Abstract
Peptide presentation is critical for immune recognition of pathogen-infected cells by CD8+ T lymphocytes. Although a limited number of immunodominant peptide epitopes are consistently observed in diseases such as HIV-1 infection, the relationship between immunodominance and antigen processing in humans is largely unknown. Here, we have demonstrated that endogenous processing and presentation of a human immunodominant HIV-1 epitope is more efficient than that of a subdominant epitope. Furthermore, we have shown that the regions flanking the immunodominant epitope constitute a portable motif that increases the production and antigenicity of otherwise subdominant epitopes. We used a novel in vitro degradation assay involving cytosolic extracts as well as endogenous intracellular processing assays to examine 2 well-characterized HIV-1 Gag overlapping epitopes presented by the same HLA class I allele, one of which is consistently immunodominant and the other subdominant in infected persons. The kinetics and products of degradation of HIV-1 Gag favored the production of peptides encompassing the immunodominant epitope and destruction of the subdominant one. Notably, cytosolic digestion experiments revealed flanking residues proximal to the immunodominant epitope that increased the production and antigenicity of otherwise subdominant epitopes. Furthermore, specific point mutations in these portable flanking sequences modulated the production and antigenicity of epitopes. Such portable epitope processing determinants provide what we believe is a novel approach to optimizing CTL responses elicited by vaccine vectors.
Introduction
Virus-specific CTLs likely play a central role in disease pathogenesis by limiting viral replication and thus are likely critical components of the protective immunity induced by candidate vaccines for chronic viral infections such as HIV and hepatitis C virus (1–5). For most viruses, far fewer potential epitopes are immunogenic than expected based on favorable motifs for class I binding (6–8). Some epitope-specific CTL responses are consistently detected at high frequency in persons sharing the restricting HLA class I molecule and are termed immunodominant whereas other epitopes elicit less frequent responses and are termed subdominant (9–20). In many of these cases, this dominance hierarchy is predictable, but the mechanisms governing it are poorly understood.
The most detailed data on CTL responses in a chronic human viral infection derive from studies of the HIV epidemic using IFN-γ ELISPOT assays. In the chronic phase of HIV infection, an average of 5 to 27 different epitopes are targeted in a given individual (9, 14, 16–18, 20). Multiple reports have shown that there is no consistent relationship between magnitude or breadth of these IFN-γ responses and viral load (9, 14, 15, 21–23), suggesting that they my differ in their antiviral potential (16, 24–27). Immunodominant responses are not necessarily protective (28) whereas some subdominant responses have been associated with lower viral loads (22, 23). Furthermore, narrow immunodominant CTL responses may select viral escape mutants, suggesting that an optimal vaccine should elicit broader CTL responses. Therefore, understanding factors contributing to HIV epitope hierarchy is necessary for rational vaccine design since it may be required to enhance protective CTL responses and to avoid stimulation of nonprotective responses.
The recognition of HIV-infected cells as well as the immunogenicity of candidate vaccines depends on the presentation of an epitopic peptide at the cell surface. This is the final step in a multistage process of intracellular antigen processing, raising the question as to whether HIV epitope hierarchy may be related to differences in kinetics or quantity of epitope production. CTLs recognize HIV epitopes that are produced via intracellular processing pathways and presented by MHC-I molecules. Thus the processing of viral peptides and their intracellular association with MHC-I molecules represent key steps leading to immune recognition — steps that are not examined in the IFN-γ ELISPOT assays currently widely used to assess antiviral immunity.
Current knowledge of viral antigen processing is largely based on congenic experimental mouse models, often with experimental antigens such as ovalbumin or with pathogens containing very limited genetic variability. These studies first identified multiple possible factors that alone or in combination are involved in the efficiency of epitope presentation and in immunodominance (29, 30). Immunodominance is due, in part, to the MHC-I–binding affinity of peptides present in the ER, since multiple peptides compete for a limited number of class I molecules (31, 32). Those with the lowest affinity are unlikely to bind MHC-I and are subsequently degraded (33). Notably, immunodominant peptides are not always those with the highest affinity for a given MHC-I molecule, as shown in murine models (29, 34, 35), an observation that has been extended to EBV (36), and HIV epitopes (23). Another factor involved in epitope hierarchy is the amount and kinetics of protein production. This has been shown for several viruses such as lymphocytic choriomeningitis virus (LCMV) (37), EBV (13), and vaccinia virus (12), in which earlier proteins are expressed, processed, and presented first, and may also be true for HIV since the most immunodominant responses are located in early accessory proteins such as Nef and highly expressed proteins like Gag (38–40). Nevertheless, within Gag and Nef lie subdominant epitopes, suggesting that other cellular factors are involved. TCR repertoire and epitope affinity for the TCR also play a role in immunodominance (29, 41–44).
In addition to the above factors, the efficiency of antigen processing has been shown to influence viral epitope hierarchy in animal models (29, 45–49). The processing of epitopes from ubiquitinylated proteins or from defective ribosomal translation products relies on sequential cleavages, mainly by the proteasome and other amino- or endopeptidases in the cytosol and in the ER (reviewed in ref. 50). Poorly processed epitopes or epitopes with a low affinity for transporter associated with antigen processing (TAP) will be less likely to enter the ER and be loaded onto the MHC (29, 34, 46). The importance of antigen-processing efficiency on epitope hierarchy is indirectly demonstrated by the effect of mutations impairing epitope production and presentation. Artificial mutations in flanking regions of mouse viral epitopes (48, 51–55) as well as naturally occurring mutations flanking HIV (56, 57) or HCV epitopes (58) have been shown to impair their processing and presentation and to lead to CTL escape. Mice deficient in an ER aminopeptidase (ERAAP/ERAP1) (59, 60) or in immunoproteasome subunits (61) involved in epitope trimming present a different pattern of viral and cellular CTL immunodominant responses than WT mice, strongly supporting the role of finely tuned antigen-processing activities in establishing immunodominance. Interestingly, alterations in immunodominance patterns in ERAP1-deficient mice were not observed with full-length ovalbumin but rather with viral epitopes, highlighting the importance of tailored studies of viral epitope processing in their natural host (60).
Despite advances in understanding antigen processing in experimental murine models and the identification of antigen-processing mutations in human pathogens, the contribution of antigen-processing efficiency to epitope hierarchy in human primary cells infected with genetically evolving natural pathogens is largely unknown (62). In particular, no study we know of has identified elements leading to efficient or inefficient epitope production, partly due to the lack of appropriate assays. Yet this information is critical in order to control epitope presentation in the context of a vaccine vector. The precise characterization of CD8+ T cell responses and viral epitopes targeted in chronic human viral infections such as HIV provides an opportunity to define the role of antigen processing in epitope production efficiency and eventually in immunodominance.
HLA-A3 is a common class I allele in humans expressed in up to 25% of people of European descent (63), and several HLA-A3–restricted HIV-1 epitopes have been identified. These include 2 overlapping Gag epitopes, only 1 of which is consistently immunodominant (18, 19, 64–66). Here we take advantage of the overlap between these 2 epitopes to examine the relationship between antigen processing and immunodominance. We have studied the processing and presentation of the dominant and subdominant epitopes using 3 independent assays: endogenous processing and presentation of epitopes in cell lines expressing HIV proteins, cells transfected with peptides by osmotic loading, and what we believe is a novel in vitro degradation assay using primary cell extracts. We show that the endogenous processing and presentation of the immunodominant epitope is more efficient than that of 2 subdominant epitopes and that the cytosolic degradation of a Gag fragment preferentially generates peptides encompassing the immunodominant epitope and lacking the subdominant epitope. Moreover, we demonstrate that the flanking regions of these dominant and subdominant epitopes can be placed adjacent to other subdominant epitopes and similarly have an impact on kinetics and magnitude of the epitopes produced. Such portable epitope processing determinants as well as point mutations we have defined within these regions that can reduce or enhance epitope production provide what we believe are novel means to modulate epitope presentation in vaccine vectors.
Results
Endogenous processing and presentation of the immunodominant HLA-A3–restricted p17 RK9 epitope is more efficient than that of subdominant Gag or RT epitopes.
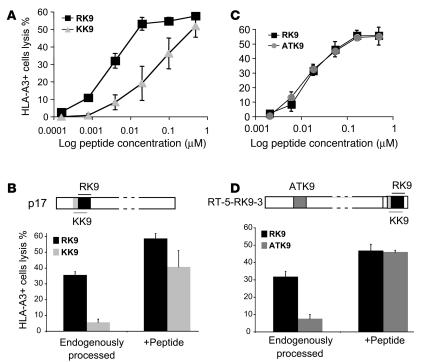
We first compared the endogenous processing and presentation of 2 well-defined and overlapping viral CTL epitopes derived from the HIV-1 Gag protein, KK9 (KIRLRPGGK, aa 18 to 26 in HXB2 sequence) and RK9 (RLRPGGKKK, aa 20 to 28). Earlier studies had shown that both epitopes are recognized following acute infection but that the CD8+ T cell response to RK9 is consistently of greater magnitude (18, 19, 64–66). HLA-A3+ HeLa cells were transiently transfected with a CMV-driven vector expressing HIV-1 LAV p17 and used as targets in a 51Cr release assay with RK9- and KK9-specific CTL clones. We first compared the killing capacity of the untransfected cells by the CTL clones. HLA-A3 HeLa cells were incubated with equal amounts of RK9 or KK9 (20 nM to 0.5 μM) peptide and used as targets in a 51Cr release assay with RK9- and KK9-specific CTLs (67) (Figure 1A). RK9-pulsed target cells were more sensitive to RK9-specific CTL lysis than KK9-pulsed cells to KK9-specific CTL lysis, likely due to a higher affinity of RK9 for HLA-A3 and/or to the TCR. Similar results were obtained with 2 additional clones from different patients (not shown). Next we examined the ability of the CTLs to lyse p17-transfected target cells and observed that lysis reached 35.5% with RK9 CTL clone whereas it was only 4.5% in the presence of the KK9 CTL clone (Figure 1B), representing much greater differential killing by the 2 clones than was observed with peptide pulsing. We next determined the amount of KK9 and RK9 peptide required to reach the levels of lysis observed in the transfected cells in order to assess whether differential processing and presentation of these epitopes contributed to these differences. The concentration of RK9 required to achieve the maximal lysis (35.5%) with RK9-specific CTL clone is 5 nM and therefore 3.3-fold greater than the 1.53 nM KK9 needed to reach 4.5% lysis. This result suggests that MHC-RK9 complexes were present in higher quantities, and that, in addition to the difference in peptide affinity, differential processing and presentation of RK9 and KK9 contribute to the lower recognition of target cells by the KK9 CTL clone.
Figure 1. The endogenous processing of an immunodominant Gag epitope is more efficient than that of subdominant epitopes.
(A) HLA-A3 HeLa cells were pulsed with decreasing amounts of p17 Gag immunodominant RK9 (squares) or p17 Gag subdominant KK9 (triangles) and used as targets in a 51Cr release assay with RK9- or KK9-specific CTLs respectively. (B) HLA-A3 HeLa cells transfected with a CMV-driven HIV-1 LAV p17 expression vector (schemed above the graph) were used as targets in a 51Cr assay with RK9- (black bars) or KK9-specific (gray bars) CTLs. The recognition of endogenously processed RK9 and KK9 epitopes by specific CTLs (left side) was compared with that of transfected cells pulsed with 0.1 μM RK9 or KK9 peptides (right side). (C) p17 immunodominant RK9 (squares) and RT subdominant ATK9 (circles) peptide titration on HeLa-A3 cells. (D) HLA-A3 HeLa cells transfected with a CMV-driven vector expressing a HIV-1 LAV RT fragment (HXB2 residues 153–560) with a C terminal p17 sequence (5-RK9-3; according to the position of RK9) (schemed above the graph) were used as targets in a 51Cr assay with RK9- (black bars) or ATK9-specific (gray bars) CTLs. The recognition of endogenously processed RK9 and ATK9 epitopes by specific CTLs (left side) was compared with that of transfected cells pulsed with 0.1 μM RK9 or ATK9 peptides (right side). Lysis of cells without peptides or with mismatched peptides was below 3%. All data represent averages of 3 experiments.
In order to better control for peptide affinity, we analyzed the endogenous processing of 2 high-affinity HLA-A3 epitopes, the immunodominant p17 RK9 and a well-defined subdominant epitope in the reverse transcriptase protein ATK9 (AIFQSSMTK), which is first presented by HLA-A3 during the chronic phase of HIV infection (68). Peptide titration experiments showed similar lysis percentages of peptide-pulsed cells by RK9- and ATK9-specific CTL clones (Figure 1C), indicating similar affinity. We next engineered an RT construct (RT-5-RK9-3) with a 17-residue C terminal extension from Gag p17 encompassing RK9 and its flanking sequences (RWEKIRLRPGGKKKYKL, designated 5-RK9-3 based on the length of sequences flanking RK9), allowing for equal expression of RT and p17 sequences in transfected cells. HLA-A3+ HeLa cells transfected with RT-5-RK9-3 were used as targets in a 51Cr release assay with RK9 and ATK9 clones (Figure 1D). Once again, the recognition of target cells by RK9 CTLs (31.6% lysis) was much higher than by ATK9 CTLs (7.5%) despite equivalent sensitivity of peptide-pulsed cells to lysis by these clones. By comparing the lysis percentage of transfected cells to that of peptide-pulsed cells (Figure 1, C and D), we estimated that there was 4.3 fold more RK9 than ATK9 displayed by HLA-A3 (4 nM ATK9 and 18.3 nM RK9 needed to reach the lysis percentage of the hybrid RT-p17 protein). This suggests that the processing and presentation of RK9 was more efficient than that of ATK9. Similar results were obtained when target cells were transfected with a vector encoding p17 with a C terminal extension of an RT fragment (5-ATK9-3) (data not shown). These data indicate that the endogenous processing and presentation of RK9 in the context of its natural flanking sequences is more efficient than that of subdominant epitopes, regardless of the position of the p17 fragment in the transfected protein.
Cytosolic processing favors the production of peptides containing an immunodominant epitope.
The above data led us to hypothesize that the processing of Gag p17 leads to more efficient production of RK9 and may thereby contribute to the immunodominance of this epitope, and these differences might be useful to identify intracellular events leading to efficient epitope presentation. The expression of peptidases involved in antigen processing varies in different mouse cell lines (69) and upon IFN-γ treatment (49, 70, 71). In order to analyze viral antigen processing in humans in a relevant in vitro experimental system, we used cytosolic or whole-cell extracts from fresh or phytohemagglutinin-activated (PHA-activated) healthy human PBMCs, which contain all the peptidases required for antigen processing in a tissue targeted by the virus. We took advantage of the overlap between dominant RK9 and subdominant KK9 to assess the contribution of antigen processing to epitope hierarchy and to quantify the production of 2 epitopes produced from the same longer peptide.
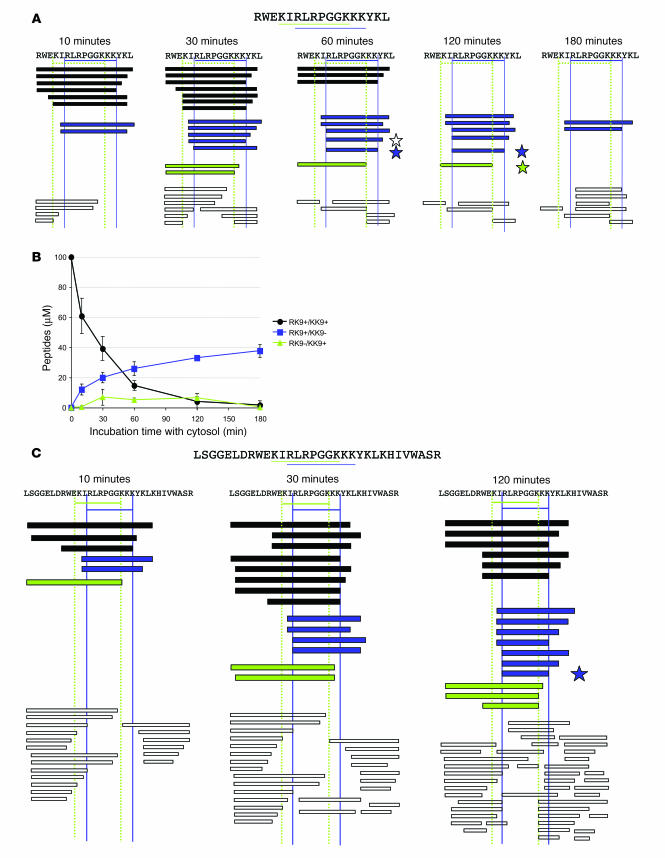
A 17-mer peptide (RWEKIRLRPGGKKKYKL) encompassing KK9 and RK9 and corresponding to the C terminal p17 tag added to RT for endogenous processing studies (Figure 1D) was incubated with PBMC-derived cytosol for increasing periods of time, and the resulting peptide products were identified by mass spectrometry (Figure 2A). Digestion products were fractionated on a C18 column and quantified by reverse phase HPLC (RP-HPLC) as described previously (56). Each peptide was identified by a unique peak at a defined elution time and quantified by the surface area under peak (Figure 2B). Over time, the 17-mer peptide was degraded into shorter fragments, some of which included both epitopes (RK9+/KK9+) whereas others contained only the RK9 (RK9+/KK9–, that is, including the entire RK9 sequence but incomplete KK9 sequence) or KK9 (RK9–/KK9+) epitope or neither epitope. Notably, after a 10-minute digestion, RK9+/KK9– peptides, lacking KK9 because of a cleavage event after position 1 or 2 within KK9, were already detected. The concentration of these RK9+/KK9– peptides increased to 25.9 μM at 60 minutes and reached a maximum of 37.6 μM at 180 minutes, at which point only RK9+/KK9– peptides were detected (Figure 2B). Optimal RK9 and KK9 epitopes were generated in cytosol after a 60- or 120-minute digestion, as previously reported for a single HLA-A3–restricted Nef peptide (72). After a 3-hour digestion in cytosol, the optimal RK9 and KK9 epitopes were no longer detected, suggesting that they were further digested by cytosolic peptidases. However, potential precursors of RK9 were still detected at this time, highlighting the stability of these epitope precursors in cell extracts. These data indicate that peptide degradation favors the production of peptides containing or limited to RK9 whereas production of KK9 was limited.
Figure 2. The cytosolic processing of Gag fragments facilitates the generation of an immunodominant epitope and the progressive loss of the subdominant one.
8 nmol of a Gag 17-mer (A) or 32-mer (C) were incubated with 40 μg PBMC cytosol for increasing periods of time. Peptides encompassing both epitopes (black bars), RK9 only (blue bars), KK9 only (green bars), or no epitope (white bars) were identified by mass spectrometry. Optimal RK9 and KK9 are indicated with blue and green stars. Another HLA-A3 epitope, RY10 (RLRPGGKKKY), codominant with RK9 in acute infection (19), was detected (white star). (B) Digestion products were fractioned on a C18 column and identified and quantified by RP-HPLC. Each peptide is identified by a unique peak at a defined elution time and quantified by the surface under peak. The column was calibrated with defined amounts of peptides covering the corresponding HIV sequence. The identity of the peptides in the digestion mix was confirmed by mass spectrometry. Peptides containing RK9 and KK9 epitopes (RK9+/KK9+, circles), RK9 only (RK9+/KK9–, squares), or KK9 only (RK9–/KK9+, triangles) were quantified over a 3-hour digestion with PBMC cytosol. Data are the average of 3 experiments performed with extracts from 3 healthy donors. The kinetics of peptide production using both the 17-mer and the 32-mer was affected by the amount of cytosol used for the experiment, with high amounts of cytosol yielding a more rapid appearance of RK9 and KK9 (not shown).
The expression of certain aminopeptidases or endopeptidases and subunits of the immunoproteasome are increased upon activation, which may induce the generation (49, 70) or destruction (71, 73) of some epitopes. We therefore compared the degradation of Gag 17-mer in cytosol prepared from either fresh PBMCs or PBMCs stimulated with PHA for 2 days. Degradation of the 17-mer was faster in PHA-activated PBMCs (100% degradation in 5 minutes with PHA-stimulated versus 30 minutes in fresh PBMC cytosol). Nevertheless, PHA-stimulated PBMC cytosol produced the same peptides, with a majority of RK9+/KK9– peptides (not shown). These results show that in both fresh and activated PBMCs, the cytosolic degradation of Gag fragments favors the production of the immunodominant RK9 epitope. Similarly, digestion experiments with whole PBMC extracts — which contain cytosol and ER extracts — also yielded a majority of RK9+/KK9– peptides (not shown). The pattern of peptides produced was independent of the HLA type of the donor (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI32047DS1). Overall, our data show that degradation of Gag sequences by cytosolic or whole-cell extracts leads to the production of many peptides that encompass the dominant RK9 epitope. Generation of peptides containing the subdominant KK9 epitope is limited, however, largely due to frequent cleavage after the first or second residue of KK9.
To confirm that the identity of the digestion products did not depend on the length of original peptide used for these experiments, we performed similar digestions using shorter p17 peptides (such as 5-RK9-2, 5-RK9-1, 5-RK9, 4-RK9-2, 4-RK9-1, 4-RK9, 3-RK9-2, 3-RK9, 2-RK9, based on the length of the N extension; not shown) and a 32-mer encompassing both epitopes (Figure 2C). Again, degradation favored RK9+/KK9– peptides, which accumulated over time. After a 2-hour digestion, RK9+/KK9– peptides previously detected during the degradation of Gag 17-mer reached 9 μM (data not shown). KK9 was frequently destroyed by a cleavage after K or I similar to what was observed during the 17-mer degradation (Figure 2C). With the 32-mer peptide, RK9 appeared after 2 hours of digestion, but KK9 remained undetectable, likely due to a slower processing of the 32-mer. Otherwise, peptides that were detected during digestion of the 17-mer, such as 5-RK9, 2-RK9, and 1-RK9-2, were found when using a 32-mer (compare Figures 2A and 2C). Some new peptides, such as 4-RK9-2 and 4-RK9, were also detected. In vitro degradation of these peptides in PBMC cytosol also led to a preferential production of peptides containing RK9 and destroying KK9 (data not shown). These results show that cytosolic processing of a Gag sequence leads to a majority of peptides encompassing the immunodominant epitope and a progressive loss of peptides that encompass the subdominant sequence, in accordance with a more efficient endogenous processing of the immunodominant epitope shown in Figure 1.
The cytosolic degradation products of a Gag sequence are preferentially targeted by the dominant RK9 CTL response over the subdominant KK9 CTL response.
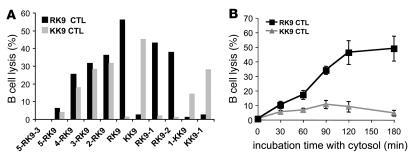
To assess the antigenicity of peptides present in the Gag 17- or 32-mer digestion products and how it compared with endogenous processing of RK9 and KK9 analyzed in Figure 1, we pulsed HLA-A3+ B cell line with the Gag digestion products and evaluated presentation to KK9- and RK9-specific CTL clones in a 51Cr release assay. We first determined conditions for which optimal epitopes and extended peptides of various lengths would elicit different CTL responses, using a panel of synthetic peptides. We measured RK9 and KK9 CTL responses elicited in a 51Cr release assay using different concentrations of peptides and CTL clones derived from 2 HIV-infected persons (67). Pulsing B cells with 0.25 μg/ml peptides allowed the elicitation of CTL responses that increased as peptide length became closer to the optimal sequence (Figure 3A). The 17-mer and peptides longer than 5-RK9 did not elicit RK9 or KK9 CTL responses (all <4% lysis). Peptides shorter than 4-RK9 and encompassing both RK9 and KK9 (4-RK9, 3-RK9, 2-RK9) elicited specific RK9 and KK9 CTL responses ranging from 15% to 45%. B cells pulsed with RK9 or KK9 at 0.25 μg/ml were lysed by corresponding CTLs (56% and 45% lysis, respectively) and not recognized by the nonspecific clone (<2.4% lysis). Lysis of peptide-pulsed cells was similar with RK9 and KK9 CTLs at 0.25 μg/ml but decreased more rapidly for KK9-specific responses when lower peptide dilutions were used due to the lower affinity of the KK9 sequence for HLA-A3 (see Figure 1). During the digestion of the 17-mer, none of the peptides used in the titration experiments represented more than 50% of the total peptides; therefore we measured the antigenicity of the 17-mer digestion products at 0.5 μg/ml. The RK9-specific CTL lysis induced by 5-RK9-3 digestion products reached 48.9% for the 3-hour time point, corresponding to the production and accumulation of RK9 and extended peptides shorter than a 12-mer encompassing RK9. In contrast, KK9-specific CTL lysis never exceeded 10.6% and decreased between 2 and 3 hours of digestion (Figure 3B). Similar results were observed when degradation products from the 32-mer were used to pulse HLA-A3 B cells although the maximum lysis was only 24% and 5% for RK9- and KK9-specific responses, respectively (not shown). In addition, we tested 2 different RK9 and KK9 CTL clones and found the same results. These data show that in vitro–digested p17 fragments elicit a strong RK9 CTL response and a weak KK9 CTL response, in accordance with results of endogenous processing of p17 presented in Figure 1.
Figure 3. The degradation products of a Gag sequence led to a strong RK9 CTL response and a weak KK9 response.
(A) 51Cr-labeled HLA-A3 B cells were pulsed with 0.25 μg/ml synthetic peptides including KK9 or RK9 with various N or C extensions and used as targets in a 51Cr release assay using RK9- (black bars) or KK9-specific CTLs (gray bars). (B) Gag 17-mer digestion products at 0.5 μg/ml were used to pulse 51Cr-labeled HLA-A3 B cells used as targets in a 51Cr release assay using RK9- (squares) or KK9-specific CTLs (triangles). Lysis percentages of peptide-pulsed or digestion product–pulsed targets were calculated after a 4-hour 51Cr release assay. Data are the average of 3 experiments performed with extracts from 3 healthy donors. Similar results were obtained with RK9 and KK9 CTL clones isolated from 2 HIV-1–infected donors (not shown).
N terminal flanking sequences are major determinants of epitope processing efficiency.
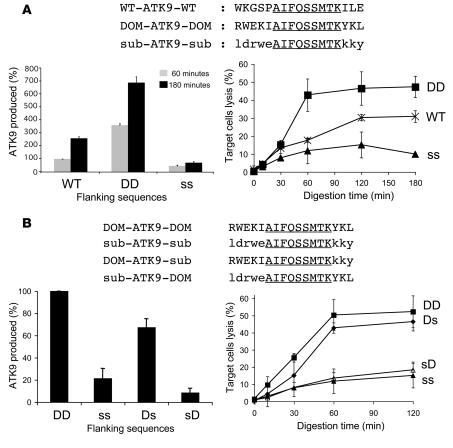
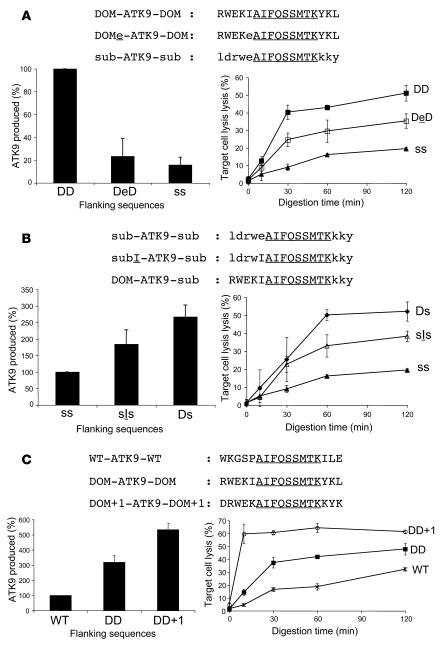
The above data suggest that the preferential production of peptides encompassing RK9 contributes to immunodominance, but other factors including affinity of peptide for HLA-A3 or for the TCR and the stability of the HLA peptide–TCR interaction may also contribute to a strong RK9 CTL response. Therefore, we sought to determine the potential contribution of flanking sequences to epitope production efficiency, independent of the binding affinity of dominant and subdominant epitopes to HLA class I or to the TCR. We designed hybrid peptides with various flanking sequences surrounding the subdominant RT epitope ATK9 (AIFQSSMTK), which exhibits less efficient endogenous processing and presentation than RK9 (Figure 1). The degradation of WT ATK9-containing peptides, such as 4-ATK9-2 (Supplemental Figure 1) and other ATK9 peptides with N and C extensions (Supplemental Figure 2 and data not shown) was similar to that observed for RK9 and KK9, with a rapid production of optimal ATK9 and extended peptides in cytosol. We compared the processing of a 17-mer WT ATK9 peptide (WT-ATK9-WT, WKGSP-ATK9-ILE) to hybrid peptides where the ATK9 epitope is flanked by sequences from the immunodominant RK9 (RWEKI-ATK9-YKL; named DOM-ATK9-DOM) or from the subdominant KK9 (ldrwe-ATK9-kky; named sub-ATK9-sub) (Figure 4A). Peptides were digested with PBMC extracts, and the identity and amount of digestion products were determined by mass spectrometry and RP-HPLC profile analysis (Figure 4A). We normalized to 100% the amount of ATK9 produced during the degradation of WT-ATK9-WT after a 1-hour digestion.
Figure 4. Identification of portable immunodominance sequences.
(A) 8 nmol of 17-mer WT-ATK9-WT or hybrid peptides DOM-ATK9-DOM and sub-ATK9-sub (where DOM and sub designate the origin [RK9 or KK9] of the flanking sequences) were incubated with 40 μg PBMC extracts for increasing periods of time (peptide sequences in top panel). The amount of ATK9 produced at time 60 (gray bars) and 180 (back bars) was analyzed by mass spectrometry and RP-HPLC (lower left panel). 100% corresponds to the amount of ATK9 produced in 1 hour during the degradation of WT-ATK9-WT. Digestion products from WT-ATK9-WT (WT, Xs), DOM-ATK9-DOM (DD, squares), or sub-ATK9-sub (ss, triangles) were used at 0.05 μg/ml to pulse 51Cr-labeled HLA-A3 B cells used as targets in a 51Cr release assay using ATK9-specific CTLs (lower right panel). (B) Similar to A except that the 4 hybrid peptides used for the digestion experiment were DOM-ATK9-DOM, sub-ATK9-sub, DOM-ATK9-sub, and sub-ATK9-DOM. 100% corresponds to the amount of ATK9 produced in 1 hour during the degradation of DOM-ATK9-DOM. Digestion products from 5R-ATK9-3R (DD, squares), sub-ATK9-sub (ss, filled triangles), DOM-ATK9-sub (Ds, diamonds) and sub-ATK9-DOM (sD, open triangles) were used at 0.05 μg/ml to pulse 51Cr-labeled HLA-A3+ B cells used as targets in a 51Cr release assay using ATK9-specific CTLs (lower right panel). Data are the average of 3 experiments performed with extracts from 3 healthy donors.
Despite large differences in sequences between WT and hybrid peptides, ATK9 was produced from all 3 peptides, as measured by RP-HPLC and mass spectrometry, suggesting that major changes in flanking sequences are tolerated as long as they do not impair peptidase activities or create intraepitope cleavage sites. After a 1-hour digestion using cytosolic extracts, DOM-ATK9-DOM yielded 3.6 times more ATK9 than WT-ATK9-WT and 8 times more ATK9 than sub-ATK9-sub (Figure 4A). After 3 hours, the amount of ATK9 produced during WT-ATK9-WT or DOM-ATK9-DOM degradation was 2.6 times higher and 6.9 times higher, respectively, than what was produced in 1 hour from WT-ATK9-WT, showing that precursors of ATK9 were being trimmed to optimal epitopes. In contrast, the amount of ATK9 generated from sub-ATK9-sub did not increase over time. The analysis of digestion products using mass spectrometry showed that the N terminal extension DOM-ATK9-DOM peptide was shortened faster than those from WT-ATK9-WT and sub-ATK9-sub (Supplemental Figure 3). In contrast, ATK9 in the context of sub-ATK9-sub was often destroyed by cleavages after position 1 and 2 in the epitope (Supplemental Figure 3), similar to the degradation of KK9 from the Gag 17-mer and 32-mer. Peptides purified from the digestion mix at various time points were used to pulse HLA-A3+ B cells and perform 51Cr release assays using ATK9-specific CTLs (Figure 4A). Over the course of 3 hours, degradation products of WT-ATK9-WT triggered an increasing amount of lysis by ATK9-specific CTLs, with a plateau of 30.2% lysis observed after a 2-hour digestion. Digestion products from DOM-ATK9-DOM yielded better ATK9 responses with 42.7% lysis after only a 1-hour digestion (compared with 17.6% for WT-ATK9-WT) and a maximum of 47.9% lysis after 3 hours. This result is consistent with a faster rate of ATK9 production in the context of DOM-ATK9-DOM degradation than with the WT flanking sequences. In contrast, sub-ATK9-sub degradation products yielded low CTL responses, indicating inefficient production of peptides containing ATK9. These data show that flanking sequences play a major role in epitope antigenicity that is independent of peptide-binding affinity to HLA-A3 or to the TCR and that this occurs by modulating epitope production.
We next determined the contribution of N and C terminal flanking sequences on epitope processing efficiency. We exchanged N and C terminal extensions of RK9 and KK9 in the reporter peptides yielding DOM-ATK9-sub and sub-ATK9-DOM (Figure 4B) and examined the degradation of these peptides in PBMC extracts. DOM-ATK9-sub degradation products contained one-third less ATK9 than DOM-ATK9-DOM but triggered a similar ATK9-specific CTL lytic response (50.3% vs. 42.7% for DOM-ATK9-DOM), likely due to the efficient production of ATK9 with a single residue N or C terminal extension that is able to stimulate ATK9 CTLs. Mass spectrometry analysis of the digestion products showed a similar cleavage pattern in the N and C terminal extensions for both peptides (not shown). sub-ATK9-DOM and sub-ATK9-sub were the least efficient peptides at producing ATK9 and yielded the lowest CTL responses (15% and 18.3% lysis, respectively) in accordance with frequent cleavages within the ATK9 sequence (Figure 4B). Overall, these results indicate that processing of the hybrid epitope was mainly determined by the N terminal extension, since peptides containing the N terminal DOM sequence consistently exhibited the highest CTL responses. Both C terminal extensions may be equally suitable to producing the exact C terminal anchor residue.
Control of epitope production through point mutations in flanking sequences.
Since the N terminal flanking sequence appears to play a determining role in reporter epitope processing, we aimed to define key residues in this region that would increase or decrease epitope production. Since the RK9 and KK9 flanking sequences overlap by 7 residues, the relevant changes are limited to 2 residues, of which 1 is an N terminal flanking residue (an isoleucine [I] in DOM-ATK9-DOM and a glutamate [E] in sub-ATK9-sub). We exchanged these residues in DOM-ATK9-DOM (yielding DOMe-ATK9-DOM) and sub-ATK9-sub (subI-ATK9-sub) and analyzed ATK9 production and the antigenicity of digestion products (Figure 5, A and B). Introducing an E upstream of ATK9 into DOM-ATK9-DOM reduced ATK9 production by 4.3-fold and reduced CTL lysis (29.6% vs. 43% after 60 minutes). The antigenicity of DOMe-ATK9-DOM degradation products was still higher than that of sub-ATK9-sub, likely due to the production of hybrid ATK9 peptides with short extensions capable of triggering CTL response. Conversely, introducing an isoleucine upstream of ATK9 into sub-ATK9-sub increased ATK9 production by 1.8-fold and doubled the CTL-induced lysis (33.1% vs. 16.1% after 60 minutes). Together these data show that a single flanking residue change can modulate epitope production and accordingly affect the CTL response.
Figure 5. Point mutations in the N terminal portable sequences alter epitope production.
(A) The in vitro degradation of 8 nmol of peptides DOM-ATK9-DOM, DOMe-ATK9-DOM (where flanking I is mutated to E), and sub-ATK9-sub was performed as described in the Figure 4 legend. The amount of ATK9 produced at time 60 was analyzed by mass spectrometry and RP-HPLC. 100% corresponds to the amount of ATK9 produced in 1 hour during DOM-ATK9-DOM degradation (lower left panel). The digestion products from DOM-ATK9-DOM (DD, filled squares), DOMe-ATK9-DOM (DeD, open squares), and sub-ATK9-sub (ss, triangles) were used at 0.05 μg/ml to pulse 51Cr-labeled HLA-A3+ B cells used as targets in a 51Cr release assay using ATK9-specific CTLs. (B) Similar to A except that the 3 peptides were sub-ATK9-sub, subI-ATK9-sub (where flanking E is replaced by I), and DOM-ATK9-sub. 100% corresponds to the amount of ATK9 produced in 1 hour during sub-ATK9-sub degradation. The digestion products from DOM-ATK9-sub (Ds, diamonds), subI-ATK9-sub (sIs, open triangles), and sub-ATK9-sub (ss, triangles) were used in a 51Cr release assay as in A. (C) Similar to A except that the 3 peptides were WT-ATK9-WT, DOM-ATK9-DOM, and DOM+1-ATK9- DOM+1 (Gag flanking sequences shifted by 1 residue). 100% corresponds to the amount of ATK9 produced in 1 hour during WT-ATK9-WT degradation. The digestion products from WT-ATK9-WT (WT, Xs), DOM-ATK9-DOM (DD, squares), and DOM+1-ATK9-DOM+1 (DD+1, circles) were used in a 51Cr release assay as in A. Data are the average of 3 experiments performed with extracts from 3 healthy donors.
The cytosolic digestion experiments showed that during the degradation of Gag 17-mer or 32-mer (Figure 2, A and C), the most frequent cleavage site is after K at position 1 in KK9, destroying KK9 and producing RK9 peptides with a 1-residue N extension. We hypothesized that we could use this specific cleavage site to increase reporter epitope presentation. We compared the degradation of DOM-ATK9-DOM (RWEKI-ATK9-KYK) to that of DRWEK-ATK9-KYK, which bears Gag flanking sequences shifted upstream by 1 residue (named DOM+1-ATK9-DOM+1) (Figure 5C). We observed that DOM+1-ATK9-DOM+1 produced 1.7-fold more ATK9 than DOM-ATK9-DOM, which already generated 3.2 times more ATK9 than WT-ATK9-WT. In addition, the processing of ATK9 from DOM+1-ATK9-DOM+1 was faster and more efficient than that of WT-ATK9-WT. Consistent with this result, the CTL response generated by DOM+1-ATK9-DOM+1 was the most efficient of all hybrid peptides, with 59.7% lysis for the 10-minute digestion products. These results provide the initial examples of portable dominance sequences that can be placed in front of a CTL epitope to increase or decrease its production as well as its antigenicity.
Portable flanking sequences modulate the endogenous processing and presentation of epitopes.
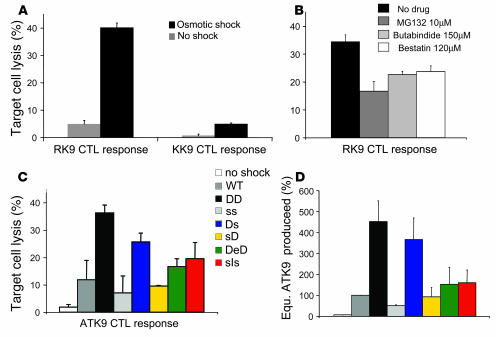
In order to test the effect of portable flanking sequences on endogenous epitope presentation, we adapted a transfection method of ovalbumin by osmotic lysis of pinosomes to our assay (74). Gag 17-mer used for in vitro epitope processing was introduced into cells by osmotic loading. The presentation of RK9 and KK9 was assessed by 51Cr assay (Figure 6A). Cells were efficiently recognized by RK9 CTLs (40.1%) and poorly by KK9 CTLs (4.9%). These results are remarkably similar to those obtained with target cells stably expressing p17 (35.5% and 4.5% lysis, Figure 1A) and to cells pulsed with in vitro degradation products of Gag 17-mer (33.3%–37.6% and 6.4%–0.3% lysis, Figure 3). Preincubation of cells with inhibitors of proteasomes (MG132), tripeptidyl peptidase II (TPPII) (butabindide), or aminopeptidases (bestatin) before transfection and throughout the experiment reduced the recognition of RK9 by 51.5, 31.9, and 31.3% respectively, showing that the processing of RK9 relies on these 3 peptidases (Figure 6B). The in vitro degradation assay of Gag 17-mer performed in the presence of various peptidase inhibitors allowed us to confirm and detail the involvement of these peptidases in the processing of RK9 and KK9 (Supplemental Figure 4). The degradation of Gag 17-mer 5-RK9-3 into 5-RK9-2 was dependent on proteasome whereas the degradation of 5-RK9-2 into shorter peptides such as 2/1RK9 relied on TPPII and aminopeptidases.
Figure 6. Transient peptide transfection endogenously replicates the effect of portable flanking sequences on epitope processing observed with in vitro degradation assays.
(A) HeLa-A3 cells were transfected with Gag 17-mer by osmotic loading (black bars) or without osmotic shock (gray bars) and used as targets with RK9- or KK9-specific CTLs. Similar results were obtained with HLA-A3 B cells (not shown). (B) Cells were preincubated with serum-free medium (black bar), MG132 (10 μM; dark gray bar), butabindide (150 μM; light gray bar), bestatin (120 μM; white bar) before transfection by osmotic loading and throughout the 51Cr assay experiment with RK9-specific CTLs. (C) HeLa A3 cells were transfected by osmotic loading with WT-ATK9-WT (WT, dark gray bar), DOM-ATK9-DOM (DD, black bar), sub-ATK9-sub (ss, light gray bar), DOM-ATK9-sub (Ds, blue bar), sub-ATK9-DOM (sD, yellow bar), DOMe-ATK9-DOM (DeD, green bar), or subI-ATK9-sub (sIs, red bar) or without osmotic loading (shown for DD, white bar). Cells were used as targets in a 51Cr assay with ATK9-specific clone. (D) The lysis percentage of transfected cells was compared with that of HeLa-A3 cells pulsed with defined amounts of ATK9 (as in Figure 1C). 100% was assigned to the amount of pulsed ATK9 required to reach the same lysis as in WT-ATK9-WT–transfected cells (5.8 nM). The equivalent ATK9 in cells transfected with hybrid sequences was calculated similarly. Data are the average of 3 experiments.
We then tested the endogenous processing and presentation of ATK9 from peptides with WT or hybrid flanking sequences introduced in cells by osmotic loading (Figure 6C). In accordance with in vitro epitope processing experiments (Figures 4 and 5), the insertion of DOM flanking sequences increased the killing of transfected cells by a factor of 3.1 (36.2% lysis vs. 11.8% for WT sequence) whereas the killing of cells transfected with sub-ATK9-sub was only 0.6-fold that of WT (7% lysis vs. 11.8% for WT sequences). The driving role of N flanking residues in efficient ATK9 production was confirmed as cells expressing DOM-ATK9-sub were more efficiently lysed than those expressing sub-ATK9-DOM (25.7 vs. 9.5%). Point mutations of N-flanking residues (DOMe-ATK9-DOM and subI-ATK9-sub) respectively decreased and increased the lysis of transfected cells compared with their WT counterparts in accordance with the critical role of this N flanking residue in ATK9 production shown in Figure 5. In order to roughly assess the number of antigenic complexes presented by cells transfected with these peptides, we calculated the amount of ATK9 required to reach the lysis percentage of transfected cells (Figure 6D). Cells transfected with DOM-ATK9-DOM and DOM-ATK9-sub displayed the most antigenic peptides (4.5- and 3.7-fold more equivalent ATK9 than from WT sequences) whereas sub-ATK9-sub produced 2 times less antigenic complexes than WT. This is in accordance with the in vitro production of ATK9 (compare ATK9 percentages in Figures 4 and 5 and Figure 6D).
These results demonstrate that portable flanking sequences can alter the endogenous processing of epitopes they surround and their subsequent antigenicity. Furthermore, they show the biological relevance of our in vitro epitope production assay by showing striking similarities in the production of epitopes and antigenicity of cells regardless of whether the epitope is endogenously produced in target cells or processed in vitro in cell extracts.
Discussion
In human viral infections, the relationship between kinetics and magnitude of antigen-processing products and immunodominance is not known. Although most current measurements of human CD8+ T cell responses rely on peptide pulsing of antigen-presenting cells, this method circumvents a number of critical steps, including epitope processing in the cytosol and ER, loading onto MHC-I molecules, and ultimately presentation at the cell surface. Here we show that the endogenous processing and presentation of the HLA-A3–restricted immunodominant Gag epitope RK9 is faster and more efficient than that of subdominant Gag KK9 and Pol ATK9 epitopes presented by the same allele (Figure 1). Using synthetic peptides containing overlapping Gag epitopes presented by HLA-A3 together with cell extracts containing natural processing enzymes, we demonstrate an association between the magnitude and kinetics of antigen processing and the immunodominance observed in vivo in a human viral infection.
Since these studies focused on overlapping epitopes derived from the same precursor peptide and presented by the same HLA class I allele, they allowed direct assessment of processing variables that are independent of HLA. Our results show the preferential production of peptides encompassing the immunodominant RK9 epitope and a progressive cytosolic degradation of the subdominant KK9 epitope. These data correlate with a stronger RK9 CTL response and a weaker KK9 CTL response induced by these digestion products and also with a more efficient endogenous processing and presentation of the immunodominant epitope. Further studies will be necessary to define the contribution of antigen processing to the immunodominance of epitopes restricted by HLA alleles other than HLA-A3 and processed through different pathways.
Mutations in N or C flanking residues affecting CTL responses in mouse models (53, 75), HCV (58), and HIV (56) have been identified, but the extent to which flanking polymorphisms may have an effect on viral epitope processing and eventually CTL responses is unknown. Our studies show differential cytosolic production of an immunodominant and subdominant epitope and establish a model system to evaluate the specific contribution of individual flanking residues in this process that can be used to define the role of individual amino acids in peptide cleavage. Using hybrid sequences that included an additional A3-restricted epitope (ATK9) and the N and C terminal flanking residues of RK9 and KK9, we show that although all of these hybrid peptides generate ATK9, there are major effects of WT and substituted flanking sequences on the quantity of antigenic peptides produced. Furthermore, peptide transfection by osmotic loading confirms that the endogenous processing of these hybrid sequences results in similar differences in epitope production and antigenicity as the in vitro degradation assay, providing strong support for the biological relevance of this in vitro degradation assay. The effects of these portable flanking sequences were extended to 2 additional subdominant epitopes, HLA-A3 QK10 and ALK9 in Nef, for which the dominant flanking sequences increased epitope production by 4.9- and 1.8-fold, respectively, and subdominant flanking decreased epitope production by 1.8-fold and 1.3-fold, respectively (Supplemental Figure 5). The modest effect of flanking sequences on ALK9 is likely due to a greater sensitivity of ALK9 sequence to peptidase degradation (not shown). Overall, these results identify portable flanking sequences altering the production of the epitopes they surround. These portable flanking sequences provide a potential means to manipulate and control the expression of epitopes in vaccine vectors.
Using PBMC extracts or peptide-transfected cells preincubated with various peptidase inhibitors, we observed that the processing of Gag 17-mer requires proteasome to cleave the last C terminal residue and that further trimming steps rely on TPPII and amino-metallopeptidases (Figure 6B and Supplemental Figure 5). Similarly, the processing of WT-ATK9-WT relies on proteasome, TPPII, and amino-metallopeptidases (not shown). The higher production of ATK9 from DOM-ATK9-DOM than from WT-ATK9-WT may correspond to the presence of residues that are better substrates for peptidases involved in ATK9 processing. TPPII cleaves preferentially after K and R (76), and we have shown that amino-metallopeptidases present in PBMC extracts cleave efficiently after K and R and not after E (Le Gall et al., unpublished observations). The degradation of the Gag 17-mer as well as DOM-ATK9-DOM shows frequent cleavages after K or R (see Figure 2, A and B, and Supplemental Figure 2). In accordance with our hypothesis about peptidase substrate preferences, the chimeric peptide producing the largest amount of ATK9 is DRWEK-ATK9-KYK, in which both N and C terminal flanking residues are K, suggesting that these flanking sequences are optimal for efficient ATK9 epitope processing whereas hybrid peptides with a flanking E yielded the lowest amount of ATK9 and antigenicity (Figure 5).
Besides the magnitude and efficiency of epitope production, the translocation of peptides produced in the cytosol into the ER through TAP may contribute to immunodominance by limiting ER access to peptides with higher affinity for TAP (34, 46). The second residue and the C terminal anchor of the peptide define its affinity for TAP (77). RK9 and KK9 have the same C terminal anchor. Since they overlap by 7 residues, many N-extended RK9 and KK9 will have the same residue at position 2 (for instance, 2-RK9 and KK9). In vitro TAP translocation experiments showed that peptides with L or I at position 2 (such as RK9, KK9, and ATK9) or W, E, and K (such as N-extended RK9 or KK9) bind to TAP with similar efficiency, suggesting that optimal and N-extended KK9 and RK9 may have similar affinity for TAP (77). Therefore it is unlikely that the affinity of optimal and extended epitopes for TAP plays a major role in RK9 immunodominance.
The immunogenicity of vaccine candidates for HIV and other human viral pathogens such as HCV will likely require efficient induction of strong antiviral CD8+ T cell responses (1, 2, 4). Because these viruses are highly variable, final selection of the actual residues that will be used in the immunogen will require careful consideration (78). Our results indicate that it will not only be important to consider the sequences of individual epitopes selected but also the impact of flanking residues on epitope processing. Whether changing the kinetics and amount of epitopes and precursors is sufficient to alter immunogenicity and dominance will need to be further evaluated in primate or humanized mouse models. Whether the candidate vaccine contains strings of WT and/or variant epitopes or immunogenic regions of the virus, assessing epitope production from vectors in a relevant human cell type will be important since some epitopes may be limited in their immunogenicity simply because of inefficient epitope processing. Shifting epitopes between viral proteins alters epitope density and immunodominance patterns in inbred mouse models (48, 51, 79). Spacers intercalated in strings of epitopes of multiepitopic vectors increase epitope presentation and immunogenicity (51, 80). In situations where subdominant responses are seen to contribute to the control of viral replication whereas some immunodominant responses may need to be eliminated (22, 28), it may become necessary to manipulate epitope production in such a way that dominance would be redirected to broader CTL responses. Yet no systematic identification of portable elements altering epitope presentation has been undertaken. Our data indicate that a single amino acid change in a flanking residue can greatly reduce the CTL response whereas others can augment kinetics and production of the relevant peptides. The definition of predictable effects of flanking regions on epitope processing and the demonstration that portable flanking sequences could be used to modulate epitope production provide an important mechanism to optimize epitope presentation. This study provides tools and rationale to further identify sequences that will alter CTL epitope processing and introduces what we believe is a novel method to modulate the immunogenicity of candidate vaccine vectors.
Methods
Peptides.
Peptides were synthesized on an AAPPPTEC Apex 396 multiple peptide synthesizer at the MGH peptide core and purified by RP-HPLC. The purity of the peptides assessed by mass spectrometry was greater than 95%. Peptide sequences were confirmed by mass spectrometry.
Cell culture.
EBV-immortalized HLA-A3 B cells were as described (56). HeLa cells stably expressing HLA-A3 (gift from J. Kessler, Leiden University Medical Center, Leiden, The Netherlands) were maintained as described (81) with 0.4 mg/ml G418 (Sigma-Aldrich). RK9-specific, KK9-specific, and ATK9-specific CTL clones were isolated by limiting dilution and maintained in the presence of 50 U/ml IL-2 (R10-IL2) using the CD3-specific mAb 12F6 and irradiated feeder cells as stimulus for T cell proliferation (56, 67, 68).
HIV-1–negative individuals enrolled at the MGH were included in this study. The MGH Institutional Review Board approved the study, and each subject gave informed consent for participation. PBMCs were separated from whole blood by Ficoll-Hypaque (Sigma-Aldrich) density gradient centrifugation. PHA-stimulated PBMCs were obtained by incubating PBMCs at 5–10 × 106/ml from healthy donors in R10-IL2 with 0.25 μg/ml PHA for 48 hours.
DNA constructs.
HIV-1 LAV p17 was amplified by PCR (5′ primer, GGTGCCACCATGGGTGCGAGAGCGTCAGTATTAAGCGGGGGAGAATTAG; 3′ primer, GCCACCGCCTCAGTAATTTTGGCTGACCTGGCT) with the following amplification conditions: 3 minutes at 94°C, 35 cycles of 30 seconds at 94°C, 30 seconds at 75°C, and 90 seconds at 72°C. The PCR product was cloned into a TA-GW8 cloning vector (Invitrogen) according to the manufacturer’s instructions and sequenced. The p17 insert was cloned into a pcDNA3 expression vector (Invitrogen) according to the manufacturer’s instructions. The p17-expressing vector was checked by restriction digestion and sequencing of the p17 insert. A fragment of HIV-1 LAV RT (residues 153–560 according to HXB2 nomenclature) with a p17 C terminal tag (RWEKIRLRPGGKKKYKL; 5-RK9-3) was cloned with a similar procedure with the following primers (5′-AGGTGGCGGCCACCATGGGATGGAAAGGATCACCAGCAATATTCCAAAGTAGCATGACAAAAATCTTAGAGCCTTTTAG; 3′-GCCACCGCCTCATAACTTGTATTTTGTCATGCTACTTTGGAATATTGCTCCGATCTTTTCCCTGGC).
Endogenous processing of HIV epitopes.
2 × 106 HeLa A3 cells were transiently transfected with 5 μg p17 or RT vector with Lipofectamine according to the manufacturer’s instructions along with 0.1 μg CMV-GFP (Invitrogen). Transfection efficiency assessed by GFP expression was 50%–60%, and dead cells were at less than 4%. Sixteen hours later, mock-, 51Cr-labeled p17- or RT-transfected HeLa-A3 were used as targets in a 51Cr release assay as described (81). RK9-specific, KK9-specific, or ATK9-specific CTL clones were added at a 4:1 ratio and incubated 4 hours with target cells. Cell lysis was calculated as [(51Cr release due to peptide – spontaneous release)/(total release – spontaneous release)] × 100. These values were compared with the lysis of HLA-A3+ B cells pulsed with 20 nM to 0.5 μM of optimal RK9, KK9, and ATK9, respectively, diluted in RPMI medium without serum (R+). RK9-specific and KK9-specific CTL clones did not recognize KK9-pulsed and RK9-pulsed cells, respectively (lysis <3%). The lysis of HLA-A3 HeLa cells in mock-transfected cells by RK9-, KK9-, or ATK9-specific CTL clones was less than 2%. Stable lines expressing p17 were derived under 10 μg/ml blasticydin selection as described in ref. 81 and similarly used as targets in a 51Cr assay (not shown).
Alternatively, B cell line or HLA-A3 HeLa cells were transfected with 17-mer HIV peptides by osmotic loading, a technique adapted from ref. 74. 2 × 106 cells were incubated at 37°C for 10 minutes with 100 to 300 ng peptides in 500 μl hyperosmotic buffer followed by 3 minutes in 15 ml hypotonic buffer (60% R+ in water). Cells were resuspended in 500 μl R+ and used as targets in a 51Cr release assay as described above (dead cells <2%). When indicated, cells were preincubated with MG132 (10 μM), butabindide (150 μM), or bestatin (120 μM) for 45 minutes before transfection and throughout the transfection and 51Cr labeling processes. Upon addition of the CTLs, the concentrations of the drugs were reduced by half and did not affect CTL killing capacity (not shown).
Cell extract preparation.
PBMCs were resuspended in a pH 7.4 buffer containing 50 mM potassium acetate (KAc), 5 mM magnesium chloride (MgCl2), 1 mM ATP, 0.5 mM EDTA, and 10% glycerol. They were broken by agitation with glass beads (75 to 85% broken cells as assessed by Trypan blue), and whole extracts were separated from nuclei, debris, and unbroken cells by a 5-minute 1,000 g spin followed by 10 minutes at 10,000 g to remove mitochondria. Cytosol was purified by 2 20-minute 105,000 g spins at 4 degrees. Protein concentration was measured with a Bio-Rad protein assay kit according to the manufacturer’s instructions. Protein content was checked by Western blot against various proteasome subunits and actin. For each preparation, proteasome and aminopeptidase activities were measured with specific fluorogenic substrates as described (82). Variations in antigen-processing activities among donors were below 15%.
HIV epitope processing assays.
8 nmol of purified peptides were incubated with 40 μg cytosol or whole-cell extracts in 100 μl pH7.4 buffer containing 137 mM KAc, 1 mM MgCl2, and 1 mM ATP. Aliquots were taken at various time points, and the reaction was stopped with 0.3% trifluoroacetic acid (TFA). The peptides present in the digestion mix were identified and quantified by RP-HPLC as described previously (56). In brief, peptides were fractionated on a Biosuite 4.6 × 50 mm 3 μm C18 column (Waters) in TFA (0.03%) and eluted on a linear gradient (0 to 40% acetonitrile and 0.02 to 0.05% TFA). The column was calibrated with defined amounts of peptides covering the corresponding HIV sequence. Each peptide, the original undigested peptide or shorter peptides produced during digestion in extracts, corresponded to a distinct peak the surface of which is proportional to the peptide concentration. The amount of peptides was calculated by the integration of peptide peaks. The identity of each peak was determined by comparing its elution time to that of known synthetic peptides covering HIV sequences. The identity of the peptides in the digestion mix was confirmed by mass spectrometry (Taplin Mass Spectrometry Facility, Harvard Medical School).
Antigenicity of the processing products.
Peptides present in the digestion mix were purified by 10% trichloroacetic acid precipitation and diluted in RPMI without serum; pH was readjusted to 7.4. 51Cr-labeled HLA-A3+ B cells were pulsed with digestion products (0.5 μg/ml for Gag 17-mer and 32-mer; 0.05 μg/ml for WT and hybrid ATK9 peptides) for 30 minutes at 37°C in the absence of serum. Killing assays with RK9-, KK9-, or ATK9-specific CTL clones at a 4:1 ratio were performed as described above. The lysis percentages were compared with the lysis of HLA-A3+ B cells pulsed with the undigested peptides (Gag 17-mer or 32-mer or hybrid peptides), optimal epitopes (KK9, RK9, or ATK9), and various extended peptides containing either epitope at concentrations ranging from 0 to 0.5 μg/ml.
Statistics.
Means and standard deviations were calculated with Microsoft Excel.
Supplementary Material
Acknowledgments
The authors would like to thank S-.C. Chang for advice on HPLC elution conditions, T. Ross for help with mass spectrometry analysis, and J. Kessler for the gift of HeLa-A3 cells. We are grateful to J. Bennink, M. Brockman, D. Kavanagh, D. Kaufmann, T. Allen, and A. Kim for their critical reading of the manuscript. This work was supported by NIH/NIAID grants AI060502-01A1 (to S. Le Gall) and AI28568-16 (to B.D. Walker), by the Harvard Center for AIDS Research (AI 060354), and by Microsoft Research funding (to S. Le Gall). B.D. Walker is a Howard Hughes Medical Institute investigator.
Footnotes
Nonstandard abbreviations used: PHA, phytohemagglutinin; TAP, transporter associated with antigen processing; TFA, trifluoroacetic acid; TPPII, tripeptidyl peptidase II.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:3563–3575 (2007). doi:10.1172/JCI32047
References
- 1.Borrow P., Lewicki H., Hahn B.H., Shaw G.M., Oldstone M.B. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper S., et al. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10:439–449. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 3.Jin X., et al. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koup R.A., et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitz J.E., et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 6.Hossain M.S., et al. Identification and characterization of HLA-A*3303-restricted, HIV type 1 Pol- and Gag-derived cytotoxic T cell epitopes. AIDS Res. Hum. Retroviruses. 2003;19:503–510. doi: 10.1089/088922203766774559. [DOI] [PubMed] [Google Scholar]
- 7.Falk K., et al. Identification of naturally processed viral nonapeptides allows their quantification in infected cells and suggests an allele-specific T cell epitope forecast. J. Exp. Med. 1991;174:425–434. doi: 10.1084/jem.174.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang K.M., et al. Identification of HLA-A3 and -B7-restricted CTL response to hepatitis C virus in patients with acute and chronic hepatitis C. J. Immunol. 1999;162:1156–1164. [PubMed] [Google Scholar]
- 9.Addo M.M., et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 2003;77:2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox A.L., et al. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology. 2005;42:104–112. doi: 10.1002/hep.20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jameson J., Cruz J., Ennis F.A. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J. Virol. 1998;72:8682–8689. doi: 10.1128/jvi.72.11.8682-8689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oseroff C., et al. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13980–13985. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pudney V.A., Leese A.M., Rickinson A.B., Hislop A.D. CD8+ immunodominance among Epstein-Barr virus lytic cycle antigens directly reflects the efficiency of antigen presentation in lytically infected cells. J. Exp. Med. 2005;201:349–360. doi: 10.1084/jem.20041542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Betts M.R., et al. Analysis of total human immunodeficiency virus (HIV)-specific CD4(+) and CD8(+) T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 2001;75:11983–11991. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao J., et al. Comprehensive analysis of human immunodeficiency virus type 1 (HIV-1)-specific gamma interferon-secreting CD8+ T cells in primary HIV-1 infection. J. Virol. 2003;77:6867–6878. doi: 10.1128/JVI.77.12.6867-6878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Draenert R., et al. Persistent recognition of autologous virus by high-avidity CD8 T cells in chronic, progressive human immunodeficiency virus type 1 infection. J. Virol. 2004;78:630–641. doi: 10.1128/JVI.78.2.630-641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frahm N., et al. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 2004;78:2187–2200. doi: 10.1128/JVI.78.5.2187-2200.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X.G., et al. Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-cell responses following acute HIV-1 infection. J. Virol. 2002;76:8690–8701. doi: 10.1128/JVI.76.17.8690-8701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altfeld M., et al. HLA class I alleles that contribute strongly to the initial CD8+ T cell responses against HIV-1 are associated with delayed progression to AIDS. PLoS Med. 2006;3:e403. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalod M., et al. Broad, intense, anti-HIV ex vivo CD8+ responses in HIV type 1-infected patients: comparison with anti-Epstein-Barr virus responses and changes during antiretroviral therapy. J. Virol. 1999;73:7108–7116. doi: 10.1128/jvi.73.9.7108-7116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards B.H., et al. Magnitude of functional CD8+-T-cell responses to the Gag protein of HIV-1 correlates inversely with viral load in plasma. . J. Virol. 2002;76:2298–2305. doi: 10.1128/jvi.76.5.2298-2305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frahm N., et al. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat Immunol. 2006;7:173–178. doi: 10.1038/ni1281. [DOI] [PubMed] [Google Scholar]
- 23.Bihl F., et al. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J. Immunol. 2006;176:4094–4101. doi: 10.4049/jimmunol.176.7.4094. [DOI] [PubMed] [Google Scholar]
- 24.Lichterfeld M., et al. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J. Exp. Med. 2004;200:701–712. doi: 10.1084/jem.20041270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loffredo J.T., et al. Tat(28-35)SL8-specific CD8+ T lymphocytes are more effective than Gag(181-189)CM9-specific CD8+ T lymphocytes at suppressing simian immunodeficiency virus replication in a functional in vitro assay. J. Virol. 2005;79:14986–14991. doi: 10.1128/JVI.79.23.14986-14991.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trimble L.A., et al. Human immunodeficiency virus-specific circulating CD8 T lymphocytes have down-modulated CD3zeta and CD28, key signaling molecules for T-cell activation. J. Virol. 2000;74:7320–7330. doi: 10.1128/jvi.74.16.7320-7330.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang O.O., et al. Impacts of avidity and specificity on the antiviral efficiency of HIV-1-specific CTL. J. Immunol. 2003;171:3718–3724. doi: 10.4049/jimmunol.171.7.3718. [DOI] [PubMed] [Google Scholar]
- 28.Kiepiela P., et al. CD8(+) T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 2006;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 29.Chen W., Anton L.C., Bennink J.R., Yewdell J.W. Dissecting the multifactorial causes of immunodominance in class I-restricted T cell responses to viruses. Immunity. 2000;12:83–93. doi: 10.1016/s1074-7613(00)80161-2. [DOI] [PubMed] [Google Scholar]
- 30.Yewdell J.W., Bennink J.R. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- 31.Chen W., Khilko S., Fecondo J., Margulies D.H., McCluskey J. Determinant selection of major histocompatibility complex class I-restricted antigenic peptides is explained by class I-peptide affinity and is strongly influenced by nondominant anchor residues. J. Exp. Med. 1994;180:1471–1483. doi: 10.1084/jem.180.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sette A., et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 1994;153:5586–5592. [PubMed] [Google Scholar]
- 33.Koopmann J.O., et al. Export of antigenic peptides from the endoplasmic reticulum intersects with retrograde protein translocation through the Sec61p channel. Immunity. 2000;13:117–127. doi: 10.1016/s1074-7613(00)00013-3. [DOI] [PubMed] [Google Scholar]
- 34.Deng Y., Yewdell J.W., Eisenlohr L.C., Bennink J.R. MHC affinity, peptide liberation, T cell repertoire, and immunodominance all contribute to the paucity of MHC class I-restricted peptides recognized by antiviral CTL. J. Immunol. 1997;158:1507–1515. [PubMed] [Google Scholar]
- 35.Mullbacher A., et al. High peptide affinity for MHC class I does not correlate with immunodominance. Scand. J. Immunol. 1999;50:420–426. doi: 10.1046/j.1365-3083.1999.00619.x. [DOI] [PubMed] [Google Scholar]
- 36.Crotzer V.L., et al. Immunodominance among EBV-derived epitopes restricted by HLA-B27 does not correlate with epitope abundance in EBV-transformed B-lymphoblastoid cell lines. . J. Immunol. 2000;164:6120–6129. doi: 10.4049/jimmunol.164.12.6120. [DOI] [PubMed] [Google Scholar]
- 37.Probst H.C., et al. Immunodominance of an antiviral cytotoxic T cell response is shaped by the kinetics of viral protein expression. J. Immunol. 2003;171:5415–5422. doi: 10.4049/jimmunol.171.10.5415. [DOI] [PubMed] [Google Scholar]
- 38.Sacha J.B., et al. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J. Immunol. 2007;178:2746–2754. doi: 10.4049/jimmunol.178.5.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ali A., et al. Impacts of epitope expression kinetics and class I downregulation on the antiviral activity of human immunodeficiency virus type 1-specific cytotoxic T lymphocytes. J. Virol. 2004;78:561–567. doi: 10.1128/JVI.78.2.561-567.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Baalen C.A., et al. Impact of antigen expression kinetics on the effectiveness of HIV-specific cytotoxic T lymphocytes. Eur. J. Immunol. 2002;32:2644–2652. doi: 10.1002/1521-4141(200209)32:9<2644::AID-IMMU2644>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 41.Campos-Lima P.O., Levitsky V., Imreh M.P., Gavioli R., Masucci M.G. Epitope-dependent selection of highly restricted or diverse T cell receptor repertoires in response to persistent infection by Epstein-Barr virus. J. Exp. Med. 1997;186:83–89. doi: 10.1084/jem.186.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trautmann L., et al. Selection of T cell clones expressing high-affinity public TCRs within Human cytomegalovirus-specific CD8 T cell responses. J. Immunol. 2005;175:6123–6132. doi: 10.4049/jimmunol.175.9.6123. [DOI] [PubMed] [Google Scholar]
- 43.Wienhold W., et al. An example of immunodominance: engagement of synonymous TCR by invariant CDR3 beta. Int. Immunol. 2000;12:747–756. doi: 10.1093/intimm/12.6.747. [DOI] [PubMed] [Google Scholar]
- 44.Pion S., Christianson G.J., Fontaine P., Roopenian D.C., Perreault C. Shaping the repertoire of cytotoxic T-lymphocyte responses: explanation for the immunodominance effect whereby cytotoxic T lymphocytes specific for immunodominant antigens prevent recognition of nondominant antigens. Blood. 1999;93:952–962. [PubMed] [Google Scholar]
- 45.Chen W., Norburry C.C., Cho Y., Yewdell J.W., Bennink J.R. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8(+) T cells at the levels of T cell repertoire and presentation of viral antigens. J. Exp. Med. 2001;193:1319–1326. doi: 10.1084/jem.193.11.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen W.M., et al. Study of antigen-processing reveals preferences explaining differential biological outcomes of two HLA-A2-restricted immunodominant epitopes from Human Immunodeficiency Virus type 1. J. Virol. 2002;76:10219–10225. doi: 10.1128/JVI.76.20.10219-10225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gileadi U., et al. Generation of an immunodominant CTL epitope is affected by proteasome subunit composition and stability of the antigenic protein. J. Immunol. 1999;163:6045–6052. [PubMed] [Google Scholar]
- 48.Mo A.X., van Lelyveld S.F., Craiu A., Rock K.L. Sequences that flank subdominant and cryptic epitopes influence the proteolytic generation of MHC class I-presented peptides. J. Immunol. 2000;164:4003–4010. doi: 10.4049/jimmunol.164.8.4003. [DOI] [PubMed] [Google Scholar]
- 49.Sewell A.K., et al. IFN-g exposes a cryptic cytotoxic T lymphocytes epitope in HIV-1 reverse transcriptase. J. Immunol. 1999;162:7075–7079. [PubMed] [Google Scholar]
- 50.Shastri N., Cardinaud S., Schwab S.R., Serwold T., Kunisawa J. All the peptides that fit: the beginning, the middle, and the end of the MHC class I antigen-processing pathway. Immunol. Rev. 2005;207:31–41. doi: 10.1111/j.0105-2896.2005.00321.x. [DOI] [PubMed] [Google Scholar]
- 51.Del Val M., Schlicht H.J., Ruppert T., Reddehase M.J., Kosinowski U.H. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell. 1991;66:1145–1153. doi: 10.1016/0092-8674(91)90037-y. [DOI] [PubMed] [Google Scholar]
- 52.Eisenlohr L.C., Yewdell J.W., Bennink J.R. Flanking sequences influence the presentation of an endogenously synthesized peptide to cytotoxic T lymphocytes. J. Exp. Med. 1992;175:481–487. doi: 10.1084/jem.175.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yellen-Shaw A.J., Eisenlohr L.C. Regulation of class I-restricted epitope processing by local or distal flanking sequence. J. Immunol. 1997;158:1727–1733. [PubMed] [Google Scholar]
- 54.Gileadi U., Gallimore A., Van der Bruggen P., Cerundolo V. Effect of epitope flanking residues on the presentation of N-terminal cytotoxic T lymphocyte epitopes. Eur. J. Immunol. 1999;29:2213–2222. doi: 10.1002/(SICI)1521-4141(199907)29:07<2213::AID-IMMU2213>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 55.Shastri N., Serwold T., Gonzalez F. Presentation of endogenous peptide/MHC class I complexes is profoundly influenced by specific C-terminal flanking residues. J. Immunol. 1995;155:4339–4346. [PubMed] [Google Scholar]
- 56.Draenert R., et al. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J. Exp. Med. 2004;199:905–915. doi: 10.1084/jem.20031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Milicic A., et al. CD8+ T cell epitope-flanking mutations disrupt proteasomal processing of HIV-1 Nef. J. Immunol. 2005;175:4618–4626. doi: 10.4049/jimmunol.175.7.4618. [DOI] [PubMed] [Google Scholar]
- 58.Seifert U., et al. Hepatitis C virus mutation affects proteasomal epitope processing. J. Clin. Invest. 2004;114:250–259. doi: 10.1172/JCI200420985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hammer G.E., Gonzalez F., James E., Nolla H., Shastri N. In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat. Immunol. 2007;8:101–108. doi: 10.1038/ni1409. [DOI] [PubMed] [Google Scholar]
- 60.York I.A., Brehm M.A., Zendzian S., Towne C.F., Rock K.L. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9202–9207. doi: 10.1073/pnas.0603095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deol P., Zaiss D.M., Monaco J.J., Sijts A.J. Rates of processing determine the immunogenicity of immunoproteasome-generated epitopes. J. Immunol. 2007;178:7557–7562. doi: 10.4049/jimmunol.178.12.7557. [DOI] [PubMed] [Google Scholar]
- 62.Yewdell J.W. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 63.Bodmer J.1996World distribution of HLA alleles and implications for disease. Ciba Found Symp. 197233–253; discussion 253–238. . [DOI] [PubMed] [Google Scholar]
- 64.Dalod M., et al. Weak anti-HIV CD8+ T-cell effector activity in HIV primary infection. . J. Clin. Invest. 1999;104:1431–1439. doi: 10.1172/JCI7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Altfeld M., et al. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J. Exp. Med. 2001;193:169–180. doi: 10.1084/jem.193.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allen T.M., et al. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J. Virol. 2004;78:7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harrer T., et al. Cytotoxic T lymphocytes in asymptomatic long-term nonprogressing HIV-1 infection. Breadth and specificity of the response and relation to in vivo viral quasispecies in a person with prolonged infection and low viral load. . J. Immunol. 1996;156:2616–2623. [PubMed] [Google Scholar]
- 68.Walker B.D., et al. Long-term culture and fine specificity of human cytotoxic T-lymphocyte clones reactive with human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 1989;86:9514–9518. doi: 10.1073/pnas.86.23.9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Butz E.A., Bevan M.J. Differential presentation of the same MHC class I epitopes by fibroblasts and dendritic cells. J. Immunol. 1998;160:2139–2144. [PubMed] [Google Scholar]
- 70.Kuckelkorn U., et al. The effect of the interferon-gamma-inducible processing machinery on the generation of a naturally tumor-associated human cytotoxic T lymphocyte epitope within a wild-type and mutant p53 sequence context. Eur. J. Immunol. 2002;32:1368–1375. doi: 10.1002/1521-4141(200205)32:5<1368::AID-IMMU1368>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 71.Morel S., et al. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity. 2000;12:107–117. doi: 10.1016/s1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]
- 72.Seifer U., et al. An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nat. Immunol. 2003;4:375–379. doi: 10.1038/ni905. [DOI] [PubMed] [Google Scholar]
- 73.Basler M., Youhnovski N., Van Den Broek M., Przybylski M., Groettrup M. Immunoproteasomes down-regulate presentation of a subdominant T cell epitope from lymphocytic choriomeningitis virus. J. Immunol. 2004;173:3925–3934. doi: 10.4049/jimmunol.173.6.3925. [DOI] [PubMed] [Google Scholar]
- 74.Moore M.W., Carbone F.R., Bevan M.J. Introduction of soluble protein into the class I pathway of antigen processing and presentation. Cell. 1988;54:777–785. doi: 10.1016/s0092-8674(88)91043-4. [DOI] [PubMed] [Google Scholar]
- 75.Beekman N.J., et al. Abrogation of CTL epitope processing by single amino acid substitution flanking the C-terminal proteasome cleavage site. . J. Immunol. 2000;164:1898–1905. doi: 10.4049/jimmunol.164.4.1898. [DOI] [PubMed] [Google Scholar]
- 76.Geier E., et al. A giant protease with potential to substitute for some functions of the proteasome. Science. 1999;283:978–981. doi: 10.1126/science.283.5404.978. [DOI] [PubMed] [Google Scholar]
- 77.Lauvau G., et al. Human transporters associated with antigen processing (TAPs) select epitope precursor peptides for processing in the endoplasmic reticulum and presentation to T cells. J. Exp. Med. 1999;190:1227–1240. doi: 10.1084/jem.190.9.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goulder P.J., Watkins D.I. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 2004;4:630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- 79.La Gruta N.L., et al. A virus-specific CD8+ T cell immunodominance hierarchy determined by antigen dose and precursor frequencies. Proc. Natl. Acad. Sci. U. S. A. 2006;103:994–999. doi: 10.1073/pnas.0510429103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sette A., et al. Optimizing vaccine design for cellular processing, MHC binding and TCR recognition. Tissue Antigens. 2002;59:443–451. doi: 10.1034/j.1399-0039.2002.590601.x. [DOI] [PubMed] [Google Scholar]
- 81.Le Gall S., et al. Distinct trafficking pathways mediate Nef-induced and clathrin-dependent major histocompatibility complex class I down-regulation. J. Virol. 2000;74:9256–9266. doi: 10.1128/jvi.74.19.9256-9266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kisselev A., Akopian T., Woo K.M., Goldberg A.L. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 1999;274:3363–3371.. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.