared to form virus particles similar to those of nodaviruses. Antigenic analysis showed that p40 was cross-reactive with anti-FHV antibody. Further studies detected that full-length RNA1 and RNA2 genomes were present in normal insect cells, suggesting that these Tn5 cells were latently infected with a nodavirus.
MATERIALS AND METHODS
Insect cells.
The insect BTI-TN-5B1-4 cell line (“High Five”), which we have designated Tn5/97, was derived from BTI-TN-5B1-4 (Tn5) and was obtained in April 1997 from a commercial supplier (Invitrogen Inc.; lot BAC0010CB). This cell line has been regularly employed for the expression of various viral capsid proteins by using recombinant baculoviruses (11, 15, 17). EX-CELL 400 or EX-CELL 405 medium (JRH Biosciences, Lenexa, KS) was used to maintain Tn5/97 cells. A different lot of the same cell line, designated Tn5/05, was obtained on 6 October 2005 from the same commercial supplier (Invitrogen Inc.; lot 1288466). Another insect cell line, Sf9, was obtained from Riken BioResource Center, Tsukuba-shi, Japan, and maintained in TC-100 medium (GIBCO BRL, Gaithersburg, MD). Drosophila melanogaster Schneider 2 (S2) cells, an insect cell line derived from D. melanogaster embryos, were obtained from Invitrogen Inc. and grown at 28°C in Schneider's insect medium supplemented with 10% heat-inactivated fetal bovine serum, 10 U of penicillin per ml, and 10 μg of streptomycin per ml. S2 cells were routinely passaged every 4 days at a 1:5 dilution.
Recombinant baculoviruses.
Wild-type baculovirus (Autographa californica nuclear polyhedrosis virus) and the following recombinant baculoviruses were used in this study: Ac[G3n13ORF2] (unpublished data), Ac[G3ORF2] (unpublished data), Ac[G1ORF2] (17), Ac[G4ORF2] (unpublished data), Ac[G1HEV] (unpublished data), Ac[G4ORF3] (unpublished data), Ac[NoroVP1] (11), Ac[BKVP1] (15), Ac[JCVP1] (unpublished data), Ac[TTVORF2] (unpublished data), Ac[polioVP1] (unpublished data), and Ac[NodaRNA2] (1,000 × g) harboring, respectively, the hepatitis E virus (HEV) genotype 3 ORF2 gene with a deletion corresponding to the N-terminal 13 amino acids of the product, full-length HEV genotype 3 ORF2, HEV genotype 1 ORF2, HEV genotype 4 ORF2, the full-length genome of HEV genotype 1, HEV genotype 4 ORF3, the norovirus genogroup I genotype 2 VP1 gene, the human BK polyomavirus VP1 gene, the human polyomavirus JC VP1 gene, transfusion-transmitted virus ORF2, the poliovirus VP1 gene, and the nodavirus RNA2, which was derived from the Tn5/97 cell line. The generation of the recombinant baculoviruses and the preparation of the seed virus were performed using Sf9 cells; protein expression was carried out with Tn5 cells.
Amplification of nodavirus RNA by reverse transcription-PCR (RT-PCR).
Total RNA from cell lysates, culture media, or purified virus particles was extracted with RNAzol-LS reagent (Invitrogen Inc., Carlsbad, CA). The extracted RNA was resuspended in 20 μl of DNase-, RNase-, and proteinase-free distilled water. Reverse transcription was performed at 42°C for 50 min and at 70°C for 15 min in a 20-μl reaction mixture containing 1 μl of Superscript II RNase H reverse transcriptase (Invitrogen Inc.), 1 μl of the primer, 1 μl of RNaseOUT, 2 μl of 0.1 M dithiothreitol, 4 μl of 5× RT buffer, 1 μl of 10 mM deoxynucleoside triphosphates, 5 μ1 of RNA, and 5 μ1 of distilled water. The reverse primer Noda-U7 (5′-ACCTCTGCCCTTTCGGGCTA-3′; nucleotides [nt] 3088 to 3107) was used for the cDNA synthesis of RNA1, and Noda-U1 (5′-ACCTTAGTCTGTTGACTTAA-3′; nt 1381 to 1400) was used for RNA2. Primers were designed on the basis of FHV (GenBank accession no. X15959).
The entire RNA1 was obtained as two overlapping fragments. The 5′-end half of the genome was amplified with the forward primer Noda-D6 (5′-GTTTTCGAAACAAATAAAACAGAAAA-3′; nt 1 to 26) and the reverse primer Noda-U9 (5′-CTTCAGGTTCAGCATCAGGA-3′; nt 2061 to 2080). The 3′-end half of the genome was amplified with the forward primer Noda-D7 (5′-AGCACTGGACTGCGAAACCA-3′; nt 421 to 440) and the reverse primer Noda-U7. The entire RNA2 was obtained similarly with a set of primers for the 5′-end half of the genome, Noda-D1 (5′-GTAAACAATTCCAAGTTCCA-3′; nt 1 to 20) and Noda-U2 (5′-GAGGATTTAATGATGGATTT-3′; nt 1133 to 1152), and a set of primers for the 3′ end of the genome, Noda-D2 (5′-CAACGAGCTCAACGCGTTGT-3′; nt 50 to 69) and Noda-U1.
PCR was performed in a 50-μl reaction mixture containing 2 μl of cDNA, 0.25 μl of ExTaq DNA polymerase (Takara Shuzo Co., Ltd., Kyoto, Japan), 1 μl of the forward primer, and 1 μl of a reverse primer. Each cycle consisted of denaturation at 95°C for 30 s, primer annealing at 55°C for 30 s, and an extension reaction at 72°C for 180 s, and the last cycle was followed by a final extension at 72°C for 7 min. The PCR products were purified using a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and cloned into the TA cloning vector pCR2.1 (Invitrogen, Carlsbad, CA).
To amplify part of the RNA2 molecule, PCR was carried out using the forward primer Noda-D4 (5′-ACATCCAGATCCGATCAAGT-3′; nt 491 to 510) and the reverse primer Noda-U4 (5′-GCCAGGAATGTTGCTTGCAA-3′; nt 1161 to 1180). Each cycle consisted of denaturation at 95°C for 30 s, primer annealing at 55°C for 30 s, and an extension reaction step at 72°C for 45 s; the procedure was completed by a final extension step at 72°C for 7 min.
The nucleotide sequencing of PCR products and plasmids was carried out with the primers used for the amplification by using an ABI 3130 genetic analyzer automated sequencer and a BigDye Terminator cycle sequencing ready reaction kit according to the instructions of the manufacturer (Applied Biosystems, Foster City, CA).
Purification of nodavirus particles from recombinant baculovirus-infected Tn5 cells.
The recombinant baculovirus-infected Tn5/97 cells were harvested at 7 days postinfection (p.i.). The medium and the cells were separated by centrifugation at 1,000 × g for 15 min at 4°C. The cells were treated with a denaturation buffer containing 50 mM sodium borate, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 5% 2-mercaptoethanol and gently rocked at room temperature for 2 h. The lysate was diluted with EX-CELL 405 and centrifuged at 31,000 rpm for 3 h in a Beckman SW32 Ti rotor, and the pellet was resuspended in EX-CELL 405. For sucrose gradient centrifugation, 100-μl samples were laid on the top of a 10 to 50% (wt/wt) gradient and centrifuged at 32,000 rpm for 2 h in a Beckman SW55 Ti rotor. For CsCl gradient centrifugation, 4.5-ml samples were mixed with 2.1 g of CsCl and centrifuged at 35,000 rpm for 24 h at 10°C in the same rotor. The gradient was fractionated into 250-μl aliquots, and each fraction was weighed in order to estimate the buoyant density and isopycnic point. Each fraction was diluted with EX-CELL 405 and centrifuged for 2 h at 50,000 rpm in a Beckman TLA55 rotor to sediment the virus particles.
Electron microscopy.
Samples containing the nodavirus particles were applied onto apioloform-coated grids, and the particles were allowed to attach to the grids for 5 min. After a rinse with distilled water, each sample was stained with 1% aqueous uranyl acetate and observed by using a Hitachi H-7000 electron microscope at 75 kV.
N-terminal amino acid sequence analysis.
The proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were visualized by staining with GelCode blue staining reagent (Pierce, Rockford, IL) and purified by using TALON resin (BD Biosciences Clontech, Palo Alto, CA). The N-terminal amino acid microsequencing was carried out using 100 pmol of the protein for Edman automated degradation on an Applied Biosystems model 477 protein sequencer.
SDS-PAGE and Western blot assay.
The proteins in the cell lysate and culture medium were separated by 5 to 20% SDS-PAGE and stained with Coomassie blue. For Western blot assays, the proteins in the SDS-polyacrylamide gel were electrophoretically transferred onto a nitrocellulose membrane. The membrane was then blocked with 5% skim milk in a solution of 50 mM Tris-HCl (pH 7.4) and 150 mM NaCl and incubated with the serum from a hepatitis E patient that was positive for anti-HEV immunoglobulin G (IgG) and IgM or with rabbit anti-FHV IgG antibody. Alkaline phosphatase-conjugated goat anti-human Ig (1:3,000 dilution; Dako A/S, Copenhagen, Denmark) or alkaline phosphatase-conjugated goat anti-rabbit Ig (1:1,000 dilution; Chemicon, Temecula, CA) was used as the secondary antibody. Nitroblue tetrazolium chloride and BCIP (5-bromo-4-chloro-3-indolylphosphate) p-toluidine were used as coloring agents (Bio-Rad Laboratories, Hercules, CA).
Infectivity of purified nodavirus particles.
Subconfluent Drosophila S2 cells (5 × 105 cells) in a 25-cm2 flask were infected with 1 ml (100 ng) of purified nodavirus particles. The virus was allowed to attach for 2 h at 28°C. The medium containing any unattached virus particles was removed, and 10 ml of fresh medium was added. Infected S2 cells were passaged every 4 days at a 1:5 dilution. Cells and supernatants were collected and assayed for nodavirus coat protein by the Western blot method using rabbit anti-FHV coat protein antibody.
Nucleotide sequence accession numbers.
The nodavirus RNA sequences determined in this study have been deposited in GenBank under accession numbers EF690537 (RNA1) and EF690538 (RNA2).
RESULTS
Induction of nodavirus virion formation by recombinant baculovirus infection.
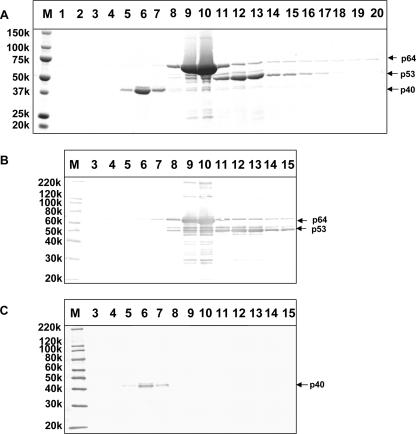
A recombinant baculovirus, Ac[G3n13ORF2], harboring the HEV genotype 3 ORF2 with the region corresponding to the N-terminal 13 amino acids of the product deleted, was used to generate HEV-LP in Tn5/97 cells. When the HEV-LP were purified by CsCl centrifugation and each fraction was analyzed by SDS-PAGE, three major proteins with molecular masses of 40 kDa (p40), 53 kDa (p53), and 64 kDa (p64) were observed (Fig. 1A). Proteins p64 and p53 appeared in fractions 9 and 10 and fractions 12 and 13, which had average densities of 1.300 g/ml3 and 1.285 g/ml3, respectively. Both p64 and p53 were reactive with the serum from a patient with acute hepatitis E, indicating that these two proteins were derived from the HEV protein (Fig. 1B). However, another protein, p40, was detected in fractions 5, 6, and 7, which had an average density of 1.350 g/ml3. A Western blot assay indicated that p40 did not react with the serum from the hepatitis E patient (Fig. 1B).
FIG. 1.
Induction of p40 protein by a recombinant baculovirus infection. (A) The HEV-LP recovered from Ac[G3n13ORF2]-infected Tn5/97 cells were purified by CsCl gradient centrifugation. The gradient was fractionated into 250-μl aliquots, and the proteins were separated by 5 to 20% SDS-PAGE and stained with Coomassie blue. (B and C) Each fraction was subjected to a Western blot assay using either the serum from a patient with acute hepatitis E (B) or rabbit anti-FHV coat protein antibody (C). M, molecular weight markers; 150k, molecular weight of 150,000.
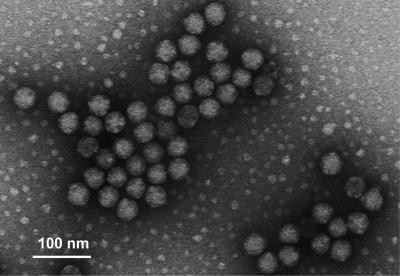
The N-terminal amino acid sequence of p40 was determined by microsequencing, and the sequence VNNSRPKRQRSQRVV was obtained. This sequence is close to the 15-amino-acid N-terminal sequence (VNNNRPRRGRAQRVV) of FHV, a nodavirus in the genus Alphanodavirus. Surprisingly, when a Western blot assay was performed with rabbit anti-FHV coat protein, p40 showed strong reactivity. A minor protein with a molecular mass of 43 kDa (p43) also appeared in fraction 6. Although p43 did not react with the serum from the hepatitis E patient, like p40, it reacted with the anti-FHV antibody (Fig. 1C). In addition, the N-terminal amino acid sequence of p43 was identical to that of p40 (data not shown). When fraction 6 was examined by electron microscopy, many small spherical particles with diameters of 35 nm were observed. These particles were similar to FHV particles (Fig. 2). These results indicated that p43 and p40 were derived from the 35-nm-diameter particles and that infecting the Tn5/97 cells with the recombinant baculovirus Ac[G3n13ORF2] induced not only the expression of the HEV ORF2 proteins but also the formation of nodavirus-like particles.
FIG. 2.
Electron microgram of nodavirus-like 35-nm particles. Fraction 6 from Fig. 1 was used for electron microscopy observations.
Amplification of nodavirus RNA2 genome from Tn5 cells.
To determine which cell and/or virus stock was contaminated with the nodavirus, total RNA was extracted from lysates and culture media from uninfected Tn5/97 cells, uninfected Sf9 cells, Ac[G3n13ORF2]-infected Tn5/97 cells, and Ac[G3n13ORF2]-infected Sf9 cells. Part of the RNA2 sequence (nt 491 to 1180) was then amplified by RT-PCR. As shown in Fig. 3, a band corresponding to 690 bp from both the lysates and culture media from uninfected Tn5/97 cells and Ac[G3n13ORF2]-infected Tn5/97 cells was detected (Fig. 3, lanes 1, 2, 5, and 6) whereas no band from either uninfected or Ac[G3n13ORF2]-infected Sf9 cells was detected (Fig. 3, lanes 3, 4, 7, and 8). These results indicated that neither the Sf9 cells nor the seed baculovirus had been contaminated with RNA2. To confirm that the nodavirus sequence was derived from the Tn5 cells, a new lot of Tn5 cells (Tn5/05) was purchased. A 200-μl aliquot of the cell suspension was removed from the original tube, the cells were separated from the culture medium, and RT-PCR was performed to determine if a region of the RNA2 sequence was present. Both the cells and the medium were positive for the 690-bp band, clearly indicating that the Tn5/05 cells had been infected, presumably latently, with a nodavirus (Fig. 3, lanes 9 and 10).
FIG. 3.
Amplification of part of the nodavirus RNA2 from insect cells and from purified nodavirus-like particles. RNA was extracted from the supernatant and lysate from uninfected Tn5/97 cells (Tn5/97-sup and Tn5/97-cell), uninfected Sf9 cells (Sf9-sup and Sf9-cell), Ac[G3n13ORF2]-infected Tn5/97 cells (Tn5/97-sup/infect and Tn5/97-cell/infect), Ac[G3n13ORF2]-infected Sf9 cells (Sf9-sup/infect and Sf9-cell/infect), and uninfected Tn5/05 cells (Tn5/05-sup and Tn5/05-cell). RNA was also extracted from nodavirus-like particles purified from an Ac[G3n13ORF2]-infected Tn5/97 cell lysate (nodavirus/Tn5). A 690-bp segment corresponding to a region of RNA2 was amplified with the primers RNA2-D4 and RNA2-U2. M, molecular weight markers; DW, distilled water.
Amplification of full-length RNA1 and RNA2 genomic RNAs from nodavirus like-particles and uninfected Tn5 cells.
The nodavirus like-particles purified from Ac[G3n13ORF2]-infected Tn5/97 cells had a density in CsCl similar to those of nodaviruses. Electron microscopy indicated that these particles contained nucleic acid(s) (Fig. 2). To determine if the particles contained RNA homologous to the nodavirus genome, RNA was extracted from purified particles and amplified by RT-PCR. Two fragments, one of 3,107 bp and the other of 1,383 bp, similar in length to FHV RNA1 and RNA2, were obtained. The nucleotide identity between the 3,107-bp fragment and FHV RNA1 was 89.3%; that between the 1,383-bp fragment and FHV RNA2 was 77.6%. No comparable amplification was observed when RNA extracted from purified HEV-LP was used (data not shown). These results indicated that the nodavirus like-particles purified from Ac[G3n13ORF2]-infected Tn5/97 cells contained a complete nodavirus genome.
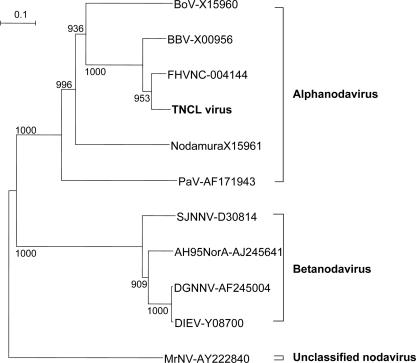
To further characterize this putative nodavirus, full-length RNA1 and RNA2 were amplified from both the lysate and the culture medium obtained from uninfected Tn5/97 cells. The nucleotide sequences of these RNAs were 100% identical to those of the RNAs extracted from the particles purified from Ac[G3n13ORF2]-infected Tn5/97 cells. These results again indicated that uninfected Tn5 cells were latently infected with a nodavirus. Phylogenetic analysis based on the amino acid sequence of the coat protein precursor, encoded in the 1,383-bp fragment corresponding to RNA2, indicated that this virus belongs to the genus Alphanodavirus in the family Nodaviridae (Fig. 4). We have designated this virus Tn5 cell line (TNCL) virus.
FIG. 4.
Phylogenetic analysis of TNCL virus on the basis of the coat protein precursor. The deduced amino acid sequence of the coat protein precursor was analyzed by the neighbor-joining method. Representative nodavirus strains and the corresponding accession numbers are indicated. The coat protein precursor from M. rosenbergii nodavirus (MrNV; accession no. AY222840) was used as an out-group. The bootstrap values correspond to 1,000 replications. BoV, Boolarra virus; BBV, black beetle virus; Nodamura, Nodamura virus; PaV, Pariacoto virus; SJNNV, striped jack nervous necrosis virus; AH95NorA, Atlantic halibut virus; DGNNV, dragon grouper nervous necrosis virus; DIEV, Dicentrarchus labrax encephalitis virus.
Induction of TNCL virus coat protein expression by recombinant baculoviruses.
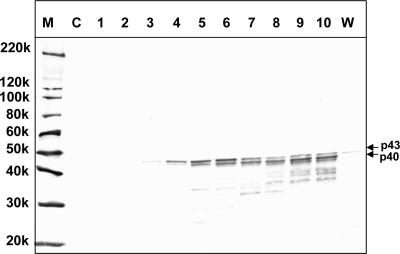
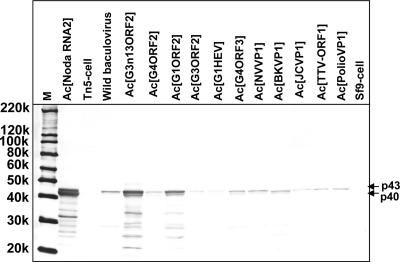
To characterize the induction of the expression of the p40 and p43 proteins, Ac[G3n13ORF2]-infected Tn5/97 cells were harvested daily for 10 days p.i. The proteins generated in infected cells were separated by SDS-PAGE and analyzed by a Western blot assay using the rabbit anti-FHV coat protein antibody (Fig. 5). Although the TNCL virus-specific protein was not detected before 2 days p.i., p43 appeared in the cells at 3 days p.i. At 5 days p.i., two bands corresponding to p40 and p43 were observed. These proteins were not apparent in uninfected Tn5/97 cells (Fig. 5, lane C), whereas a faint band corresponding to p43 was observed in wild-type baculovirus-infected cells (Fig. 5, lane W). The p43 was thought to be the coat protein precursor, and p40 was the mature coat protein. To confirm that p43 and p40 were derived from RNA2, a recombinant baculovirus, Ac[NodaRNA2], comprising the entire TNCL virus RNA2 genome was generated and used to infect Sf9 cells. In Ac[NodaRNA2]-infected Sf9 cells, both p43 and p40 were detected by a Western blot assay after 5 days p.i. (Fig. 6).
FIG. 5.
TNCL virus coat protein synthesis in Ac[G3n13ORF2]-infected Tn5 cells. Tn5/97 cells grown in a 25-cm2 flask were infected with Ac[G3n13ORF2] at a multiplicity of infection of 10 and incubated at 26.5°C. On days 1 through 10 p.i. (lanes 1 through 10, respectively), the cell lysates were separated by 5 to 20% SDS-PAGE and analyzed by a Western blot assay with rabbit anti-FHV antibody. M, molecular weight markers; C, uninfected Tn5/97 cells (control); W, wild-type baculovirus-infected Tn5/97 cells; 220k, molecular weight of 220,000.
FIG. 6.
Induction of TNCL virus coat protein synthesis by various recombinant baculoviruses. Tn5/97 cells were infected with the indicated 11 recombinant baculoviruses and wild-type (wild) baculovirus. The cells were harvested at 5 days p.i., and the cell lysates were analyzed by Western blot assays using rabbit anti-FHV coat protein antibody. M, molecular weight markers; Ac[NodaRNA2], cell lysate from Ac[NodaRNA2]-infected Sf9 cells; Tn5-cell, uninfected Tn5/97 cells; Sf9-cell, uninfected Sf9 cells; 220k, molecular weight of 220,000.
In addition to Ac[G3n13ORF2] and Ac[NodaRNA2], we examined other recombinant baculoviruses, including Ac[G1ORF2], Ac[G3ORF2], Ac[G4ORF2], Ac[G1HEV], Ac[G4ORF3], Ac[NoroVP1], Ac[BKVP1], Ac[JCVP1], Ac[TTVORF2], and Ac[polioVP1]. The recombinant baculovirus-infected Tn5/97 cells were harvested at 5 days p.i., and the various cell lysates were analyzed by Western blot assays (Fig. 6). The nodavirus coat protein was detected in the wild type-infected and all the recombinant baculovirus-infected cell lysates but not in uninfected Tn5 and Sf9 cell lysates (Fig. 6, lanes Tn5-cell and Sf9-cell). We also infected Sf9 cells with the above-named recombinant baculoviruses, but no coat protein was detected (data not shown). These results indicated that the expression of TNCL virus coat protein was induced only when Tn5 cells were infected with either the wild-type or a recombinant baculovirus.
TNCL virus is infectious in Drosophila S2 cells.
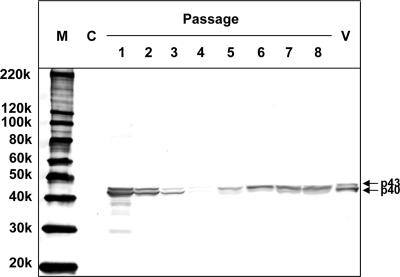
To determine whether TNCL virus particles purified from Tn5 cells were infectious, Drosophila S2 cells were infected with purified TNCL virus particles and harvested at each of eight passages. The presence of TNCL virus coat protein was assayed by Western blotting. As shown in Fig. 7, coat protein was detected at passages 1 through 8 but was not detected in uninfected S2 cells. The amount of coat protein decreased from passage 1 through passage 4 but then increased from passage 5 through passage 8. Coat protein was not detected in the supernatants (data not shown). If TNCL virus was not infectious, then the amount of the coat protein in the cells should have decreased steadily with repeated passaging because of the 1:5 dilution at each passage. The coat protein observed in passage 1 through 4 was presumably derived from the remaining TNCL virus used for the infection. However, the amount of the coat protein obviously increased from passage 5 through passage 8, clearly indicating that TNCL virus was capable of infecting S2 cells. These results indicated that at least some TNCL virus particles were infectious.
FIG. 7.
Infectivity of TNCL virus particles purified from Tn5 cells. Drosophila S2 cells were infected with purified TNCL virus particles and passaged every 4 days at a 1:5 dilution. The lysates from uninfected S2 cells (control [C]) and infected S2 cells (passages 1 through 8) were separated by 5 to 20% SDS-PAGE. The coat protein of TNCL virus was detected by a Western blot assay with rabbit anti-FHV coat protein antibody. M, molecular weight markers; V, purified TNCL virus particles; 220k, molecular weight of 220,000.
DISCUSSION
A latent infection of a widely used Trichoplusia ni-derived insect cell line, Tn5, by a previously undescribed nodavirus, TNCL virus, was accidentally disclosed following the infection of the cells with a recombinant baculovirus. A similar infection of a D. melanogaster cell line by a nodavirus had been reported previously (7), so such unapparent infections with nodaviruses may not be uncommon. Since these infections may present no obvious symptoms in the cell cultures, detection may occur only serendipitously. Such infections may go undetected for years.
Phylogenetic analysis of the virus described here (TNCL virus) indicates that it belongs to the genus Alphanodavirus. Wild-type baculovirus itself was capable of inducing the TNCL virus, so the foreign virus genes contained within the recombinant baculovirus genomes were not in themselves responsible for the induction. However, the inducing ability seems to be dependent on the inserted foreign genes as well as the expressed protein(s) (Fig. 6). As TNCL virus coat protein was detected only when wild-type or recombinant baculovirus was used to infect the Tn5 cells and other stresses like heat shock and cold shock did not induce the expression of the virus coat protein (data not shown), the translation and replication of the nodavirus genome seemed to be activated by the baculovirus infection. However, the mechanism by which baculovirus infection activates TNCL virus replication is unclear at moment. Mutant baculoviruses may provide a clue to the mechanism. At this time, we do not know the gene(s) or the mechanism for the stimulated expression of p40 and p43 and the particle formation.
Full-length genomic RNA1 and RNA2 of TNCL virus were detected in the two lots of uninfected Tn5 cells, one obtained in 1997 (Tn5/97) and subcultured since then and the second obtained in 2005 (Tn5/05), suggesting that these commercially available Tn5 cells had been infected latently with this virus. The nucleotide sequence of TNCL virus was closest to that of FHV among the alphanodaviruses (Fig. 4), and Western blot assays demonstrated that the coat protein was cross-reactive with anti-FHV antibody, suggesting that TNCL virus was antigenically similar to FHV. Nodamura virus, black beetle virus, FHV, and Boolarra virus have been shown to be cross-reactive with one another by double-diffusion immunoprecipitation tests; however, they represent different serotypes (25). Whether TNCL virus is serologically different from other alphanodaviruses is not clear. The nucleotide sequence homologies between the genomes of TNCL virus and FHV were 89.3% for RNA1 and 77.6% for RNA2. The amino acid homologies between proteins A and the coat protein precursors alpha were 83.36% and 89.68%, respectively. In addition, when the nucleotide sequences of TNCL virus and FHV were compared, we found that the RNA2 of TNCL virus lacked 17 bases present at the end of the RNA2 of FHV corresponding to the C terminus (nt 1248 to 1264). These results suggest that the TNCL virus is a new species of the genus Alphanodavirus.
The recombinant baculovirus expression system is routinely used for the production of foreign proteins. Two insect cell lines, derived from Spodoptera frugiperda (Sf9) and Trichopulsia ni (Tn5 [BTI-Tn-5B1-4]), are frequently used (9, 31). Tn5 cells have better secretion, which results in a higher productivity of recombinant proteins with the baculovirus expression system. Initial reports demonstrated that Tn5 cells produce at least 20-fold more secreted alkaline phosphatase than the Sf9 or Sf2l cell lines (6). The recombinant baculovirus expression system has also been employed for the efficient production of virus-like particles from different sources. Because virus-like particles closely mimic the properties of native virions, virus-like particles are attractive molecular candidates for recombinant vaccines. In fact, virus-like particles derived from HEV, hepatitis C virus, human BK polyomavirus, human polyomavirus JC, and human papillomavirus have been prepared using this expression system (4, 13, 16, 18).
Commercially available Tn5 cells were found to be latently infected with TNCL virus, and the infectious virus was induced by recombinant baculovirus infection. Unfortunately, this means that there is a serious risk of contamination by TNCL virus when virus-like particles are produced using this system. Although no alphanodavirus infections of humans have been reported, the type virus, Nodamura virus, is unique in being capable of infecting suckling mice and suckling hamsters (8, 23, 24), with resulting paralysis and death. When virus-like particles are considered for use as a vaccine, it is essential that the nodavirus, present as the result of a latent, unapparent infection, be removed. All cell lines derived from the insect Tn5 cell line should be examined for the presence of such nodaviruses. Because of the ability of nodaviruses to establish latent and unapparent infections, all insect cell lines should be screened routinely for the presence of virus.
Acknowledgments
We thank Tomoko Mizoguchi and Satoko Ogawa for secretarial work.
The study was supported in part by grants including Research on Emerging and Re-emerging Infectious Diseases, Research on Hepatitis, and Research on Food Safety from the Ministry of Health, Labor and Welfare, Japan.
Footnotes
Published ahead of print on 8 August 2007.
REFERENCES
- 1.Bailey, L., and H. A. Scott. 1973. The pathogenicity of Nodamura virus for insects. Nature 241:545. [DOI] [PubMed] [Google Scholar]
- 2.Ball, L. A., D. A. Hendry, J. E. Johnson, R. R. Rueckert, and P. D. Scotti. 2000. Nodavirus RNA replication: mechanism and harnessing to vaccinia virus recombinants. Academic Press, San Diego, CA. [DOI] [PubMed]
- 3.Ball, L. A., and K. L. Johnson. 1998. Nodaviruses of insects. Plenum, New York, NY.
- 4.Baumert, T. F., J. Vergalla, J. Satoi, M. Thomson, M. Lechmann, D. Herion, H. B. Greenberg, S. Ito, and T. J. Liang. 1999. Hepatitis C virus-like particles synthesized in insect cells as a potential vaccine candidate. Gastroenterology 117:1397-1407. [DOI] [PubMed] [Google Scholar]
- 5.Dasmahapatra, B., R. Dasgupta, A. Ghosh, and P. Kaesberg. 1985. Structure of the black beetle virus genome and its functional implications. J. Mol. Biol. 182:183-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, T. R., T. J. Wickham, K. A. McKenna, R. R. Granados, M. L. Shuler, and H. A. Wood. 1993. Comparative recombinant protein production of eight insect cell lines. In Vitro Cell Dev. Biol. Anim. 29A:388-390. [DOI] [PubMed] [Google Scholar]
- 7.Friesen, P., P. Scotti, J. Longworth, and R. Rueckert. 1980. Black beetle virus: propagation in Drosophila line 1 cells and an infection-resistant subline carrying endogenous black beetle virus-related particles. J. Virol. 35:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garzon, S., H. Strykowski, and G. Charpentier. 1990. Implication of mitochondria in the replication of Nodamura virus in larvae of the Lepidoptera, Galleria mellonella (L.) and in suckling mice. Arch. Virol. 113:165-176. [DOI] [PubMed] [Google Scholar]
- 9.Granados, R. R., C. G. D. Anja, and G. D. Kathleen. 1986. Replication of the Trichoplusia ni granulosis and nuclear polyhedrosis viruses in cell cultures. Virology 152:472-476. [DOI] [PubMed] [Google Scholar]
- 10.Guarino, L. A., A. Ghosh, B. Dasmahapatra, R. Dasgupta, and P. Kaesberg. 1984. Sequence of the black beetle virus subgenomic RNA and its location in the viral genome. Virology 139:199-203. [DOI] [PubMed] [Google Scholar]
- 11.Hansman, G. S., K. Natori, H. Shirato-Horikoshi, S. Ogawa, T. Oka, K. Katayama, T. Tanaka, T. Miyoshi, K. Sakae, S. Kobayashi, M. Shinohara, K. Uchida, N. Sakurai, K. Shinozaki, M. Okada, Y. Seto, K. Kamata, N. Nagata, K. Tanaka, T. Miyamura, and N. Takeda. 2006. Genetic and antigenic diversity among noroviruses. J. Gen. Virol. 87:909-919. [DOI] [PubMed] [Google Scholar]
- 12.Hosur, M. V., T. Schmidt, R. C. Tucker, J. E. Johnson, T. M. Gallagher, B. H. Selling, and R. R. Rueckert. 1987. Structure of an insect virus at 3.0 A resolution. Proteins 2:167-176. [DOI] [PubMed] [Google Scholar]
- 13.Ishii, Y., S. Ozaki, K. Tanaka, and T. Kanda. 2005. Human papillomavirus 16 minor capsid protein L2 helps capsomeres assemble independently of intercapsomeric disulfide bonding. Virus Genes 31:321-328. [DOI] [PubMed] [Google Scholar]
- 14.Kaesberg, P., R. Dasgupta, J. Y. Sgro, J. P. Wery, B. H. Selling, M. V. Hosur, and J. E. Johnson. 1990. Structural homology among four nodaviruses as deduced by sequencing and X-ray crystallography. J. Mol. Biol. 214:423-435. [DOI] [PubMed] [Google Scholar]
- 15.Li, T. C., N. Takeda, K. Kato, J. Nilsson, L. Xing, L. Haag, R. H. Cheng, and T. Miyamura. 2003. Characterization of self-assembled virus-like particles of human polyomavirus BK generated by recombinant baculoviruses. Virology 311:115-124. [DOI] [PubMed] [Google Scholar]
- 16.Li, T. C., N. Takeda, T. Miyamura, Y. Matsuura, J. C. Wang, H. Engvall, L. Hammar, L. Xing, and R. H. Cheng. 2005. Essential elements of the capsid protein for self-assembly into empty virus-like particles of hepatitis E virus. J. Virol. 79:12999-13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, T. C., Y. Yamakawa, K. Suzuki, M. Tatsumi, M. A. Razak, T. Uchida, N. Takeda, and T. Miyamura. 1997. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J. Virol. 71:7207-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuo, E., H. Tani, C. Lim, Y. Komoda, T. Okamoto, H. Miyamoto, K. Moriishi, S. Yagi, A. H. Patel, T. Miyamura, and Y. Matsuura. 2006. Characterization of HCV-like particles produced in a human hepatoma cell line by a recombinant baculovirus. Biochem. Biophys. Res. Commun. 340:200-208. [DOI] [PubMed] [Google Scholar]
- 19.Munday, B. L., J. Kwang, and N. Moody. 2002. Betanodavirus infections of teleost fish. J. Fish Dis. 25:127-142. [Google Scholar]
- 20.Newman, J. F., and F. Brown. 1976. Absence of poly (A) from the infective RNA of Nodamura virus. J. Gen. Virol. 30:137-140. [DOI] [PubMed] [Google Scholar]
- 21.Newman, J. F., and F. Brown. 1978. Further physicochemical characterization of Nodamura virus. Evidence that the divided genome occurs in a single component. J. Gen. Virol. 38:83-95. [DOI] [PubMed] [Google Scholar]
- 22.Reinganum, C., J. B. Bashiruddin, and G. F. Cross. 1985. Boolarra virus: a member of the Nodaviridae isolated from Oncopera intricoides (Lepidoptera: Hepialidae). Intervirology 24:10-17. [DOI] [PubMed] [Google Scholar]
- 23.Scherer, W. F., and H. S. Hurlbut. 1967. Nodamura virus from Japan: a new and unusual arbovirus resistant to diethyl ether and chloroform. Am. J. Epidemiol. 86:271-285. [DOI] [PubMed] [Google Scholar]
- 24.Scherer, W. F., J. E. Verna, and W. Richter. 1968. Nodamura virus, an ether- and chloroform-resistant arbovirus from Japan: physical and biological properties, with ecologic observations. Am. J. Trop. Med. Hyg. 17:120-128. [DOI] [PubMed] [Google Scholar]
- 25.Schneemann, A., L. A. Ball, C. Delsert, J. E. Johnson, and T. Nishizawa (ed.). 2004. Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 26.Schneemann, A., V. Reddy, and J. E. Johnson. 1998. The structure and function of nodavirus particles: a paradigm for understanding chemical biology. Adv. Virus Res. 50:381-446. [DOI] [PubMed] [Google Scholar]
- 27.Schneemann, A., W. Zhong, T. M. Gallagher, and R. R. Rueckert. 1992. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 66:6728-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scotti, P. D., S. Dearing, and D. W. Mossop. 1983. Flock House virus: a nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeidae). Arch. Virol. 75:181-189. [DOI] [PubMed] [Google Scholar]
- 29.Sri Widada, J., S. Durand, I. Cambournac, D. Qian, Z. Shi, E. Dejonghe, V. Richard, and J. R. Bonami. 2003. Genome-based detection methods of Macrobrachium rosenbergii nodavirus, a pathogen of the giant freshwater prawn, Macrobrachium rosenbergii dot-blot, in situ hybridization and RT-PCR. J. Fish Dis. 26:583-590. [DOI] [PubMed] [Google Scholar]
- 30.Wery, J. P., V. S. Reddy, M. V. Hosur, and J. E. Johnson. 1994. The refined three-dimensional structure of an insect virus at 2.8 A resolution. J. Mol. Biol. 235:565-586. [DOI] [PubMed] [Google Scholar]
- 31.Wickham, T. J., and G. R. Nemerow. 1993. Optimization of growth methods and recombinant protein production in BTI-Tn-5B1-4 insect cells using the baculovirus expression system. Biotechnol. Prog. 9:25-30. [DOI] [PubMed] [Google Scholar]
- 32.Zeddam, J. L., J. L. Rodriguez, M. Ravallec, and A. Lagnaoui. 1999. A noda-like virus isolated from the sweetpotato pest Spodoptera eridania (Cramer) (Lep.; Noctuidae). J. Invertebr. Pathol. 74:267-274. [DOI] [PubMed] [Google Scholar]