Abstract
Persistence in canine distemper virus (CDV) infection is correlated with very limited cell-cell fusion and lack of cytolysis induced by the neurovirulent A75/17-CDV compared to that of the cytolytic Onderstepoort vaccine strain. We have previously shown that this difference was at least in part due to the amino acid sequence of the fusion (F) protein (P. Plattet, J. P. Rivals, B. Zuber, J. M. Brunner, A. Zurbriggen, and R. Wittek, Virology 337:312-326, 2005). Here, we investigated the molecular mechanisms of the neurovirulent CDV F protein underlying limited membrane fusion activity. By exchanging the signal peptide between both F CDV strains or replacing it with an exogenous signal peptide, we demonstrated that this domain controlled intracellular and consequently cell surface protein expression, thus indirectly modulating fusogenicity. In addition, by serially passaging a poorly fusogenic virus and selecting a syncytium-forming variant, we identified the mutation L372W as being responsible for this change of phenotype. Intriguingly, residue L372 potentially is located in the helical bundle domain of the F1 subunit. We showed that this mutation drastically increased fusion activity of F proteins of both CDV strains in a signal peptide-independent manner. Due to its unique structure even among morbilliviruses, our findings with respect to the signal peptide are likely to be specifically relevant to CDV, whereas the results related to the helical bundle add new insights to our growing understanding of this class of F proteins. We conclude that different mechanisms involving multiple domains of the neurovirulent A75/17-CDV F protein act in concert to limit fusion activity, preventing lysis of infected cells, which ultimately may favor viral persistence.
Canine distemper virus (CDV) is a negative-stranded RNA virus, a morbillivirus of the Paramyxoviridae family, closely related to measles virus. The CDV genome is 15,690 nucleotides long and contains six genes. Two nonstructural and six structural proteins are encoded by six separately capped and polyadenylated mRNAs. CDV infection induces high morbidity and mortality in dogs and numerous other domestic and wild species (1, 16, 27), which frequently result from damage to the nervous system (10, 14, 15, 34, 40).
Brain infection with the wild-type strain A75/17-CDV is persistent and produces a chronic demyelinating disease in the central nervous system of dogs and other carnivores. This type of demyelination is considered a model for multiple sclerosis in humans (36). In dog brain cell cultures, this virus produces a noncytolytic, persistent infection with selective virus spread along cell processes (43), mimicking the in vivo situation in the central nervous system (37). Wild-type CDV infection in brain cells is characterized by little virus budding and lack of cell destruction, which may allow the virus to escape immune surveillance (3, 21, 22), in this way favoring persistence. Lack of cytolysis in persistently infected dog brain cell cultures is associated with very limited cell-cell fusion, in contrast to the highly attenuated Onderstepoort CDV vaccine strain (OP), which produces cytolytic infection resulting from extensive cell-cell fusion (44). Thus, the restriction of cell-cell fusion induced by wild-type CDV is thought to be correlated with viral persistence.
The CDV fusion (F) protein is a classical type I glycoprotein consisting of 662 amino acids that is essential for virus entry and spread (17). Previous studies have suggested that translation of the F protein starts at either the first start codon, AUG1, or at the second codon, AUG61. Translation initiation from these start codons yields the primary translation precursor products, designated pre-F0 AUG1 and pre-F0 AUG61 (9, 38). Both precursors are translocated into the lumen of the endoplasmic reticulum (ER) and subsequently are cleaved between amino acids 135 and 136 by a cellular signal peptidase (SPase) (38), thus producing an unusually long signal peptide (SP) of 75 or 135 amino acids, depending on the translation initiation codon used, and the second immature F0 precursor. The ER is also the location where oligomerization of F0 into a homotrimer is thought to occur (26). The F0 homotrimer then is glycosylated and cleaved into the three disulfide-linked F1 and F2 subunits by furin, a cellular serine protease acting in the Golgi compartment (4). Targeting of the folded and processed F protein to the plasma membrane represents the actual potentially fusion-active structure of the protein. In a receptor- and hemagglutinin (H)-dependent manner, the F protein then undergoes a cascade of conformational alterations, which finally leads to membrane merging (18).
Combined with the previously described crystal structures of two paramyxovirus F proteins in the postfusogenic hairpin form (7, 41), the recently determined X-ray structure of the simian virus 5 (SV5) F protein in its native prefusogenic conformation has provided a unique opportunity to more accurately study the regulation of the paramyxovirus membrane fusion activity (42). Besides two heptad repeat regions (HRA and HRB) in the F1 ectodomain, which are well known to play critical roles in paramyxovirus membrane fusion (2, 11, 30, 39), other regions are only poorly characterized for their specific function in the fusion process itself. Interestingly, recently it has been demonstrated that a conserved region, located between both heptad repeats (HRA and HRB) of SV5 and Hendra virus F proteins, was important for regulating F protein expression and activity (13). In addition, single mutations in the globular head and outside both HR domains of SV5 and Newcastle disease virus (NDV) have been shown to allow the F protein to activate the fusion process in the absence of the receptor-binding protein (HN) (20, 29).
Our previous work suggested that some viral determinants limiting cell-cell fusion in persistent A75/17-CDV infection were residing in the F protein (23). In the present study, we have further investigated which domains of the A75/17 F protein (referred to as F-A75/17) were controlling fusion efficiency. To this aim, we first generated chimeric F proteins by swapping SP and F0 trimer domains with one of the highly fusogenic vaccine F proteins. While in F-A75/17 both regions were implicated in limiting fusogenicity, we further demonstrated that the SP played a critical role in modulating intracellular protein expression. Moreover, by serially passaging a poorly syncytium-forming virus in Vero cells, a fusogenic variant was selected. We identified the mutation L372W as being responsible for this increase in cell-cell fusion activity. Residue L372 potentially resides in the helical bundle (HB) domain, located just carboxyl terminal to the HRA domain of the F protein ectodomain. On introducing the mutation into both CDV strains, F proteins drastically increased their fusion-inducing capacities in an SP-independent manner. This work demonstrated, for the first time, how two different domains of the CDV F protein acted in concert to limit fusogenicity, providing new insights into our understanding of virulent CDV-induced restricted fusion activity, which is believed to be related to viral persistence.
MATERIALS AND METHODS
Cell cultures and viruses.
Vero African green monkey kidney cells were grown in Dulbecco's modified Eagle's medium (Gibco, Invitrogen) with 10% fetal calf serum (FCS) at 37°C in the presence of 5% CO2. BHK-derived Bsr-T7/5 cells expressing constitutively the T7 polymerase (a gift from K. Conzelmann, Federal Research Center for Virus Diseases of Animals, Germany) were grown in the same manner as the Vero cells, except that the medium was supplemented with 2% FCS and 0.5 mg/ml of G-418. The wild-type A75/17-CDV strain (a gift from M. Appel, Cornell University, Ithaca, NY) was adapted to Vero cells by 17 serial passages and was designed A75/17-V. The modified vaccinia virus Ankara (MVA)-T7 recombinant vaccinia virus was used for a quantitative cell-cell fusion assay (see below) and was obtained from B. Moss, NIH, Bethesda, MD.
Construction of expression plasmids.
The plasmids pF-A75/17, pF-OP, pH-A75/17, and pH-OP cloned in the mammalian expression vector pCI (Promega) were described previously (8). By using PCR and recombinant PCR techniques (Pfu Turbo DNA polymerase; Stratagene), two hybrid constructs were generated from genes of F-A75/17 and the F protein of the OP strain of CDV (F-OP) in which the DNA sequences encoding the SP sequence (nucleotides 1 to 490) were replaced by the corresponding sequences from the F-OP and F-A75/17 genes, respectively. This generated pF-OA (F0 derived from F-A75/17 fused to the OP-CDV F SP) and pF-AO (F0 derived from F-OP fused to F-A75/17 SP). The SP of the human secretory component (hsc) protein also was fused to the F0 part of both F-OP and F-A75/17. This led to the generation of phscFA (F0 derived from F-A75/17 fused to the hsc SP) and phscFO (F0 derived from F-OP fused to the hsc SP). The four F hybrid genes were cloned into BamHI/XbaI-cleaved pCI plasmid. All plasmid sequences were confirmed by automated nucleotide sequence analysis.
Transfections and quantitative fusion assay.
Vero cells, in 6-well plates at 90% confluence, were cotransfected with 2 μg of the CDV F protein constructs and 1 μg of pCI-HOP using 9 μl of Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Pictures were taken 24 h posttransfection with a Nikon Coolpix 990 digital camera on a Nikon Eclipse TS100 microscope.
The quantitative fusion assay was performed essentially as described previously (19). For measles virus, the fusion inhibitory peptide (FIP) has been used to inhibit F/H-protein-induced syncytium formation during the first period of incubation (25). As the FIP did not have the desired effect in transfection experiments using the CDV F gene, Vero cells cotransfected with the F protein and H protein expression plasmids and 0.1 μg of pTM-CAT were incubated in the presence of a CDV antiserum (VMRD, Inc.). In parallel, separate 6-well plates of Vero cells at 30% confluence were infected with the recombinant vaccinia virus MVA-T7 expressing the T7 RNA polymerase (35) at a multiplicity of infection of 1. Sixteen hours posttransfection or postinfection, transfected Vero cells were subcultured in duplicate with the MVA-T7-infected Vero cells. After incubation at 37°C for 6 h, cells were lysed and chloramphenicol acetyltransferase (CAT) production was determined using an enzyme-linked immunosorbent assay kit (Roche Biochemicals).
Western blotting.
Vero cells seeded into 6-well plates were transfected with the different F protein constructs using Lipofectamine 2000 (Invitrogen) and were incubated at 37°C for 24 h. For the analysis of total cellular proteins, cells were washed twice with ice-cold phosphate-buffered saline before the addition of 150 μl of lysis buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 1% deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]) with complete protease inhibitor (Roche Biochemicals). After incubation for 20 min at 4°C, the lysates were cleared by centrifugation at 5,000 × g for 15 min at 4°C, and the supernatant was mixed with an equal amount of 2× Laemmli sample buffer (Bio-Rad) containing 100 mM dithiothreitol, subsequently boiled at 95°C for 5 min, and fractionated on SDS-8 or 10% polyacrylamide gels under denaturing conditions. Separated proteins were transferred to nitrocellulose membranes by electroblotting, and the membranes then were soaked in TBS-Tween (25 mM Tris-HCl, pH 7.5, 137 mM NaCl, 3 mM KCl, 0.1% Tween 20) containing 5% nonfat dry milk. The membranes then were incubated with the polyclonal rabbit anti-CDV F antiserum (8). Following incubation with a peroxidase-conjugated secondary antibody, the membranes were subjected to an enhanced chemiluminescence kit (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Surface biotinylation.
Vero cells seeded into 6-well plates were transfected with 4 μg of plasmid DNA encoding CDV F protein variants using 12 μl of Lipofectamine 2000 (Invitrogen). Twenty-four hours posttransfection and after being washed in cold PBS, cells were incubated in PBS with 0.5 mg/ml sulfosuccinimidyl-2-(biotinamido)ethyl-1,3 dithiopropionate (Pierce) for 20 min at 4°C, washed three times with 0.5 M glycine in PBS, and further incubated in 1 ml of 0.5 M glycine-PBS for 20 min at 4°C to quench the excess of biotin. Samples then were scraped into 150 μl of lysis buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 1% deoxycholate, 1% Triton X-100, 0.1% SDS) containing protease inhibitors (complete mix; Roche), and the lysates were cleared by centrifugation for 20 min at 20,000 × g and 4°C. The biotinylated proteins were adsorbed to Sepharose-coupled streptavidin (Amersham Pharmacia Biotech) overnight at 4°C and washed three times in lysis buffer, and then 30 μl of 2× Laemmli sample buffer (Bio-Rad) containing 100 mM dithiothreitol was added. The samples then underwent Western blot analysis as described above.
RESULTS
F-A75/17 is efficiently expressed, processed, and cell surface targeted.
We previously described that the wild-type F-A75/17 markedly reduced fusogenicity compared to that of F-OP both in transient transfection experiments and in the context of a viral infection (23). It was therefore of interest to determine the molecular mechanisms underlying this observation.
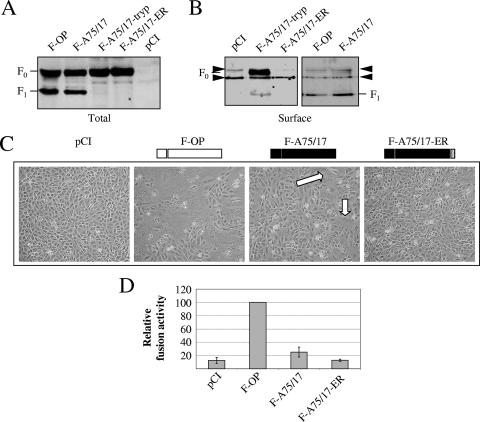
To assess whether the F-A75/17 phenotype was related to a lack of intracellular transport competence and/or to a loss of F protein functionality, we first carried out Western blotting on either total intracellular or plasma membrane-bound proteins of F-protein-transfected Vero cells. Our results showed that F-A75/17 was not associated with a defect in protein synthesis or F0 processing into F1 and F2, since a pattern was observed that was similar to that of the fusion-competent F-OP protein (Fig. 1A). In a comparison of surface expression levels, again both F1 subunits efficiently reached the cell surface (Fig. 1B). In fact, 24 h posttransfection, there was even a slight increase in the A75/17 F1 subunit reaching the cell surface compared to the level for F-OP. As a control for cell surface targeting, a variant of the wild-type F protein containing an ER retention motif fused to the C-terminal part of the cytoplasmic tail (26), designated F-A75/17-ER, was analyzed. This showed that despite high intracellular expression, no proteins could be detected at the cell surface, presumably due to its ER retention localization (Fig. 1A and B). Furthermore, in F-A75/17-ER-transfected cells, F0 was not cleaved into the F1/F2 subunits, which confirmed that F-A75/17-ER did not enter the Golgi compartment, where the furin protein resides. As expected, F-A75/17-ER did not induce membrane fusion activity (Fig. 1C and D). F-A75/17-tryp is a mutant that is no longer dependent on furin but rather on trypsin for processing into F1 and F2 subunits. Therefore, in the absence of trypsin, only the immature F0 form of the protein will be expressed and targeted to the cell surface. This mutant not only confirmed the F0 bands in all constructs but also showed that F-A75/17-ER F0 precursor is indeed not transported to the plasma membrane (Fig. 1A and B).
FIG. 1.
F protein of the neurovirulent A75/17-CDV is efficiently expressed, processed, and cell surface targeted, but it induces limited cell-cell fusion. (A) For F protein expression and processing analyses, Western blot analyses were performed from lysates of Vero cells transfected with the F protein expression plasmids as indicated. Twenty-four hours posttransfection, the lysates were separated by reducing SDS-polyacrylamide gel electrophoresis and blotted onto nitrocellulose membranes. The F proteins were revealed with an anti-F protein ectodomain serum. (B) Surface biotinylation of cells expressing different F proteins to determine F protein plasma membrane steady-state levels. Biotinylated proteins of F-protein-transfected (and derivative-transfected) cells were precipitated, and Western blot analyses were performed as indicated above. Black arrowheads highlight two unspecific bands precipitated in the assay and recognized by our anti-F protein serum. (C) Syncytium formation after cotransfection of the cells with plasmid DNA encoding CDV H-OP and F-A75/17, F-OP, or F-A75/17-ER. Mock-transfected cells (pCI) received CDV H-OP-encoding plasmid and empty vector; representative fields of view were photographed 24 h posttransfection. (D) For quantitative fusion assays, Vero cells either were infected with MVA-T7 (multiplicity of infection of 1) or were transfected with the different F proteins, a plasmid encoding H-OP, and a plasmid containing the CAT reporter gene under the control of the T7 promoter. Twelve hours after transfection, both cell populations were mixed and seeded into fresh plates. After 6 h at 37°C, fusion was quantified by measuring the amount of CAT protein produced. For each experiment, the value for the F-OP/H-OP combination was set to 100%. Means and standard deviations from three independent experiments performed in duplicate are shown.
Using a reporter gene content mix assay to more accurately quantify fusogenicity, we next confirmed previous data showing the limited cell-cell fusion induced by F-A75/17 (Fig. 1D). The content mix assay confirmed the microscopic images of the various F-protein- and H-induced syncytium formations (Fig. 1C and D). In this report, we studied fusion efficiency by coexpressing the H-OP with the various F mutants in Vero cells. Indeed, we recently obtained evidence that this environment led to the most obvious alterations in cell-cell fusion efficiency (23). This cell system therefore was used, since it allowed us to easily analyze subtle fusogenic differences.
These experiments showed that F-A75/17 was correctly folded, processed, and transported to the plasma membrane. While differences in cellular transport kinetics remain possible, we showed that the steady-state level 24 h posttransfection of cell-surface-expressed F-A75/17 was constantly slightly increased compared to the level of F-OP. Nevertheless, despite higher cell surface expression 1 day posttransfection, F-A75/17 was reproducibly characterized by a reduced membrane fusion activity compared to that of F-OP.
Limited cell-cell fusion of F-A75/17 depends on both the SP and the F1/F2 homotrimer.
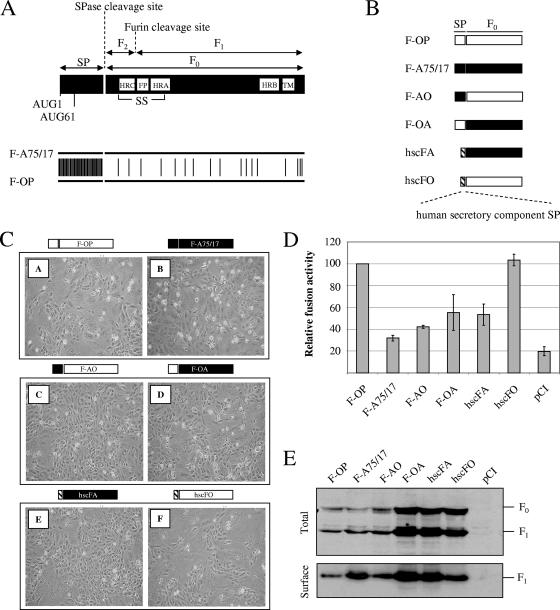
Comparison between F-A75/17 and F-OP amino acid sequences showed considerable differences, with 41 and 17 substitutions located in the SP and the F1/F2 trimer domains, respectively (Fig. 2A). To investigate which domains regulate F-A75/17-limited fusogenicity, we generated hybrid F proteins with swapped or heterologous SP. We thus fused the F-OP SP to F0 of A75/17 (F-OA) and generated the opposite construct (F-AO). Figure 2B summarizes the different F protein variants and their respective names. A reporter gene content mix assay next was used to quantify their fusion-inducing capacities. Interestingly, targeting F-A75/17 to the plasma membrane-bound pathway through the OP-CDV SP showed a slight increase in cell-cell fusion compared to that of F-A75/17 (Fig. 2C, image D, and D). In sharp contrast, F-AO was characterized by a significant reduction in cell-cell fusion compared to parental F-OP-mediated fusion efficiency (40% relative to that of F-OP) (Fig. 2C, image C, and D). This demonstrates an inhibiting role of the F-A75/17 SP in syncytium formation. Furthermore, when both F proteins were fused to the hsc SP (hscFO and hscFA), a clear difference in cell-cell fusion was observed between both fusion-active F proteins (Fig. 2C, images E and F, and D). Thus, independently of the SP, the F1/F2 domain of the neurovirulent CDV F protein was reduced by about 50% relative to the level of F-OP expression (Fig. 2D).
FIG. 2.
Multiple domains of F-A75/17 are implicated in restricting membrane fusion activity. (A) The upper portion shows a linear drawing of the CDV F protein. The protein is translated as a long precursor, which is targeted to the ER via the unusually long SP. The SP then is cleaved prior to cleavage activation of the F0 precursor into the disulfide-linked (SS) F1 and F2 subunits. The main HR, the fusion peptide (FP), and the transmembrane domain (TM) are represented by white boxes. Amino acid positions of the two potential initiation codons also are highlighted (AUG1 and AUG61). The lower portion shows a schematic representation of the amino acid differences (horizontal bars) between F-A75/17 and F-OP. (B) Schematic representation of the various F constructs and their attributed names. The SP domain and the immature precursor (F0) are shown. F-A75/17 domains are drawn in black. F-OP domains are drawn in white. The heterologous SP of the hsc is indicated in hatched boxes. (C) Syncytium formation after cotransfection of the cells with plasmid DNA encoding CDV H-OP and F proteins of both CDV strains and their respective chimeric derivatives; representative fields of view were photographed 24 h posttransfection. (D) Quantitative fusion assay of the different F proteins. The assay was done as described in the legend to Fig. 1D. For each experiment, the value for the F-OP/H-OP combination was set to 100%. Means and standard deviations from three independent experiments performed in duplicate are shown. (E) Western blot analyses of F-protein-antigenic materials of total lysates (Total) and surface-exposed (Surface) protein expression were performed as described in the legends to Fig. 1A and B.
To verify whether all F protein variants were correctly expressed, processed, and surface targeted, Western blotting on total intracellular and surface-exposed proteins was performed. As shown in Fig. 2E, all mutant proteins showed efficient processing and transport competence to the plasma membrane. Interestingly, it appeared that in the absence of the CDV SP, a higher expression level of both CDV F proteins was observed. Consequently, their steady-state levels at the cell surface were increased. The same phenotype was seen when F-A75/17 was targeted to the plasma membrane via the SP of F-OP (Fig. 2E). For these three F protein mutants (hscFA, hscFO, and F-OA), a higher expression level of F1/F2 at the cell surface always was reflected by an increase in membrane fusion activity, as determined by the reporter gene content mix assay (Fig. 2D). Note that the difference in fusogenicity between F-OP and hscFO was less drastic than that for F-A75/17. Indeed, we believe that very efficient fusion was achieved with F-OP in this cell system, which obviously saturated the quantification of more fusogenic F protein variants.
These results demonstrated that the SP of both CDV F proteins controlled limited intracellular and indirectly surface-exposed protein expression, which subsequently led to a decrease in membrane fusion activity. Interestingly, whereas the F-OP SP lost the capacity to decrease protein expression when fused to F-A75/17, the F-A75/17 SP retained this function independently of the F0 protein with which it is fused. However, although it appeared that the SP could indirectly modulate fusion activity, the cell-cell fusion efficiency induced by the different F protein SP variants never reached the fusogenicity of F-OP. It appeared, therefore, that the plasma membrane-bound F-A75/17 F1/F2 trimer contained a critical inherent determinant(s) in limiting syncytium formation.
Residues upstream of AUG61 of the F-A75/17 SP are sufficient to restrict protein expression.
We next wished to map the specific SP sequence responsible for limiting protein expression. Previous works showed that the F proteins of both CDV strains possess two potential translation initiation codons, which therefore would lead to the generation of precursors differing in their respective SP size (8). Interestingly, it has been reported that in CDV the length of the SP modulates cell-cell fusion by some unknown mechanism (38). In agreement with these findings, we reasoned that the SP sequence modulating protein expression might be located in a common region between the F proteins of both CDV strains and that differential translation initiation would eliminate the down-regulating domain from the F-OP protein.
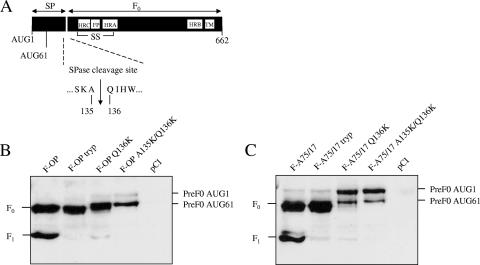
In order to assess from which start codon translation is initiated in both F proteins, residue 136 or residues 135 and 136 were mutated into lysines (Fig. 3A). Indeed, previously it had been suggested that the SPase cleaves in between these specific amino acids (38). In F-OP- or F-A75/17-transfected Vero cells, precursors with the SP still fused are only weakly detectable by Western blotting, presumably because they are constantly cleaved by the SPase. However, when the various SPase F protein mutants were transfected, pre-F0 precursors were detected efficiently by the F protein polyclonal antiserum. Note that in F protein mutants with only one residue mutated (Q136K), a faint band corresponding to F1 was still present, showing that the SPase cleavage site was impaired but not completely inactivated. More importantly, whereas in the case of F-OP there was a clear initiation at AUG61, F-A75/17 was preferentially translated from AUG1 (Fig. 3B and C). Thus, we showed for the first time that, depending on the viral strain, the length of the SP was indeed different for the two F proteins.
FIG. 3.
Differences in initiation codon usage between F proteins of both CDV strains. (A) Linear drawing of the CDV F protein. The various highlighted boxes are as described in the legend to Fig. 1A. The SPase cleavage site, as suggested by means of computer analysis, also is represented. (B and C) Western blot analyses of total intracellular F protein expression were performed as described in the legend to Fig. 1A. SS, disulfide-linked F1 and F2 subunits; TM, transmembrane.
Since SPase mutants are not processed and transported to the cell surface (data not shown), we could not directly assess the SP length dependence in syncytium formation. To overcome this problem, F proteins mutated individually in their respective potential translation initiation AUGs were generated (Fig. 4A). As expected, fusion activity induced by F-OP with AUG1 mutated (F-OP AUG61) was as efficient as that of the parental F-OP (120% relative to that of F-OP) (Fig. 4B and C). This was not surprising, since this mutant represented the main precursor expressed by F-OP (Fig. 3B). In contrast, F-OP AUG1 showed only 50% of the fusion activity of F-OP, confirming that in the context of the OP-CDV F protein, a longer SP inhibits fusion (Fig. 4B and C). However, regardless of whether AUG1 or AUG61 was mutated in the genetic background of F-A75/17, the rate of fusion remained mostly unaltered (Fig. 4B and C). Indeed, it appeared clearly that a shorter F-A75/17 SP did not significantly increase syncytium formation.
FIG. 4.
Shorter F-A75/17 SP does not increase cell-cell fusion. (A) Linear drawing of the CDV F protein. The various highlighted boxes are as described in the legend to Fig. 1A. The two potential initiation codons also are represented. SS, disulfide-linked F1 and F2 subunits; TM, transmembrane. (B) Syncytium formation after cotransfection of the cells with plasmid DNA encoding CDV H-OP and F protein of both CDV strains and their respective derivatives; representative fields of view were photographed 24 h posttransfection. (C) Quantitative fusion assay of the different F proteins. The assay was done as described in the legend to Fig. 1D. For each experiment, the value for the F-OP/H-OP combination was set to 100%. Means and standard deviations from three independent experiments performed in duplicate are shown. (D) Western blot analyses of F-protein-antigenic materials of total lysates (Total) and surface-exposed (Surface) protein expression were performed as described in the legends to Fig. 1A and B. (E) High levels of cell surface expression of F-A75/17 compared to those of F-OP are not responsible for the reduction in membrane fusion activity. Values shown between both panels represent decreasing amounts of F-A75/17-transfected DNA. Western blot analyses of F-protein-antigenic materials of total lysates and surface-exposed protein expression were performed as described in the legends to Fig. 1A, B, and D, respectively. For F-A75/17-mediated cell-cell fusion quantification, the numbers of syncytia induced in 10 random areas were counted. Means and standard deviations are shown.
To determine cell surface expression of F protein mutants, Western blot analysis of intracellular and surface-exposed proteins was performed. Interestingly, whereas F-OP AUG61 was more synthesized than the parental protein, F-OP AUG1 was clearly less expressed than the parental F-OP protein (Fig. 4D, left panel). In turn, the steady-state levels at the cell surface and, consequently, membrane fusion activity were increased for AUG61 and were reduced for AUG1 (Fig. 4D, left panel). In contrast, shortening F-A75/17 SP did not alter protein expression (Fig. 4D, right panel), which reflects the results obtained using the content mix assay showing no major differences between all F-A75/17 mutants in inducing membrane fusion (Fig. 4C).
We repeatedly observed a slightly higher level of expression of F-A75/17 at the cell surface than that of F-OP. To investigate whether higher F-A75/17 expression at the plasma membrane was not responsible for the observed reduction in membrane fusion activity, decreasing amounts of F-A75/17 plasmid DNA were transfected in Vero cells. Smaller amounts of DNA in transfection mixtures yielded lower levels of intracellular, surface-exposed, and, subsequently, membrane fusion activity (Fig. 4E). This result confirmed that a higher level of expression of F-A75/17 at the plasma membrane was not responsible for the limited cell-cell fusion phenotype.
These results showed that the determining SP sequence responsible for modulating protein expression was surprisingly different in the F proteins of the two CDV strains. While in the F-OP SP the first 60 amino acids were critical for limiting protein expression, in the genetic background of F-A75/17 even the shorter form of the SP (residues 61 to 135) sustained this function. Interestingly, although protein translation in F-OP was preferentially initiated at AUG61, the longer precursor (translated from AUG1), which was only poorly expressed, appeared to be responsible for the generally observed reduction in intracellular protein expression.
Residue L372, potentially located in the HB of the F globular head, plays a critical role in controlling F1/F2-mediated limited fusogenicity.
Since there were 17 amino acid differences in the F1/F2 complex between the F proteins of the two CDV strains, we decided to use an evolutive approach in order to investigate regions controlling membrane fusion activity. For this purpose, a recombinant nonfusogenic virus was passaged several times in Vero cells, and a fusogenic variant was selected (data not shown). Sequence analysis of the F glycoprotein gene before and after passaging showed only one nucleotide modification, which also leads to a change in the predicted amino acid sequence. The analysis pointed out the mutation L372W. It is reasonable to assume that analogous residues of the paramyxovirus F proteins form similar structures. Thus, with respect to the recently determined SV5 and human parainfluenza virus type 3 (hPIV3) F protein crystal structures, residue L372 potentially lies in a short α-helix of the HB. The HB is a subdomain of the globular head (native, SV5) or neck (hairpin, hPIV3) F protein structures (7, 41, 42). Interestingly, this domain undergoes only limited secondary-structure rearrangements between both F protein conformations (Fig. 5A). When the CDV sequences of the HB were aligned to those of other F proteins of paramyxovirus members, this specific residue appeared to be almost completely conserved in all genera. Overall, the HB region showed significant conservation through the different paramyxovirus sequences analyzed (Fig. 5A). Interestingly, 2 of the 17 mutations located in the F1/F2 complex between F-A75/17 and F-OP also resided in the HB (S337I and N366S), of which only N366S is putatively located in or close to the α-helix where L372W resides (Fig. 5A).
FIG. 5.
Mutation of residue L372 in the F-A75/17 trimer enhances fusogenicity despite the presence of the SP. (A) Linear drawing of the CDV F protein. The various highlighted boxes are as described in the legend to Fig. 1A. The three domains (DI to DIII) of the globular F protein head are shown. The HB F protein subdomain (HB) also is represented. For the sequence alignment of various paramyxoviruses, residues in black are completely conserved amino acids; residues in gray are conserved across most members; residues in light blue are similar amino acids; residues in red are positions of the mutation obtained after virus passages; residues in orange are 2 of the 17 mutations between F-A75/17 and F-OP trimers also located in the HB. SS, disulfide-linked F1 and F2 subunits; TM, transmembrane. (B and C) Syncytium formation after cotransfection of the cells with plasmid DNA encoding CDV H-OP and F protein of both strains and their respective derivatives; representative fields of view were photographed 24 h posttransfection. (D) Quantitative fusion assay of the different F proteins. The assay was done as described in the legend to Fig. 1D. For each experiment, the value for the F-OP/H-OP combination was set to 100%. Means and standard deviations from three independent experiments performed in duplicate are shown. (E) Western blot analyses of F-protein-antigenic materials of total lysates (Total) and surface-exposed (Surface) protein expression were performed as described in the legends to Fig. 1A and B.
To investigate a potential role of the HB in fusogenicity, we introduced the L372W change in the F proteins of both CDV strains and studied their phenotypes in transfection experiments. Remarkably, in the presence of H-OP, F-A75/17 L372W displayed a hyperactive fusion activity (Fig. 5B), which was confirmed using the content mix assay (Fig. 5D). Although less drastic, the identical change in F-OP also contributed to an increase in cell-cell fusion (Fig. 5C and D). As described above, this phenomenon probably is due to an already-saturating effect in membrane fusion activity induced by F-OP in Vero cells. This was confirmed by transfecting F-OP and F-OP L372W in Bsr-T7 cells, a cell line in which F-OP induced only limited cell-cell fusion. In these cells, F-OP L372W showed a drastic increase in membrane fusion activity compared to that of F-OP (data not shown).
Western blot analysis of these mutant proteins showed no major differences in expression and processing compared to those of both F-OP and F-A75/17 (Fig. 5E). These results suggest that this specific residue in the HB domain is involved in one of the different steps leading the F protein from its native to its highly stable six-helix bundle postfusion conformation. To further investigate a role of the small α-helix of the HB in F protein functionality, we introduced the N366S mutation in F-A75/17. However, this mutation was not able to render F-A75/17 hyperfusogenic but nevertheless slightly increased syncytium formation (Fig. 5B and D).
Taken together, these results showed that a single mutation in the globular head (native) and/or neck (hairpin) F protein structures, potentially located in a small α-helix of the HB, was able to render F-A75/17 highly fusogenic despite the presence of the SP.
Residue L372 in the core of the F protein globular head structure regulates membrane fusion activity independently of the SP.
The above results demonstrated an important role of residue L372 in membrane fusion control. However, while the mutation L372W might influence the fusion process itself, it remained possible that this specific change solely overcame the effect of the F-A75/17 SP described above. To discriminate between both functions, we introduced the mutation into the genetic background of F proteins of both CDV strains bearing the heterologous hsc SP. Indeed, these mutants allowed us to analyze the effect of residue L372 specifically in F1/F2-mediated membrane fusion activity.
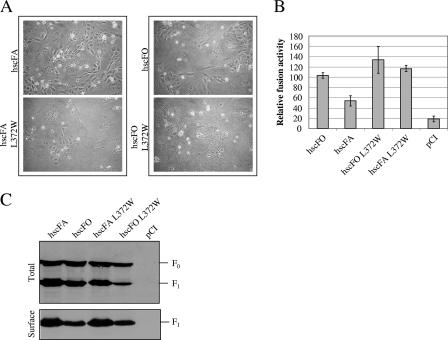
While Western blot analysis showed that both F protein variants (hscFA L372W and hscFO L372W) were not impaired in expression, processing, and cell surface targeting (Fig. 6C), both the microscopic and the content mix assays demonstrated a clear fusion-enhancing effect of F proteins bearing the L372W mutation (Fig. 6A and B). Thus, it appeared that a single-amino-acid substitution in the HB of the F protein globular head rendered F-A75/17 hyperfusogenic in the presence or absence of the unusually long SP and confirmed a role of residue L372 in modulating the fusion process itself.
FIG. 6.
L372W mutation increases F1/F2-induced fusogenicity independently of the CDV SP. (A) Syncytium formation after cotransfection of the cells with plasmid DNA encoding CDV H-OP and F protein mutants; representative fields of view were photographed 24 h posttransfection. (B) Quantitative fusion assay of the different F proteins. The assay was done as described in the legend to Fig. 1D. For each experiment, the value for the F-OP/H-OP combination was set to 100%. Means and standard deviations of three independent experiments in duplicate are shown. (C) Western blot analyses of F-protein-antigenic materials of total lysates (Total) and surface-exposed (Surface) protein expression were performed as described in the legend to Fig. 1A and B.
Hyperfusogenic F protein mutants are functional only in the presence of the receptor-binding H protein.
It has been shown that some mutations in the F protein ectodomain of NDV and SV5 may be responsible for allowing the protein to be activated in the absence of the receptor-binding proteins (20, 29). It was therefore of interest to investigate whether the hyperfusogenic F protein variants bearing L372W also could induce membrane fusion in the absence of the CDV H protein. However, it appeared that neither F-A75/17 L372W nor F-OP L372W, nor their respective variants bearing the hsc SP (hscFA L372W and hscFO L372W), was capable of inducing syncytium formation without H protein coexpression (data not shown).
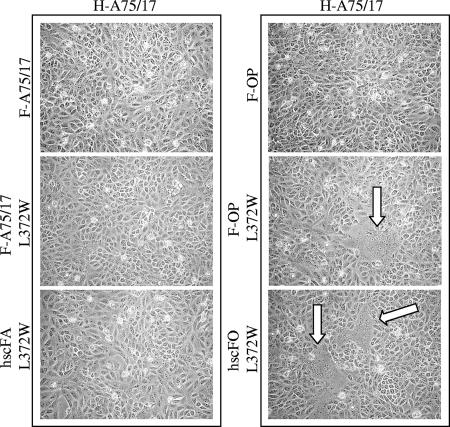
Nevertheless, as stipulated above, we always coexpressed H-OP with the different F proteins, since we previously have shown that H-A75/17 did not support the fusion process in this cell line (23). Interestingly, overlaying H-A75/17/F-A75/17- or H-A75/17/F-OP-coexpressing Vero cells into Vero-SLAM cells showed efficient fusion activity with both F/H protein combinations (data not shown). This confirmed that at least some lateral interactions occurred in heterologous F/H protein pairing and that the absence of fusion in F/H-A75/17-coexpressing Vero cells presumably was due to impaired interactions with the unknown CDV cell surface receptor. However, we speculated that minimal interactions of H-A75/17 with the Vero cell receptor occurs, since A75/17-CDV was able, although very inefficiently, to enter Vero cells (data not shown). To investigate whether the H-A75/17-receptor interaction was sufficient to activate F protein variants, we coexpressed L372W-bearing F proteins together with H-A75/17. While H-A75/17 did not support fusion activity in combination with the F protein of either CDV strain, it did so when coexpressed with the mutated F-OP L372W and hscFO L372W (Fig. 7). In the case of the wild parental and mutant F proteins, it appeared that any or all of the 17 mutations in the homotrimer were responsible for further limiting membrane fusion activity, since no syncytia were observed when combined with the homologous H-A75/17 protein (Fig. 7). Nevertheless, results obtained with the F-OP variants confirmed that H-A75/17-receptor interactions did indeed occur in Vero cells. Thus, while responsible for a hyperfusogenic phenotype, mutant F proteins bearing the L372W mutation could not circumvent the H-dependent fusion support function.
FIG. 7.
H-protein-dependent membrane fusion activity. Syncytium formation after cotransfection of the cells with plasmid DNA encoding CDV H-A75/17 and various F protein genes; representative fields of view were photographed 24 h posttransfection.
DISCUSSION
A75/17-CDV leads to persistent infection in the central nervous system of dogs and other species (36, 37), which is responsible for the high mortality of this disease. Understanding the mechanism of viral persistence in neurological distemper is critical for unraveling the pathogenesis of the disease. Among wild-type CDV field isolates, A75/17-CDV is unique regarding its lineage. Indeed, A75/17-CDV has never been passaged in any cultured cells, and its genome was sequenced directly from infected lymph nodes (24). Based on recombinant chimeric viruses, we have previously shown that F-A75/17 clearly reduced the extent of cell-cell fusion (23). In this study, we used transient transfection of F-A75/17 and derivative proteins as well as comparison with the attenuated OP-CDV F protein to analyze the mechanisms underlying F-A75/17 functionality.
It has been demonstrated that, in the genetic background of F-OP, shortening the SP led to an increased cell-cell fusion activity, suggesting that the length of the SP modulates fusion efficiency (38). The present paper supports and extends these reported findings. Here, we have shown for the first time that F-A75/17 is translated preferentially from AUG1 and F-OP is translated preferentially from AUG61. Thus, at first sight, the association between the longer SP of the wild F protein and its lower level of fusogenicity seems to be in line with the findings of von Messling and Cattaneo (38). However, we showed that, while lengthening the SP of F-OP by forcing the protein to initiate translation only at AUG1 did indeed decrease cell-cell fusion, shortening F-A75/17 SP did not significantly enhance membrane fusion activity, demonstrating fundamental differences between wild and attenuated CDV strains. Moreover, while exchanging the CDV F SP with that of the hsc protein, we found that the simultaneous presence of the SP of either CDV strain decreased intracellular F protein expression. While the sequence of the SP modulating F-OP expression maps to the first 60 amino acids, the 75 amino acids located upstream from AUG61 of the shortened F-A75/17 SP seem critical for modifying F-A75/17-mediated cell-cell fusion. Thus, it appeared that different regions of both CDV F protein SPs were responsible for modulating protein expression. However, the mechanism by which both SPs control limited membrane fusion activity seems to be identical in both F proteins. Indeed, the presence of the two SPs reduced total protein expression and, in turn, cell surface protein expression. Due to the unique structure of the SP even among morbilliviruses, its function is likely to be relevant only for CDV.
Intriguingly, we noticed that, irrespective of whether the F-OP trimer was transported to the plasma membrane via the A75/17-CDV (F-AO) or the OP-CDV (F-OP) SP, the steady-state level of the F-OP trimer at the cell surface remained mostly unaltered. However, despite a similar cell surface expression, F-AO constantly achieved reduced fusion activity compared to that of F-OP. Inhibition in trans of the highly fusogenic F-OP protein by cotransfecting the F-A75/17 SP alone or fused to green fluorescent protein did not reduce cell-cell fusion (data not shown). Thus, in addition to targeting nascent protein in the ER and the newly discovered function that limits protein expression, a third putative role specific for the SP of the virulent CDV F protein might be taken into consideration. We speculate that upon cleavage from the pre-F0 precursor, the F-A75/17 CDV SP still somehow interacts with the native F protein structure. Indeed, the cytoplasmic tail of the paramyxovirus F protein has been suggested to modulate fusogenicity by an inside-out mechanism (12, 31, 33, 39). Interactions of the F-A75/17 SP with the cytoplasmic tail domain might stabilize and hence increase the energy barrier required to activate F. Additional work will be necessary to understand the relevance and the molecular mechanisms underlying this observation.
While we clearly demonstrated that the F protein SP of the persistent CDV strain indirectly induced a limited cell-cell fusion activity by reducing total protein expression, exchanging the SP of F-A75/17 and F-OP with the shorter hsc SP still showed important differences in cell-cell fusion activity between the two F trimers. These findings indicate that the F-A75/17 F1/F2 trimer has an inherent limited cell-cell fusion capacity, which is independent of the SP. Based on our findings, the L372W mutation appears to be a key determinant in this respect.
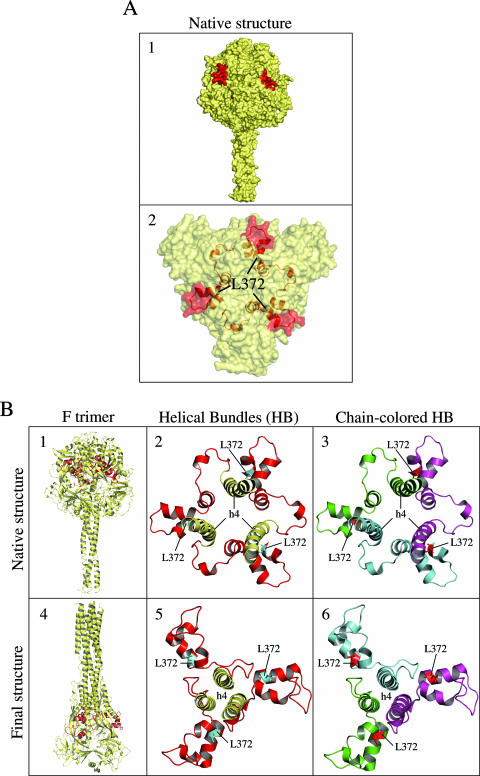
After the F protein is triggered by the receptor-binding protein, a series of conformational changes are believed to occur in a defined sequence. First, the HRB coiled coil melts, pulling the transmembrane apart; second, the crumpled HRA structure extends into a coiled coil, propelling the fusion peptide into the target membrane; and finally, HRB swings around the globular head to form the stable six-helix bundle with HRA. However, no biochemical data are available to date for a function of the HB in any or all of these different structural transitions. In the published native SV5 (42) and hPIV3 hairpin (41) F structures, the HB is part of the globular head (native) or neck (hairpin) conformation, respectively (Fig. 8B, images 1 and 4). When represented with a Connolly-derived surface model, it appeared that, while part of the HB points to the exterior of the structure (Fig. 8A, image 1), it is mostly composed of turns and short α-helices lying inside the F protein globular head (Fig. 8A, image 2). It is reasonable to assume that analogous residues in both SV5 and CDV F proteins form similar structures. Thus, in the native conformation, residue L372 would lie in the fourth α-helix of the HB, which is located in the core of the F protein head, in close proximity to the h4 helix (Fig. 8B, image 2). The latter is the basal α-helix of the HRA coiled coil generated during the fusion process (28). Interestingly, when the F protein chains were highlighted with different colors, it appeared clearly that the HB undergoes significant spatial rearrangements between both F protein conformations (Fig. 8B, images 3 and 6). It is thus tempting to speculate that residue L372 may be one of the crucial residues involved in stabilizing the F protein trimer globular head by interacting with other subdomains located in the core of the protein before the HB undergoes structural transitions during membrane fusion. The L372W substitution would affect these interactions and consequently would decrease the energy barrier required to activate F protein, thus facilitating the HB and other domains to transit from their prefusion to final postfusion structures. Due to the location of the HB in the native F protein form, its action in the fusion process might not be related to HRB melting but rather to the initiation of the drastic refolding of spring-loaded residues. Nevertheless, it may be possible that the HB also affects other steps in the fusion process, such as serving as a hinge region allowing HRB to swing around the F protein head. Additional work is needed to determine exactly in which stages of membrane fusion the HB is involved.
FIG. 8.
Structure of the paramyxovirus F protein. (A) Image 1 shows a Connolly-surface model of the structure of the PIV5 F protein ectodomain in its uncleaved, prefusion form. In red are residues of the HB in the F protein globular head pointing to the exterior. Image 2 shows the position of the corresponding CDV L372 residue in the PIV5 native structure (L250). (B) The red in images 1 and 4 shows the representation and localization of the HB in the native PIV5 and hairpin hPIV3 F structures. Images 2, 3, 5, and 6 provide close-up views of the HB and the h4 helix. Images 3 and 6 are representations of the HB with the three chains of F protein highlighted in three different colors. (Adapted from reference 42, using PyMOL 0.99 and color coding for clarification.).
Our data showed that highly fusion-active F protein L372W variants induced cell-cell fusion only in the presence of H protein. Although the F protein of a closely related morbillivirus (peste-des-petits-ruminants virus) has been shown to induce residual fusion in the absence of H (32), the H protein fusion support function may be an absolute requirement to activate the CDV F protein. In the presence of H-OP, it is clear that F-OP already is highly fusogenic, even in the absence of the mutation in the HB. On the other hand, introducing the N366S mutation in the F-A75/17 genetic background, which is 1 of the 17 amino acid differences between the F proteins of both CDV strains and is located in the HB domain, did not drastically increase fusion activity. These findings confirm that one or several of the differences between the F-A75/17 homotrimer and F-OP contributed to its limited membrane fusion activity. In agreement with this conclusion, only the OP-CDV L372W F protein mutants induced fusion activity in the presence of H-A75/17, supporting evidence that, in addition to the HB, other subdomains in the virulent CDV F protein are responsible for limiting fusion activity, although presumably through additional molecular mechanisms.
Taken together, our observations suggest that the highly neurovirulent CDV strain evolved so as to conserve a poorly fusion-active F protein. We hence speculate that this property is a clear advantage in the establishment of a persistent infection, a key event in the pathogenesis of the disease. However, although both F domains also were shown to play a role in the context of a viral infection, other components modulating the overall phenotype of a natural infection must be taken into consideration. Indeed, the matrix protein very likely plays an additional role in modulating the extent of cell-cell fusion, as suggested for measles virus (5, 6). Furthermore, the H protein/receptor affinity is certainly of critical importance, since we have previously shown that A75/17-CDV induces a fusogenic or nonfusogenic infection depending on the presence or absence of the cellular receptor SLAM (23). Our results obtained from comparisons of F/H-A75/17- and F/H-OP-coexpressing Vero cells supported this hypothesis.
In conclusion, we showed in this report that the presence of the SP indirectly affects the fusion activity of the F protein of virulent CDV by limiting intracellular and, consequently, surface protein expression, which in turn results in reduced cell-cell fusion. In addition, we identified a substitution of a conserved leucine residue potentially located in the HB of the head of the F protein that dramatically enhanced fusogenicity. Our observations add new aspects to our understanding of the role of different domains of the F protein and support growing evidence that the neurovirulent CDV field isolate evolved to restrict cell-cell fusion, which may favor the establishment of a long-term persistent infection.
Acknowledgments
We are grateful to Ernst Peterhans (Institute of Veterinary Virology, University of Bern, Switzerland), Ruth Parham, and members of the Zurbriggen laboratory for critically reviewing the manuscript.
We thank the Swiss National Science Foundation, which supported this study with grant 31-58657.99 to R.W. and A.Z.
Footnotes
Published ahead of print on 8 August 2007.
REFERENCES
- 1.Appel, M. J., and B. A. Summers. 1995. Pathogenicity of morbilliviruses for terrestrial carnivores. Vet. Microbiol. 44:187-191. [DOI] [PubMed] [Google Scholar]
- 2.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 3.Barben, G., M. Stettler, A. Jaggy, M. Vandevelde, and A. Zurbriggen. 1999. Detection of IgM antibodies against a recombinant nucleocapsid protein of canine distemper virus in dog sera using a dot-blot assay. Zentbl. Vetmed. A 46:115-121. [DOI] [PubMed] [Google Scholar]
- 4.Bolt, G., and I. R. Pedersen. 1998. The role of subtilisin-like proprotein convertases for cleavage of the measles virus fusion glycoprotein in different cell types. Virology 252:387-398. [DOI] [PubMed] [Google Scholar]
- 5.Cathomen, T., B. Mrkic, D. Spehner, R. Drillien, R. Naef, J. Pavlovic, A. Aguzzi, M. A. Billeter, and R. Cattaneo. 1998. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J. 17:3899-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, L., P. M. Colman, L. J. Cosgrove, M. C. Lawrence, L. J. Lawrence, P. A. Tulloch, and J. J. Gorman. 2001. Cloning, expression, and crystallization of the fusion protein of Newcastle disease virus. Virology 290:290-299. [DOI] [PubMed] [Google Scholar]
- 8.Cherpillod, P., K. Beck, A. Zurbriggen, and R. Wittek. 1999. Sequence analysis and expression of the attachment and fusion proteins of canine distemper virus wild-type strain A75/17. J. Virol. 73:2263-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherpillod, P., L. Zipperle, R. Wittek, and A. Zurbriggen. 2004. An mRNA region of the canine distemper virus fusion protein gene lacking AUG codons can promote protein expression. Arch. Virol. 149:1971-1983. [DOI] [PubMed] [Google Scholar]
- 10.Domingo, M., L. Ferrer, M. Pumarola, A. Marco, J. Plana, S. Kennedy, M. McAliskey, and B. K. Rima. 1990. Morbillivirus in dolphins. Nature 348:21. [DOI] [PubMed] [Google Scholar]
- 11.Doyle, J., A. Prussia, L. K. White, A. Sun, D. C. Liotta, J. P. Snyder, R. W. Compans, and R. K. Plemper. 2006. Two domains that control prefusion stability and transport competence of the measles virus fusion protein. J. Virol. 80:1524-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutch, R. E., and R. A. Lamb. 2001. Deletion of the cytoplasmic tail of the fusion protein of the paramyxovirus simian virus 5 affects fusion pore enlargement. J. Virol. 75:5363-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner, A. E., K. L. Martin, and R. E. Dutch. 2007. A conserved region between the heptad repeats of paramyxovirus fusion proteins is critical for proper F protein folding. Biochemistry 46:5094-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griot, C., M. Vandevelde, M. Schobesberger, and A. Zurbriggen. 2003. Canine distemper, a re-emerging morbillivirus with complex neuropathogenic mechanisms. Anim. Health Res. Rev. 4:1-10. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy, S. 1998. Morbillivirus infections in aquatic mammals. J. Comp. Pathol. 119:201-225. [DOI] [PubMed] [Google Scholar]
- 16.Likhoshway, Y. V., M. A. Grachev, V. P. Kumarev, Y. Solodun, O. A. Goldberg, O. I. Belykh, F. G. Nagieva, V. G. Nikulina, and B. S. Kolesnik. 1989. Baikal seal virus. Nature 339:266. [DOI] [PubMed] [Google Scholar]
- 17.Merz, D. C., A. Scheid, and P. W. Choppin. 1980. Importance of antibodies to the fusion glycoprotein of paramyxoviruses in the prevention of spread of infection. J. Exp. Med. 151:275-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison, T. G. 2003. Structure and function of a paramyxovirus fusion protein. Biochim. Biophys. Acta 1614:73-84. [DOI] [PubMed] [Google Scholar]
- 19.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paterson, R. G., C. J. Russell, and R. A. Lamb. 2000. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 270:17-30. [DOI] [PubMed] [Google Scholar]
- 21.Pearce-Kelling, S., W. J. Mitchell, B. A. Summers, and M. J. Appel. 1990. Growth of canine distemper virus in cultured astrocytes: relationship to in vivo persistence and disease. Microb. Pathog. 8:71-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearce-Kelling, S., W. J. Mitchell, B. A. Summers, and M. J. Appel. 1991. Virulent and attenuated canine distemper virus infects multiple dog brain cell types in vitro. Glia 4:408-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plattet, P., J. P. Rivals, B. Zuber, J. M. Brunner, A. Zurbriggen, and R. Wittek. 2005. The fusion protein of wild-type canine distemper virus is a major determinant of persistent infection. Virology 337:312-326. [DOI] [PubMed] [Google Scholar]
- 24.Plattet, P., C. Zweifel, C. Wiederkehr, L. Belloy, P. Cherpillod, A. Zurbriggen, and R. Wittek. 2004. Recovery of a persistent canine distemper virus expressing the enhanced green fluorescent protein from cloned cDNA. Virus Res. 101:147-153. [DOI] [PubMed] [Google Scholar]
- 25.Plemper, R. K., and R. W. Compans. 2003. Mutations in the putative HR-C region of the measles virus F2 glycoprotein modulate syncytium formation. J. Virol. 77:4181-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plemper, R. K., A. L. Hammond, and R. Cattaneo. 2001. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J. Biol. Chem. 276:44239-44246. [DOI] [PubMed] [Google Scholar]
- 27.Roelke-Parker, M. E., L. Munson, C. Packer, R. Kock, S. Cleaveland, M. Carpenter, S. J. O'Brien, A. Pospischil, R. Hofmann-Lehmann, H. Lutz, et al. 1996. A canine distemper virus epidemic in Serengeti lions (Panthera leo). Nature 379:441-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sergel, T. A., L. W. McGinnes, and T. G. Morrison. 2000. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J. Virol. 74:5101-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sergel-Germano, T., C. McQuain, and T. Morrison. 1994. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J. Virol. 68:7654-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seth, S., A. L. Goodman, and R. W. Compans. 2004. Mutations in multiple domains activate paramyxovirus F protein-induced fusion. J. Virol. 78:8513-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seth, S., and M. S. Shaila. 2001. The fusion protein of Peste des petits ruminants virus mediates biological fusion in the absence of hemagglutinin-neuraminidase protein. Virology 289:86-94. [DOI] [PubMed] [Google Scholar]
- 33.Seth, S., A. Vincent, and R. W. Compans. 2003. Mutations in the cytoplasmic domain of a paramyxovirus fusion glycoprotein rescue syncytium formation and eliminate the hemagglutinin-neuraminidase protein requirement for membrane fusion. J. Virol. 77:167-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Summers, B. A., H. A. Greisen, and M. J. Appel. 1984. Canine distemper encephalomyelitis: variation with virus strain. J. Comp. Pathol. 94:65-75. [DOI] [PubMed] [Google Scholar]
- 35.Sutter, G., M. Ohlmann, and V. Erfle. 1995. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 371:9-12. [DOI] [PubMed] [Google Scholar]
- 36.Vandevelde, M., and A. Zurbriggen. 2005. Demyelination in canine distemper virus infection: a review. Acta Neuropathol. (Berlin) 109:56-68. [DOI] [PubMed] [Google Scholar]
- 37.Vandevelde, M., A. Zurbriggen, R. J. Higgins, and D. Palmer. 1985. Spread and distribution of viral antigen in nervous canine distemper. Acta Neuropathol. (Berlin) 67:211-218. [DOI] [PubMed] [Google Scholar]
- 38.von Messling, V., and R. Cattaneo. 2002. Amino-terminal precursor sequence modulates canine distemper virus fusion protein function. J. Virol. 76:4172-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waning, D. L., C. J. Russell, T. S. Jardetzky, and R. A. Lamb. 2004. Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. Proc. Natl. Acad. Sci. USA 101:9217-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winters, K. A., L. E. Mathes, S. Krakowka, and R. G. Olsen. 1983. Immunoglobulin class response to canine distemper virus in gnotobiotic dogs. Vet. Immunol. Immunopathol. 5:209-215. [DOI] [PubMed] [Google Scholar]
- 41.Yin, H. S., R. G. Paterson, X. Wen, R. A. Lamb, and T. S. Jardetzky. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA 102:9288-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin, H. S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zurbriggen, A., H. U. Graber, and M. Vandevelde. 1995. Selective spread and reduced virus release leads to canine distemper virus persistence in the nervous system. Vet. Microbiol. 44:281-288. [DOI] [PubMed] [Google Scholar]
- 44.Zurbriggen, A., H. U. Graber, A. Wagner, and M. Vandevelde. 1995. Canine distemper virus persistence in the nervous system is associated with noncytolytic selective virus spread. J. Virol. 69:1678-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]