Abstract
The dominant paradigm of protein engineering is structure-based site-directed mutagenesis. This rational approach is generally more effective for the engineering of local properties, such as substrate specificity, than global ones such as allostery. Previous workers have modified normally unregulated reporter enzymes, including β-galactosidase, alkaline phosphatase, and β-lactamase, so that the engineered versions are activated (up to 4-fold) by monoclonal antibodies. A reporter that could easily be “reprogrammed” for the facile detection of novel effectors (binding or modifying activities) would be useful in high throughput screens for directed evolution or drug discovery. Here we describe a straightforward and general solution to this potentially difficult design problem. The transcription factor p53 is normally regulated by a variety of post-translational modifications. The insertion of peptides into intrinsically unstructured domains of p53 generated variants that were activated up to 100-fold by novel effectors (proteases or antibodies). An engineered p53 was incorporated into an existing high throughput screen for the detection of human immunodeficiency virus protease, an arbitrarily chosen novel effector. These results suggest that the molecular recognition properties of intrinsically unstructured proteins are relatively easy to engineer and that the absence of crystal structures should not deter the rational engineering of this class of proteins.
Cells generally employ sensor proteins (also called “biosensors” or “switches”) to detect chemical stimuli and activate downstream components of signal transduction systems. We sought to fabricate artificial molecular sensors by engineering proteins that are specifically activated when bound or modified by novel effectors. Such sensors have practical utility in high throughput screens for drug discovery or directed protein evolution. They have also proved to be useful as research reagents. For example, two-hybrid systems (1, 2) and protein fragment complementation assays (3) couple the interactions of fusion proteins within transgenic cells to the production of signals. Cleverly designed sensors based upon fluorescent resonance energy transfer between green fluorescent protein analogues have also enabled the observation of intracellular protein modification events (4), including protein phosphorylation (5) and proteolysis (6). We expect that the utility of engineered protein sensors will continue to increase as they are deployed as diagnostic reagents (7) and pathogen-activated biotherapeutics (8, 9).
Rational protein design is generally synonymous with structure based site-directed mutagenesis (10). Reporter proteins are usually selected as starting points for sensor design because their structure-have been solved and because their activities are amenable to high throughput screening. Previous workers have inserted peptide epitopes into β-galactosidase (11), alkaline phosphatase (12), or β-lactamase (13). This approach has generally produced catalytically compromised enzymes that are activated up to 4-fold by antibody binding (14), presumably through allosteric mechanisms (7). It nevertheless remains difficult to predict whether the insertion of any peptide epitope into a particular position of a protein will generate the desired antibody-dependent activity. In contrast, natural selection has no bias in favor of proteins that crystallize readily or those with spectroscopically detectable activities. It has in effect generated vast numbers of proteins that are regulated through modification or binding, presumably through parsimonious evolutionary pathways. We therefore considered nature’s solutions to the problem of sensor design before formulating our own strategies.
By choosing globular and normally unregulated reporter enzymes such as β-galactosidase and alkaline phosphatase, protein engineers may be undertaking unnecessarily difficult design problems. In contrast, natural proteins that participate in signal transduction and gene expression tend to be intrinsically unstructured (15). The unbound forms of these proteins have been described as “beads on a flexible string,” where the beads are domains (often molecular recognition elements) connected by linkers (16). We hypothesized that intrinsically unstructured proteins are easy to engineer because of their inherent modularity and relative absence of functional constraint.
Our strategy was therefore to fabricate novel sensors by engineering an intrinsically unstructured protein. Sensors that are activated are preferable to those that are inactivated because the latter are more likely to produce “false positives” during high throughput screens. Lim and coworkers (17) previously reprogrammed the effector dependence and gating behavior of the neuronal Wiskott-Aldrich syndrome protein (N-WASP), which also contains unstructured regions (according to the DisProt data base, Ref. 18). N-WASP is modular in design and easily reprogrammed, but its activity (actin polymerization) is not particularly convenient to assay.
We chose the transcription factor p53 as a starting point for several reasons. It is an important tumor suppressor, so its structure and function are well understood (Fig. 1). Regions within the N and C termini of p53 are thought to be intrinsically unstructured (19, 20) and are therefore likely to accommodate almost any insertion. The wild-type p53 remains inactive in vitro until the C-terminal 30 amino acids are bound by an antibody, deleted, or phosphorylated (21). The exact mechanism of activation remains unclear (22) but can nevertheless be exploited. The sequence-specific DNA binding activity of p53 can be detected in vitro using labeled DNA probes or in vivo using reporter genes within yeast or mammalian cells (Fig. 1b).
FIGURE 1. p53 as a universal translator.
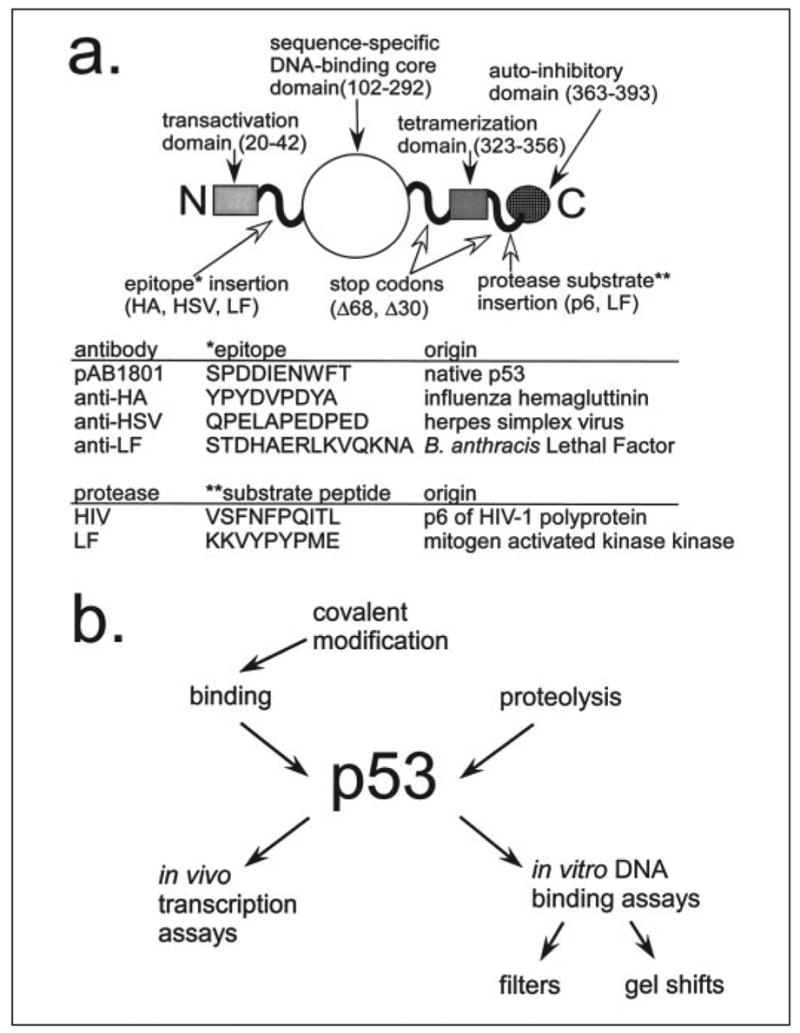
a, the domain structure of the wild-type p53 protein is shown with the unstructured regions represented as curvy lines. The peptide sequences inserted into the p53 protein are shown in the table below.b, the versatility of p53 as a sensor is represented as a flow chart.
p53 binds double-stranded DNA with two adjacent copies of the consensus sequence 5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′ (23), and its transcription activation activity in cultured cells is dependent on DNA binding (24). We and others employ the artificial p53CON target sequence, 5′-GGACATGCCCGGGCATGTCC-3′, which was isolated from a pool of random sequence double-stranded oligonucleotides (25). Active p53 protein can be expressed in Escherichia coli and is thus inexpensive and easy to produce. The high affinity of p53 for p53CON (KD = 5 ×10−10 m, (26)) enables highly sensitive assays. Active and latent p53 proteins differ greatly in sequence-specific DNA binding activity. In fact, the “activation factor” we observed of p53 (as high as ~100-fold increase in signal) greatly exceeds those of textbook allosteric enzymes (<70% increase, (27)). The signal produced by p53 in vitro remains constant once it reaches equilibrium, so assays are less time dependent and labor intensive than those that employ enzymes.
Here we have demonstrated the versatility of p53-based molecular sensors. Site-directed insertion mutagenesis and heterologous expression were used to fabricate p53 variants that display peptides recognized by proteases (human immunodeficiency virus (HIV)3 protease, Bacillus anthracis Lethal Factor) or monoclonal antibodies (three different epitopes). All of the p53 variants were specifically activated by their designated effectors in in vitro assays. These sensors have immediate utility in high throughput screens and could potentially be used in other applications (see “Discussion”). More importantly, this work has demonstrated a simple but effective alternative to structure-based site-directed mutagenesis for the fabrication of artificial sensors.
EXPERIMENTAL PROCEDURES
Materials
Expression vectors pET20b+, pET28a+, and pCDF Duet were from Novagen (Madison, WI). The p1+IQ HIV protease expression vector (ATCC number 68352) and the human p53 cDNA (ATCC number 57254) were obtained from the American Type Culture Collection. The Lethal Factor expression vector, pLF, was a gift from Dr. Stephen Leppla. E. coli strain BL21(DE3) Gold/pLysS was from Stratagene (La Jolla, CA). E. coli strain InvαF′was from Invitrogen. γ-labeled P-32 ATP was from MP Biomedicals (Irvine, CA) and Butterfly nitrocellulose membranes from Schleicher and Schuell. Oligonucleotides were synthesized by IDT (Coralville, IA); the IRD-700-labeled oligo was from LiCor (Lincoln, NE). The anti-p53 p53 monoclonal antibody (pAb1801) and the anti-Lethal Factor antibody (BAL0105) were from AbCam (Cambridge, UK). The HA antibody was from Covance (Princeton, NJ); the HSV antibody and purified Lethal Factor protease were from EMD Biosciences (San Diego, CA). The BigDye 3.1 DNA sequencing and GeneAmp XL long PCR kits were from Applied Biosystems (Foster City, CA).
Construction of p53 Variants
The human p53Δ30 gene from vector p53Δ30-pET20b+ (28) was fused to a sequence encoding an N-terminal His6 tag by subcloning p53Δ30 into pET28a+ (Novagen) using restriction enzymes NdeI and Hind III. The His6-p53Δ30 was then subcloned back into pET20b+ using XbaI and Hind III. The TEM-1 β-lactamase gene of the resulting His6-p53Δ30-pET20b+ was replaced with the kanamycin phosphotransferase gene from pET28a+ using BspHI, thereby creating His6-p53Δ30-pET20b+ (kanR). The 3′-end of the full-length wild-type p53 from pHp53B was subcloned into His6-p53Δ30-pET20b + using NcoI and BamHI, thereby creating the His6-wild-type p53-pET20b+ (kanR) vector.
The p53 to detect HIV protease cleavage (p53/p6) was created by replacing amino acids 360–369 with the p6 site by whole plasmid PCR (29) using the following 5′-phosphorylated primers (p6 encoding sequence underlined): 5′-CCTCAGATCACTCTGAAGTCCAAAAAGGGTCAGTCTACC-3′ (p53 p6–1) and 5′-GAAGTTAAAGCTTACTGGCTCCTTCCCAGCCTGGGCATC-3′ (p53 p6–2). The p53/-LF10 expression vector was similarly created using site-directed insertion of the recognition sequence at p53 codon 364. The primers used were 5′-CCGTATCCGATGGAAGCTCACTCCAGCCACCTGAAGTCCAA-3′ (LF p53) and 5′-CTCGAGATACACTTTCTTCCTGCTCCCCCCTGGCTCCTTCC-3′ (LF p53rev).
The p53-Δ 68 mutant was created by deleting the sequence encoding the tetramerization and activation domains of p53 in a whole plasmid PCR (29), using primers 5′-TAGGACGTCGAAGCCGAATTCCAGCACACTGGCG-3′ (p53Δ68) and 5′-ATCCAGTGGTTTCTTCTTTGGCTGGGG-3′ (p53Δ68rev). All antibody epitopes were inserted in place of the native pAb1801 epitope (codons 46 –55) by whole plasmid PCR as described above. The constructs were created using the following primers (epitope is underlined): His6-p53-HSV-Δ68 5′-CCGGAAGATCCGGAAGATGAAGACCCAGGTCCAGATGAAGCTCC-3′ (HSVp53out) and 5′-CGCCAGTTCCGGCTGCATCAAATCATCCATTGCTTGGGACGG-3′ (HSVp53rev); His6-p53-LF(ab)- Δ 68 5′-GCGTTCCGCATGATCGGTGCTCAAATCATCCATTGCTTGGGACGGCAA-3′ LFp53rev(Ab) and 5′-CTGAAAGTGCAGAAAAACGCGCCAGGTCCAGATGAAGCTCCCAGAATG-3′ (LFp53out(ab)).
Construction of Protease Expression Vectors
The inducible HIV protease expression vector, p1+IQ (lacZ−) was constructed as follows. The lacZ gene was excised from expression vector p1 +IQ (30) using BamHI; the remaining DNA was purified, self-ligated, and used to transform E. coli strain InvαF′. The PBAD-HIV PR-pCDF expression vectors were constructed in two stages. First, we made the PBAD-pCDF expression vector by subcloning the araC repressor and PBAD promoter from pBAD myc His A into pSL1180 using SphI and NcoI; the PBAD promoter was then subcloned from PBAD-pSL1180 into pCDF Duet using NcoI and XbaI. Second, the HIV protease gene in p1+IQ was PCR amplified, subcloned into pET28a+ using NdeI and Hind III, and sequenced to confirm its wild-type identity. The subcloning fused DNA encoding a hexahistidine tag to its 5′-end; this tag does not affect enzyme activity (31). The inactivating D25N mutation was introduced into His6-HIVPR-pET28 by whole circle PCR using primers 5′-GAAGCTCTATTAAATACAGGAGCAGATG-3′ (HIVPR-D25N-62) and 5′-CTTTAGTTGCCCCCCTATCTTTATTGTG-3′ (HIVPR-62out). The wild-type and D25N variants of the His6-HIV PR gene were subcloned from their respective His6-HIV PR-pET28 plasmids into PBAD-pCDF using NcoI and XhoI.
The Lethal Factor protease gene was cloned from the pLF7 vector. The signal peptide and an internal NcoI site was removed using two-step cloning (based on Park and Leppla, Ref. 32). The first PCR reaction used the primers 5′-AAAAAAACCATGGCGGGCGGTCATGGTGATGTAGG-3′ (5′-LF NcoI) and 5′-TTGAAGGTCCATGCAGTAATATAGAACGG-3′ (LF 2088rev). The second PCR reaction used primers 5′-CCGTTCTATATTACTGCATGGACCTTCAA-3′ (LF 2126) and 5′-TTTTTGGGCCCGGATCCTTATGAGTTAATAATGAAC-3′ (3′LF BamHI). The two products were combined in a third PCR reaction in which the entire Lethal Factor gene amplified using the external 5′-LF NcoI and 3′-LF BamHI primers. The Lethal Factor gene was then cloned into the pCDF Duet vector (Novagen) for the in vivo assays. All variants were sequenced using the Applied Biosystems Big Dye protocol at the Center for Fundamental and Applied Molecular Evolution (Emory University).
Protein Purification
E. coli BL21 (DE3) cells containing the plasmid pLysS were transformed with constructs that expressed the wild-type or engineered p53 genes fused to N-terminal six-histidine tags. The transformants were grown at 37 °C to mid-log (A600 = 0.3) and then induced with 0.5 mM isopropyl-thio-galactopyranoside (IPTG) and shaken at 23 °C for 4 h. The cells were spun down and stored as a pellet at −80 °C. The cells were lysed by sonication and the insoluble fraction removed by centrifugation. The protein was purified as described in the pET manual (Novagen) except that the binding, wash, and elution buffers were replaced with p53 binding buffer (10 mM Tris-HCl, pH 7.6, 50 mM NaCl, 10% glycerol, 0.1% Triton X-100, and 5 mM β-mercaptoethanol) without β-mercaptoethanol, plus 450 mM NaCl and 0, 60, and 1000 mM imidazole, respectively. The eluted p53 proteins were dialyzed in p53 binding buffer (plus 450 mM NaCl) and stored in p53 binding buffer (plus 45% glycerol and 450 mM NaCl) at −20 °C. The total protein concentration was quantified using the Bradford protein assay (Bio-Rad). HIV protease was purified and refolded according to an established protocol (31) and protein concentration also quantified using the Bradford protein assay. We used a commercially available fluorogenic substrate to show that our HIV protease was as active as those described in the literature (33).
The p53/p6 expression vector was co-expressed with HIV protease by induction with 0.5 mM IPTG for 4 h at 23 °C, whereas the p53/LF10 expression vector was co-expressed with Lethal Factor by induction with 0.5 mM IPTG for 18 h at 18 °C. The wild-type and engineered p53 proteins were expressed, either alone or co-expressed with HIV protease (from p1+IQ (30)) or LF protease (from LF-pCDF), and purified by immobilized metal affinity chromatography (IMAC) as described above. The purified proteins (2.0 μM) were incubated with 1.0 pM N-terminal infrared-labeled oligo p53CON, 50 ng of pLS1180, and 10 μg of bovine serum albumin in a final volume of 20 μl. After a 30-min incubation on ice, the mixtures were loaded onto 4% non-denaturing acrylamide gels and evaluated by electrophoretic mobility shift assay (EMSA) as described below.
EMSA Assays
The EMSAs were performed as described (34) except p53 binding buffer was used. The purified proteins (50 nM p53-Δ30, 10 nM p53-Δ68, 10 nM p53-HA-Δ68, 10 nM p53-LF(ab)- Δ68, 20 nM p53-HSV-Δ68) were incubated with 1.0 pM 5′-IRD-700-labeled oligo p53CON (5′-ATGGACATGCCCGGGCATGTCC-3′ (25)) and 0.25–1.0 μg of monoclonal antibodies in a total volume of 20 μl. After a 30-min incubation on ice, the mixtures were loaded onto 4% acrylamide gels (19:1 acrylamide to bisacrylamide, 0.33×TBE, 0.1% Triton X-100) and run at 200 V for 1 h at 4 °C. The gels were scanned using the LiCor Odyssey Infrared Imager; the intensities of the pixels within each band were quantified with the associated Odyssey software (version 1.1). The activation factors were calculated by dividing the intensities of the antibody/engineered p53/p53CON complexes by the intensity of the engineered p53/p53CON complex.
The in vitro protease assays were performed using purified p53 (p53/p6, p53/LF10, p53Δ30, or wild-type) and protease (HIV protease (31) or Lethal Factor) proteins. The purified p53 proteins (2 μM) were reacted with the HIV protease (10 μM) or LF (2 μM) proteases (EMD Biosciences) for 48 h in p53 binding buffer at 4 °C. Following the incubation, the p53 activity was determined by EMSA as described above.
High Throughput Assay
The screen for p53 function is based on a method developed by Singh et al. (35). E. coli strain BL21(DE3) cells carrying the T7 lysozyme expression vector, pLysS, were transformed with the p53 expression constructs and plated on Luria Broth (LB) plates containing 34 μg/ml chloramphenicol and 100 μg/ml kanamycin (LB-kan/chl). After 16 h of growth at 37 °C, the colonies were adsorbed onto a nitrocellulose filter and transferred colony-side-up to LB-kan/chl plates containing 0.5 mM IPTG to induce expression of p53. The colonies adsorbed to the nitrocellulose filter were induced at 23 °C for 4 h. The cells remaining on the original plate were regrown into full colonies by a further 8 h of incubation at 37 °C.
The cell membranes of the p53-expressing colonies were disrupted with chloroform gas for 15 min, giving the intracellular T7 lysozyme access to the peptidoglycan. The remaining manipulations were carried out in p53 binding buffer (described above) at 23 °C. The filters were treated with 2.5 units/ml DNase I (in p53 binding buffer plus 10 mM MgCl2) for 15 min, blocked with 5% nonfat dry milk (in p53 binding buffer plus 40 mM Tris-HCl, pH 7.6) for an hour, washed three times in binding buffer for 5 min each, and probed with 20 nM radiolabeled oligonucleotide containing the p53CON sequence (underlined) 5′-GTGGACATGCCCGGGCATGTCC (25) (plus 5 μg/ml denatured salmon sperm DNA) for 1 h. The fluorescent IRD700-p53CON probe apparently interacts non-specifically with endogenous E. coli proteins (data not shown) and is therefore unsuitable for the colony lift screen. The filters were washed four times more for 7.5 min each, and the quantity of probe bound to each filter was measured using a BAS-1000 Bio-imaging Analyzer System (Fujifilm Medical Systems USA, Stamford, CT).
RESULTS
Protease Activation of p53
In Vivo Activation
We first engineered p53 variants that were specifically activated by HIV protease or the B. anthracis Lethal Factor (a metalloprotease). These effectors were novel because p53 does not ordinarily recognize or respond to them. Both proteases catalyze the hydrolysis of peptide substrates but do not overlap in sequence or conformation specificity (36, 37). Site-directed insertion mutagenesis was applied to replace p53 codons 360–369, which encode an unstructured spacer upstream of the C-terminal autoinhibitory domain (Fig. 1), with a sequence encoding the HIV protease substrate (p6, VSFNFPQITL). Similarly the sequence encoding the Lethal Factor substrate (LF10, KKVYPYPME (37)) was inserted at codon 364. The engineered p53 variants (designated p53/p6 and p53/LF10) and the wild-type p53 gene were separately co-expressed in E. coli with either HIV protease, LF, or no protease.
The hexahistidine-tagged p53 proteins were purified by IMAC and analyzed by SDS-PAGE. The engineered protein migrated more quickly than the wild-type (at the same rate as the p53Δ30 control, which lacks its C-terminal domain), but only after co-expression with protease (data not shown). Equimolar quantities of the purified proteins were incubated with a double-stranded p53CON target sequence (25) conjugated to a near-infrared dye (IRD700-p53CON); the sequence-specific DNA binding activity of each was measured in an EMSA. The bands at the top of the gel reflect protein-DNA complexes, and their intensities are a measure of activation. Protease co-expression increased the apparent activity of the engineered p53 variants by ~30-fold for p53/p6 and a factor of >100-fold for p53/LF10. (Fig. 2a).
FIGURE 2. p53 is activated in vivo and in vitro by HIV and Lethal Factor protease.
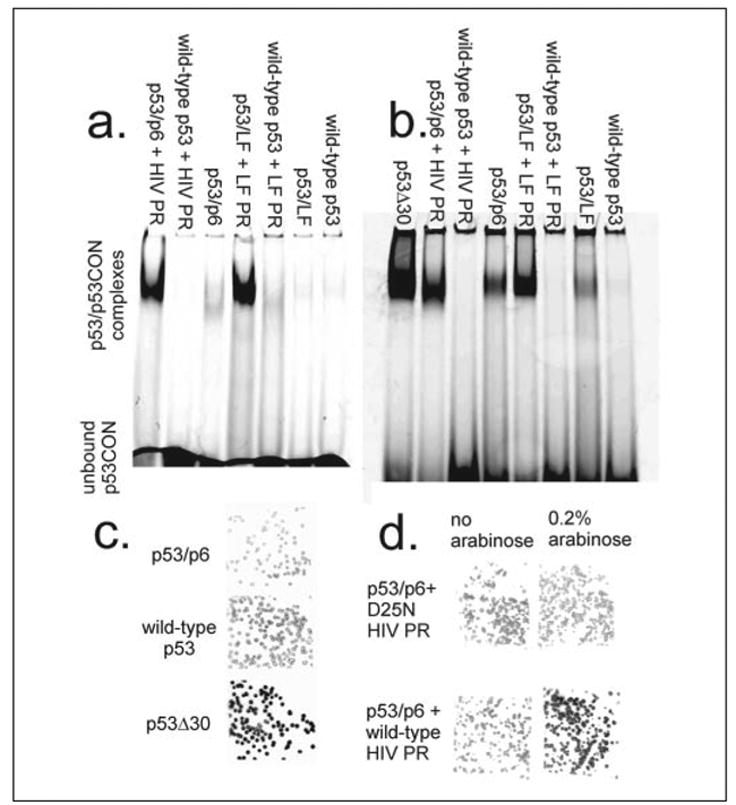
a, the engineered p53 variant or a wild-type control was co-expressed in E. coli with (or without) HIV or LF protease. The p53 proteins were purified by IMAC, and their sequence-specific DNA binding activity was measured in an EMSA. b, the engineered and wild-type p53 proteins were separately expressed in E. coli and purified by IMAC. Each of the full-length p53 proteins was reacted with purified HIV protease (31) or purified LF protease for 48 h in p53 binding buffer at 4 °C. The DNA binding activity of each variant was determined in an EMSA. c, the wild-type, activated (p53Δ30) and engineered (p53/p6) genes were induced within E. coli colonies. The colonies were lysed and probed with a radiolabeled double-stranded oligonucleotide encoding a p53 binding site (p53CON). d, E. coli were co-transformed with a p53/p6 expression vector and either PBAD-wild-type HIV PR-pCDF (bottom row) or PBAD-D25N HIV PR-pCDF (top row). The resulting colonies were induced with 0.5 mM IPTG (p53/p6 only, left column) or 0.5 mM IPTG plus 0.2% L-arabinose (p53/p6 and HIV PR, right column).
In Vitro Protease Activation
We confirmed that the proteases directly activated the engineered p53 variants through in vitro assays using purified proteins. The p53/p6, p53/LF10, and wild-type p53 proteins were separately expressed in E. coli and purified by IMAC; HIV protease was also separately expressed, mostly in inclusion bodies, solubilized in urea, purified by IMAC, and refolded (31). The purified p53 proteins were reacted with either purified HIV protease or Lethal Factor (EMD Biosciences); SDS-PAGE analysis confirmed the expected differences in migration after reactions with the proteases. EMSA analysis showed that Lethal Factor restored the activity of the engineered p53/ LF10 variant to that of p53Δ30, whereas the HIV protease elicited more modest ~2-fold activation in p53/p6 (Fig. 2b). The latter activation factor is apparently worse than that of the comparable in vivo reaction, most likely because the pH of in vitro assay (7.6) was optimized for p53 rather than HIV protease (38).
High Throughput Protease Screen
Several high throughput p53 assays have been reported (see “Discussion”), and here we have demonstrated the utility of the p53/p6 variant within a semi-in vivo filter-lift screen (28). E. coli BL21(DE3) Gold/ pLysS cells were separately transformed with p53/p6, p53Δ30, and wild-type p53 expression vectors. The transformed colonies were filter lifted onto plates supplemented with 0.5 mM IPTG. Expression of the p53 genes was induced at room temperature for 4 h; the colonies were lysed by exposure to chloroform gas and probed with a radiolabeled p53CON oligonucleotide. Phosphorimaging analysis showed that the p53Δ30 protein exhibited ~2-fold greater activity (comparing photoluminescence/mm2) than the wild-type or p53/p6 proteins (Fig. 2c), consistent with published in vitro results (34).
Next we co-transformed E. coli BL21(DE3) Gold/pLysS with the p53/p6 expression vector and either PBAD-HIV PR-pCDF or PBAD- D25N-HIV PR-pCDF. The latter vectors are identical except that the first produces the wild-type protease when induced with L-arabinose, whereas the other produces the catalytically inactive D25N mutant. The double transformants were propagated on LB-kan/spectinomycin/chl plates and then filter lifted onto plates containing 0.5 mM IPTG or 0.5 mM IPTG plus 0.2% L-arabinose. Colonies expressing the wild-type HIV PR exhibited ~2-fold greater p53 activity than those expressing the inactive D25N protease variant (Fig. 2d). We have already used this screen to detect and isolate p53 mutants with increased activity (28); the precision of the assay is more than sufficient to detect a 2-fold difference in activity. This result demonstrates the compatibility of the engineered p53 variants with existing high throughput screens. Both p53 and HIV protease are largely insoluble when expressed in E. coli, so the visualization of both activities within individual colony remnants indicates the acute sensitivity of p53-based sensors.
Antibody Activation of p53
We also designed p53 variants that are activated by monoclonal antibodies rather than proteases. These sensors would have immediate utility in high throughput screens for antibodies and other binding effectors. Our strategy was to engineer monomeric p53 variants that are dimerized by antibodies. The exploitation of this second activation mechanism would further demonstrate the modularity of p53. Engineered p53 monomers, which lack the entire tetramerization domain, exhibit 1000-fold less affinity for their DNA targets than does the tetrameric wild-type protein. Engineered p53 dimers bind their DNA targets with one-sixth the affinity of the wild-type tetramer (39). Induced dimerization of the monomer should therefore be easy to detect in EMSA and other binding assays (Fig. 1b).
To create a monomeric form of p53, we used site-directed deletion mutagenesis to remove the tetramerization and autoinhibitory domains (amino acids 324–393) (40). The resulting p53-Δ68 gene was subcloned into pET28, thus fusing it to DNA encoding an N-terminal hexahistidine tag. We then replaced the pAb1801 epitope (wild-type p53 amino acids 46–55) of His6-p53-Δ68 with a sequence encoding the HA epitope (YPYDVPDYA), the HSV epitope (QPELAPEDPED), or an epitope to the Lethal Factor (LF(Ab)-STDHAERLKVQKNA) (Fig. 1). All four His6-p53-Δ68 variants were separately expressed in E. coli and purified by IMAC.
The purified p53-Δ68 proteins were separately reacted with pAb1801 or the anti-epitope monoclonal antibodies and the IRD700-p53CON probe. The antibody-p53-DNA complexes were separated from the free DNA by EMSA (Fig. 3a). The p53-Δ68 variant had an ~80-fold increase in activity upon the addition of the pAb1801 Ab. p53-HA-Δ68 had an ~30-fold increase after adding the HA antibody, p53-HSV-Δ68 had an ~100-fold increase, and p53-LF(Ab)- Δ68 had a 3-fold increase in activity (Fig. 3b). We were not surprised to see two bands at the top of the gels, as both p53CON and the antibodies are capable of multimeric binding. We assayed our p53 variants at concentrations that were barely detectable in the absence of antibody and would have obtained even higher activation factors if we had employed the sensor at lower concentrations. The differences in the activation factors might be because of differences in the binding affinity of the antibody to the inserted tag. These values are significantly better than previously reported antibody sensors (14) and should enable assays with broader dynamic range.
FIGURE 3. Engineered p53 monomers are specifically activated by monoclonal antibodies.
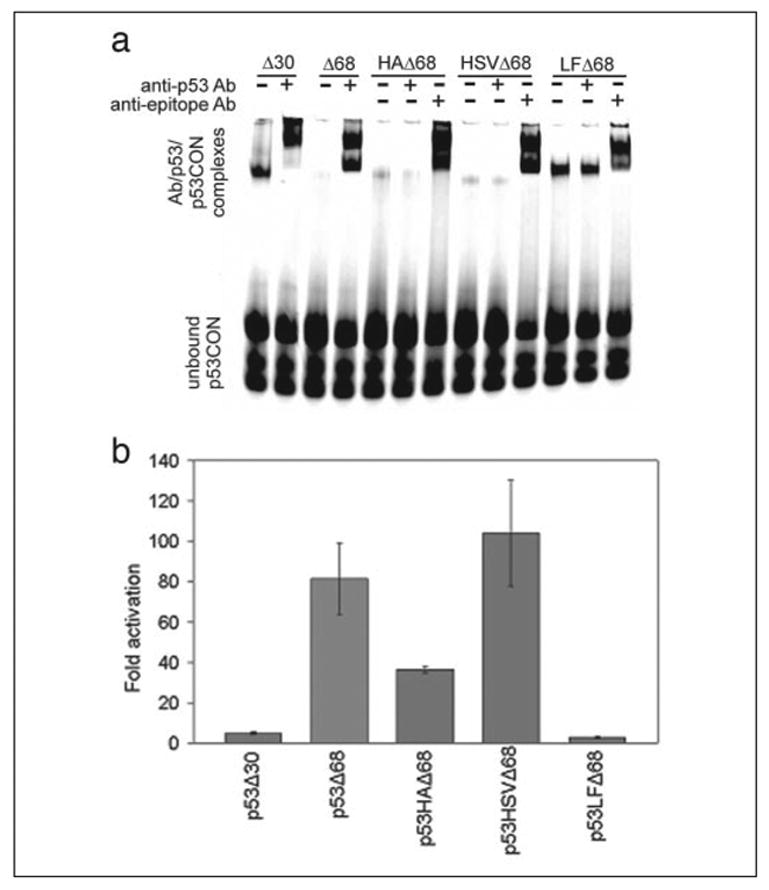
a, the monomeric p53-Delta;68 variants were expressed with N-terminal hexahistidine tags in E. coli. We then replaced the pAb1801 epitope (wild-type p53 codons 46–55) of His6-p53-Delta;68 with a sequence encoding the HA epitope, the HSV epitope, or an epitope to the Lethal Factor from B. anthracis (see “Experimental Procedures”). These p53 variants were purified and reacted with pAb1801 or the anti-HA, HSV, or LF monoclonal antibodies and an IRD700-labeled double-stranded p53 target sequence (p53CON). The antibody-p53-DNA complexes were separated from the free DNA by EMSA. b, quantification of the activation factors (n = 3). The pixel intensities of bands containing antibody/engineered p53/p53CON complexes were divided by those of bands associated with the corresponding engineered p53/p53CON (no antibody) complex.
DISCUSSION
We have designed p53 variants that are specifically activated by HIV protease, Lethal Factor, or monoclonal antibodies specific to the HA, HSV, and LF epitopes. These p53-based sensors are versatile with respect to both input molecular recognition and output signals. All of these inserted peptides differ significantly in amino acid sequence and conformation (Fig. 1a). HIV protease recognizes bent, hydrophobic peptides (36); Lethal Factor recognizes straight, basic peptides (37). Yet the p53/p6 and p53/LF10 proteins were specifically recognized, cleaved, and activated by HIV protease and Lethal Factor, respectively. We therefore believe that p53 could easily be reprogrammed for the detection of any protease or antibody. The latter is significant because modification-specific antibodies should enable p53-based high throughput screen for kinases, acetylases, and other protein-modifying activities.
With regard to applications, we showed here that p53 can be detected in EMSA assays and in high throughput colony lift assays. These results underscore the versatility of p53, because it is relatively difficult to control the amount of p53 expression within individual E. coli colonies. The wild-type p53 protein functions in vivo as a sensor of DNA damage, hypoxia, ribosome biogenesis, rNTP depletion, spindle damage, temperature shock, nitric oxide, and oncogene activation; these signals are mediated by an array of upstream regulatory proteins (41). In vivo p53 assays based on reporter gene activation have been developed in transgenic mammalian (42) and yeast (43) cells. The co-expression of p53-based sensors and engineered p53-binding proteins should similarly enable in vivo screens and selections for a wide variety of effectors.
The HIV protease-activated p53 variant also has potential as a therapy or “intracellular vaccine” for AIDS. Expression of the wild-type p53 protein normally leads to repression of transcription from the HIV-1 long terminal repeat (viral promoter; Ref. 44), as well as G1 growth arrest or premature apoptosis. The virus normally overcomes these pleiotropic effects by making a protein, Tat, that represses transcription of p53 (45). Expression of the HIV protease-activated variant in HIV-infected cells would have the following virtues as a gene therapy for AIDS. First, it is unlikely that a naturally occurring protein that requires activation by HIV-1-encoded factors will cause side effects. Second, p53 activity has graduated effects (repression of viral transcription, G1 growth arrest, apoptosis) that are less drastic than those of other “Trojan horse” therapies (8, 9). Third, these effects are mediated by cellular factors that inhibit viral replication, so it unlikely that HIV-1 could evolve immunity against the engineered p53.
We have also shown that p53 can display a variety of peptide sequences at two different locations. Previous workers have reprogrammed the effector dependence of N-WASP (17). We are therefore very optimistic about the prospects for the rational design of intrinsically unstructured proteins in general. An estimated 25–40% of all amino acid residues are thought to reside in unstructured domains (46), and many play important regulatory roles (16). We were initially reluctant to attempt rational design in the absence of a crystal structure but now encourage others to take advantage of the modularity and structural permissiveness of this functionally important class of proteins.
Acknowledgments
T. L. O. provided purified HIV protease; K. K. W. constructed the Lethal Factor expression vector and did the in vivo Lethal Factor experiments; I. M. constructed the HIV protease-activated p53 alleles and the HIV protease expression vectors and developed the screen (Fig. 2, c and d). We thank Dr. Stephen Leppla for providing the Lethal Factor gene and Dr. Andy Ellington for his ideas. We are grateful to Dr. Justin Gallivan, Shawn Desai, Dr. Marc Ostermeier, and Dr. Wayne Patrick for reading the manuscript.
Footnotes
This work was supported by NIAID, National Institutes of Health Grant 1 R21AI054602-01.
The abbreviations used are: HIV, human immunodeficiency virus; IMAC, immobilized metal affinity chromatography; EMSA, electrophoretic mobility shift assay; IPTG, isopropyl-thio-galactopyranoside; HA, hemagglutinin; HSV, herpes simplex virus; LF, lethal factor.
References
- 1.Baker K, Bleczinski C, Lin H, Salazar-Jimenez G, Sengupta D, Krane S, Cornish VW. Proc Natl Acad Sci U S A. 2002;99:16537–16542. doi: 10.1073/pnas.262420099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drees BL. Curr Opin Chem Biol. 1999;3:64–70. doi: 10.1016/s1367-5931(99)80012-x. [DOI] [PubMed] [Google Scholar]
- 3.Michnick SW. Curr Opin Biotechnol. 2003;14:610–617. doi: 10.1016/j.copbio.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Tsien RY. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 5.Violin JD, Zhang J, Tsien RY, Newton AC. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heim R, Tsien RY. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 7.Villaverde A. FEBS Lett. 2003;554:169–172. doi: 10.1016/s0014-5793(03)01160-8. [DOI] [PubMed] [Google Scholar]
- 8.Liu S, Redeye V, Kuremsky JG, Kuhnen M, Molinolo A, Bugge TH, Leppla SH. Nat Biotechnol. 2005;23:725–730. doi: 10.1038/nbt1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vocero-Akbani AM, Heyden NV, Lissy NA, Ratner L, Dowdy SF. Nat Med. 1999;5:29–33. doi: 10.1038/4710. [DOI] [PubMed] [Google Scholar]
- 10.Brannigan JA, Wilkinson AJ. Nat Rev Mol Cell Biol. 2002;3:964–970. doi: 10.1038/nrm975. [DOI] [PubMed] [Google Scholar]
- 11.Feliu JX, Villaverde A. FEBS Lett. 1998;434:23–27. doi: 10.1016/s0014-5793(98)00943-0. [DOI] [PubMed] [Google Scholar]
- 12.Brennan CA, Christianson K, La Fleur MA, Mandecki W. Proc Natl Acad Sci U S A. 1995;92:5783–5787. doi: 10.1073/pnas.92.13.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Legendre D, Soumillion P, Fastrez J. Nat Biotechnol. 1999;17:67–72. doi: 10.1038/5243. [DOI] [PubMed] [Google Scholar]
- 14.Feliu JX, Ferrer-Miralles N, Blanco E, Cazorla D, Sobrino F, Villaverde A. Biochim Biophys Acta. 2002;1596:212–224. doi: 10.1016/s0167-4838(02)00226-1. [DOI] [PubMed] [Google Scholar]
- 15.Tompa P. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 16.Dyson HJ, Wright PE. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 17.Dueber JE, Yeh BJ, Chak K, Lim WA. Science. 2003;301:1904–1908. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- 18.Vucetic S, Obradovic Z, Vacic V, Radivojac P, Peng K, Iakoucheva LM, Cortese MS, Lawson JD, Brown CJ, Sikes JG, Newton CD, Dunker AK. Bioinformatics. 2005;21:137–140. doi: 10.1093/bioinformatics/bth476. [DOI] [PubMed] [Google Scholar]
- 19.Bell S, Klein C, Muller L, Hansen S, Buchner J. J Mol Biol. 2002;322:917–927. doi: 10.1016/s0022-2836(02)00848-3. [DOI] [PubMed] [Google Scholar]
- 20.Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, Chang J, Sung YC, Choi KY, Han KH. J Biol Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- 21.Hupp TR, Lane DP. Curr Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 22.Ahn J, Prives C. Nat Struct Biol. 2001;8:730–732. doi: 10.1038/nsb0901-730. [DOI] [PubMed] [Google Scholar]
- 23.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 24.Kern SE, Pietenpol JA, Thiagalingam S, Seymour A, Kinzler KW, Vogelstein B. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 25.Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall AR, Milner J. Oncogene. 1995;10:561–567. [PubMed] [Google Scholar]
- 27.Berg JM, Tymoczko JL, Stryer L. Biochemistry. 5. W. H. Freeman and Company; New York: 2002. p. 266. [Google Scholar]
- 28.Matsumura I, Ellington AD. Protein Sci. 1999;8:731–740. doi: 10.1110/ps.8.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geddie ML, Matsumura I. J Biol Chem. 2004;279:26462–26468. doi: 10.1074/jbc.M401447200. [DOI] [PubMed] [Google Scholar]
- 30.Baum EZ, Bebernitz GA, Gluzman Y. Proc Natl Acad Sci U S A. 1990;87:10023–10027. doi: 10.1073/pnas.87.24.10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vickrey JF, Logsdon BC, Proteasa G, Palmer S, Winters MA, Merigan TC, Kovari LC. Protein Expression Purif. 2003;28:165–172. doi: 10.1016/s1046-5928(02)00650-2. [DOI] [PubMed] [Google Scholar]
- 32.Park S, Leppla SH. Protein Expression Purif. 2000;18:293–302. doi: 10.1006/prep.2000.1208. [DOI] [PubMed] [Google Scholar]
- 33.Matayoshi ED, Wang GT, Krafft GA, Erickson J. Science. 1990;247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 34.Hupp TR, Meek DW, Midgley CA, Lane DP. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 35.Singh H, Clerc RG, LeBowitz JH. BioTechniques. 1989;7:252–261. [PubMed] [Google Scholar]
- 36.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. Structure (Camb) 2002;10:369–381. doi: 10.1016/s0969-2126(02)00720-7. [DOI] [PubMed] [Google Scholar]
- 37.Turk BE, Wong TY, Schwarzenbacher R, Jarrell ET, Leppla SH, Collier RJ, Liddington RC, Cantley LC. Nat Struct Mol Biol. 2004;11:60–66. doi: 10.1038/nsmb708. [DOI] [PubMed] [Google Scholar]
- 38.Louis JM, Wondrak EM, Kimmel AR, Wingfield PT, Nashed NT. J Biol Chem. 1999;274:23437–23442. doi: 10.1074/jbc.274.33.23437. [DOI] [PubMed] [Google Scholar]
- 39.Weinberg RL, Veprintsev DB, Fersht AR. J Mol Biol. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 40.Hansen S, Hupp TR, Lane DP. J Biol Chem. 1996;271:3917–3924. doi: 10.1074/jbc.271.7.3917. [DOI] [PubMed] [Google Scholar]
- 41.Harris SL, Levine AJ. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 42.Zhang WW, Labrecque S, Azoulay E, Dudley R, Matlashewski G. J Biotechnol. 2001;84:79–86. doi: 10.1016/s0168-1656(00)00330-8. [DOI] [PubMed] [Google Scholar]
- 43.Ishioka C, Frebourg T, Yan YX, Vidal M, Friend SH, Schmidt S, Iggo R. Nat Genet. 1993;5:124–129. doi: 10.1038/ng1093-124. [DOI] [PubMed] [Google Scholar]
- 44.Subler MA, Martin DW, Deb S. J Virol. 1992;66:4757–4762. doi: 10.1128/jvi.66.8.4757-4762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li CJ, Wang C, Friedman DJ, Pardee AB. Proc Natl Acad Sci U S A. 1995;92:5461–5464. doi: 10.1073/pnas.92.12.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tompa P. BioEssays. 2003;25:847–855. doi: 10.1002/bies.10324. [DOI] [PubMed] [Google Scholar]


