Abstract
In this issue of the JCI, two separate studies on MHC-bound peptides reopen the debate on the utility of peptides for the purposes of vaccination and treatment of autoimmune diseases. In the first study, by Wahlström et al., peptides bound to HLA-DR17 from bronchoalveolar lavage cells of sarcoidosis patients were analyzed in order to identify target antigens of the autoimmune response (see the related article beginning on page 3576). In the second study, by Le Gall et al., the modulation of epitope immunodominance and the processing and presentation of HIV peptides for MHC class I recognition were shown to be dependent on flanking residues that were N terminal to the natural epitopes (see the related article beginning on page 3563). Both studies highlight the tremendous therapeutic potential of MHC-bound peptides. They also emphasize that technical issues are still plaguing this field and hindering our understanding of MHC presentation in vivo.
The mid-1980s were the best of days for antigen presentation. In 1987, the determination of the structure of the human MHC class I molecule HLA-A2 struck the imagination of all immunologists (1). It provided the structural basis for the fact that T cells recognize peptides derived from antigens bound to MHC molecules and not antigens themselves (2). The conclusions that one could draw from this paradigm were to drive research for the next 20 years.
MHC class I peptides derive from cytoplasmic degradation by the proteasome of abortive translation products, viral proteins, and normal cytoplasmic proteins. Composed of 8 or 9 amino acids, these peptides are translocated into the ER by the transporter associated with antigen processing (TAP) and loaded onto nascent MHC class I polypeptides to allow their proper folding.
Peptides that appear in the groove of MHC class II molecules have a similar binding core (8–9 amino acid residues) but are extended at both ends, protruding outside the peptide-binding groove of the MHC class II molecule by 1–5 residues that are sometimes essential for either MHC binding and stability and/or T cell recognition. The production of MHC class II–binding peptides is almost exclusively limited to endosomal compartments and as such involves polypeptides that have reached this location through endocytosis, e.g., soluble proteins, receptor-bound molecules, viral particles, and opsonized bacteria. The simple fact that T cell recognition could be achieved simply via the detection of these short peptides was seen as the answer to vaccine design and antigen-specific immunomodulation. Countless ventures were initiated with these very aims, but none have brought to market a successful peptide-based immunotherapy. Why has this approach not worked?
Moving peptides into the therapeutic arena
At first sight, peptides are seductive — they are exquisitely specific for MHC binding and T cell recognition, easy and inexpensive to manufacture in large, good manufacturing practice–grade quantities, water soluble, and stable in solution. On their dark side, one would argue that their pharmacological properties are poor — they possess a short half-life due to proteolytic cleavage, show poor tissue distribution, and show poor subcellular targeting. However, peptides still appear to be the only practical vehicle to target T cell specificity and, although we all know about dramatic practical failures of their use, we still dream of using a peptide to modulate our favorite autoimmune disease and suppress the effects of aggressive T cells. Toward this goal, the search for antigens that are targets of self-reactive T cells is still very active.
A new approach in the search for the sarcoidosis antigen
In the current issue of the JCI, Wahlström et al. (3) describe a new technique for the analysis of HLA peptides in patients with the autoimmune disease sarcoidosis, an inflammatory condition characterized by formation of granulomas, most often in the lungs or lymph nodes, with the hope of identifying the specific antigen or antigens that trigger the excessive proliferative response of CD4+ T cells in patient lungs. Linkages between particular subtypes of the MHC class II cell surface receptor HLA-DR (e.g., HLA-DRB1*0301, investigated in the current study) and patient subgroups of this granulomatous lung disease have been identified (4). Sarcoidosis patients with the DRB1*0301 genotype have been shown to possess a distinct local immune response, which includes expansion of a limited set of CD4+ T cells expressing a particular TCR α chain variable gene segment (AV2S3) in their lungs (5). This observation strongly suggested that a limited number of peptides might be responsible for the selection and expansion of these particular T cells. Here comes the tricky part: how does one analyze the peptide repertoire responsible for this phenomenon? The lung offers a unique opportunity for study, given the possibility of recovering lung cells via bronchoalveolar lavage (BAL). However, BALs recover only a limited number of cells, and in the current study by Wahlström et al., BAL fluid cells from 16 patients had to be pooled in order to perform a single experiment, and no control group could be provided. Indeed, the analysis of MHC-bound peptides described in their report follows a long, multistep procedure that requires a minimum of 109 MHC-expressing cells. This large number of cells indicates how inefficient this technology still is. The most wasteful step in the process is the purification of MHC molecules. Affinity chromatography with anti-MHC antibodies is the only way to proceed and does not recover more than 50% of the target molecules. In addition, some peptides will be lost during the procedure because they have low or medium affinity for MHC or they have hydrophobic sequences that favor exchange in the presence of detergents. The final step, comprising capillary liquid chromatography followed by mass spectrometry (6), also leads to some attrition because some peptides will be lost or poorly separated by the chromatography or not ionized properly in the mass spectrometer chamber. Even though the theoretical and sometimes practical limits of detection of mass spectrometry have reached the femtomolar range, in instances like the present one, in which the starting material is highly heterogeneous and unambiguous sequence information is desired, much higher concentrations are required to allow sample reruns and definitive identifications. That series of limitations can only be compensated for by increasing the amount of starting material, which Wahlström et al. have done. The application of such an approach would be difficult to envisage in the case of examination of pancreatic tissue from patients with immune-mediated diabetes or of skin in individuals with psoriasis but could be carried out in individuals with rheumatoid arthritis using pannus cells isolated from patients undergoing surgery. Despite these challenges, in their current report, Wahlström et al. utilized this technique to identify some well-known autoantigens in the BAL fluid of sarcoidosis patients, such as vimentin, lysyl-tRNA synthase, and β actin, but the results did not reveal new potential candidates that could be the specific target of the autoimmune response in sarcoidosis. Prediction algorithms for MHC binding can be used to provide an indication that peptides identified by MHC purification/mass spectrometric techniques actually bind the HLA of interest, but a final confirmation can only come from direct binding studies with purified MHC molecules. Ultimately, the series of peptides purified from BAL fluid will have to be tested for their ability to stimulate the AV2S3-positive T cells present in the lung of patients with sarcoidosis. These studies could very well reveal that one of the peptides identified in the present study drives the autoimmune response in sarcoidosis and as such may be a target for immune intervention.
Flanking sequences alter HIV epitope processing
The report by Le Gall et al. (7), also in this issue of the JCI, which investigates the processing and presentation of HIV peptides for MHC class I recognition, did not present the technical difficulties of the Wahlström et al. (3) study. MHC class I epitopes have been classified as immunodominant or subdominant, depending on the magnitude of the immune response they generate. In HIV research, the factors governing epitope processing, presentation, and immunodominance have garnered great interest as these factors likely have an impact on recognition of HIV-infected cells by the immune system and also affect the immunogenicity of candidate HIV vaccines. A small number of immunodominant epitopes have been observed in HIV infection (8, 9); however, the existence of a relationship between epitope immunodominance and antigen processing has remained unclear. In their study, Le Gall and colleagues used synthetic peptides to study the presentation and processing to CD8+ T lymphocytes of epitopes originating from the HIV-1 Gag protein. Using a combination of synthesis, in vitro processing using cytoplasmic extracts, and antigen presentation, the authors established that N terminal–flanking residues were important for the efficient production of immunodominant epitopes whereas subdominant epitopes were often destroyed by the proteasome and that the artificial modification of flanking residues could transform a subdominant epitope into a dominant one.
The role of flanking residues in MHC class II–restricted antigen presentation has been long recognized and the focus of many studies (Figure 1A). Because the groove of MHC class II molecules is open at both ends, flanking sequences are free and exposed and can interact with either the MHC molecule and/or the TCR. Because the groove of MHC class I is self limiting, with closed ends, and acts as a caliper for 8- or 9-mer peptides, one would assume that flanking residues are unimportant for MHC class I epitopes. Wrong! Wrong for at least three reasons (Figure 1B). The first reason is the one that is illustrated in the study by Le Gall et al. (7). Both N and C termini of the epitope are critical for processing by the proteasome. C termini are often important for the initial binding and proteolytic cleavage by the proteasome, but the N terminus is also important to limit proteolysis. After proteasomal digestion, MHC class I peptides often have a definitive C terminal residue but not a final N terminus (10). The latter will be produced by aminopeptidases present in the cytoplasm and/or the ER, such as ERAP (11). The second important function of the termini of a peptide is for its translocation from the cytoplasm to the ER and preferential binding to the TAP transporter. Indeed, the three N terminal positions and the C terminal residue of cytoplasmic peptides are critical in the selection of substrates by TAP and their efficient transport (12). The final role of the termini of a peptide is in the process of its binding to MHC class I molecules. At 8, 9, and even 10 residues in length, peptide binding to MHC class I can be optimal and satisfy anchoring to pockets (13). Beyond this length, MHC does not exclude binding altogether, but binding becomes suboptimal by allowing only the extremities of the peptide to be associated with the MHC molecule. The peptide–MHC class I complexes that are produced by 12-, 13-, or even 14-mer peptides (14) are structurally unique with a central bulge that will be recognized and flattened by the T cell receptor upon binding (15). Thus, the flanking residues of an MHC class I epitope can influence processing, transport, MHC binding, and T cell recognition. These considerations are critical for the design of peptide-based vaccines, but their prediction is often difficult and requires in vivo models to be confirmed.
Figure 1. Role of flanking amino acid residues in MHC class I and class II peptide biology.
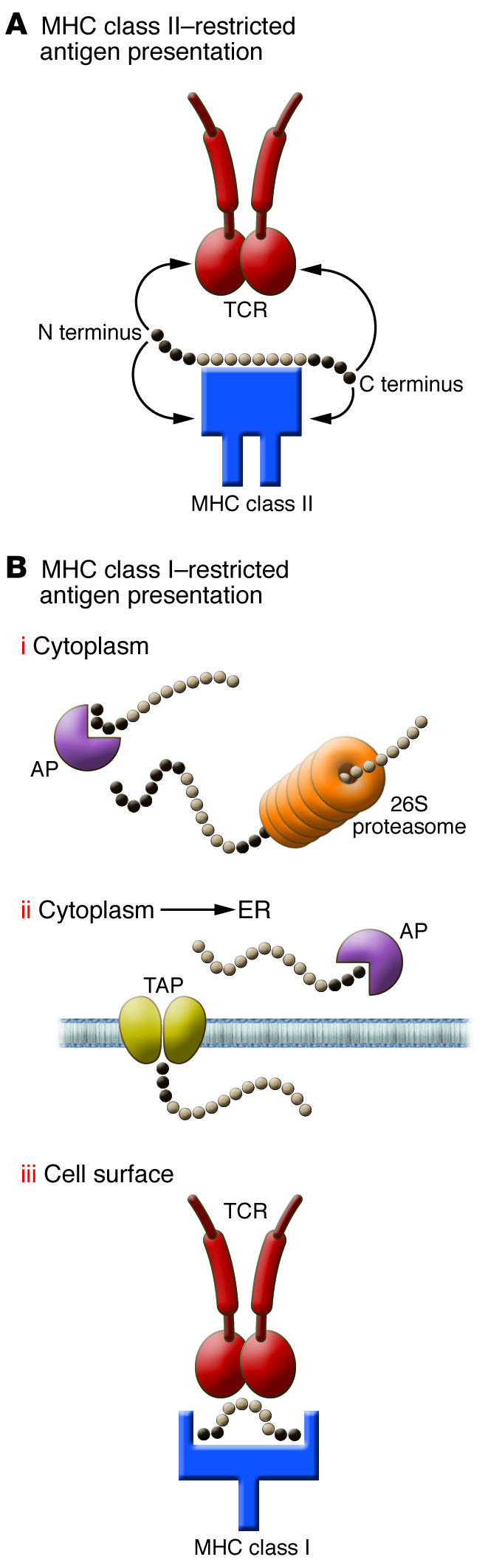
For MHC class II molecules (A), N and C terminal flanking residues protrude outside the MHC class II groove and are sometimes involved in MHC binding and/or TCR recognition. For MHC class I residues (B), amino acids N and C terminal of the 8- to 9-mer core peptide have multiple functions: (i) binding and processing by the proteasome and cytoplasmic aminopeptidases (AP); (ii) binding and transport into the ER by the TAP heterodimer and further digestion by ER aminopeptidases; and (iii) T cell recognition when a longer peptide is tethered by its extremities and bulging outside of the MHC class I groove. The observation made by Le Gall et al. (7) in this issue of the JCI is likely to be linked to steps i and ii, in which the N terminal sequence of the peptide will control the accessibility of aminopeptidases and the overall abundance of the final MHC class I peptide.
It is clear from the studies by Wahlström et al. (3) and Le Gall et al. (7) that appear in this issue of the JCI that the lessons we can learn from antigen presentation are some of the most challenging to translate into therapeutic approaches, but nobody should be afraid of a challenge that might bring huge returns.
Acknowledgments
L. Teyton is supported by the NIH.
Footnotes
Nonstandard abbreviations used: AV2S3, α chain variable gene segment 2.3; BAL, bronchoalveolar lavage; TAP, transporter associated with antigen processing.
Conflict of interest: The author has declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:3164–3166 (2007). doi:10.1172/JCI33997.
See the related article beginning on page 3576.
References
- 1.Bjorkman P.J., et al. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 2.Townsend A.R., et al. The epitopes of influenza nucleoprotein recognized by cytotoxic T lymphocytes can be defined with short synthetic peptides. Cell. 1986;44:959–968. doi: 10.1016/0092-8674(86)90019-x. [DOI] [PubMed] [Google Scholar]
- 3.Wahlström J., et al. Identification of HLA-DR–bound peptides presented by human bronchoalveolar lavage cells in sarcoidosis. J. Clin. Invest. 2007;117:3576–3582. doi: 10.1172/JCI32401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlin M., Fogdell-Hahn A., Olerup O., Eklund A., Grunewald J. HLA-DR predicts the prognosis in Scandinavian patients with pulmonary sarcoidosis. Am. J. Respir. Crit. Care Med. 1997;156:1601–1605. doi: 10.1164/ajrccm.156.5.9704069. [DOI] [PubMed] [Google Scholar]
- 5.Katchar K., Wahlstrom J., Eklund A., Grunewald J. Highly activated T-cell receptor AV2S3(+) CD4(+) lung T-cell expansions in pulmonary sarcoidosis. Am. J. Respir. Crit. Care Med. 2001;163:1540–1545. doi: 10.1164/ajrccm.163.7.2005028. [DOI] [PubMed] [Google Scholar]
- 6.Hunt D.F., et al. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255:1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 7.Le Gall S., Stamegna P., Walker B.D. Portable flanking sequences modulate CTL epitope processing. J. Clin. Invest. 2007;117:3563–3575. doi: 10.1172/JCI32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi H., et al. Structural requirements for class I MHC molecule-mediated antigen presentation and cytotoxic T cell recognition of an immunodominant determinant of the human immunodeficiency virus envelope protein. J. Exp. Med. 1989;170:2023–2035. doi: 10.1084/jem.170.6.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaul R., et al. CD8+ lymphocytes respond to different HIV epitopes in seronegative and infected subjects. . J. Clin. Invest. 2001;107:1303–1310. doi: 10.1172/JCI12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cascio P., Hilton C., Kisselev A.F., Rock K.L., Goldberg A.L. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001;20:2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan J., et al. In vivo role of ER-associated peptidase activity in tailoring peptides for presentation by MHC class Ia and class Ib molecules. J. Exp. Med. 2006;203:647–659. doi: 10.1084/jem.20052271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uebel S., et al. Requirements for peptide binding to the human transporter associated with antigen processing revealed by peptide scans and complex peptide libraries. J. Biol. Chem. 1995;270:18512–18516. doi: 10.1074/jbc.270.31.18512. [DOI] [PubMed] [Google Scholar]
- 13.Fremont D.H., Matsumura M., Stura E.A., Peterson P.A., Wilson I.A. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science. 1992;257:919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- 14.Probst-Kepper M., et al. Conformational restraints and flexibility of 14-meric peptides in complex with HLA-B*3501. J. Immunol. 2004;173:5610–5616. doi: 10.4049/jimmunol.173.9.5610. [DOI] [PubMed] [Google Scholar]
- 15.Tynan F.E., et al. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat. Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]


