Abstract
Human SFMBT (hSFMBT) is postulated to be a Polycomb (PcG) protein. Similar to other PcG proteins, we found that hSFMBT displays robust transcriptional repressor activity. In addition, hSFMBT localized to the nucleus where it strongly associates with chromatin by directly and selectively binding the N-terminal tail of histone H3. Importantly, we discovered that the four tandem MBT repeats of hSFMBT were sufficient for nuclear matrix-association, N-terminal tail H3 binding, and required for transcriptional repression. These findings indicate that the tandem MBT repeats form a functional structure required for biological activity of hSFMBT and predict similar properties for other MBT domain-containing proteins.
Keywords: SFMBT, Polycomb, transcription, chromatin, histone H3
1. Introduction
The ability to regulate the transcription of specific sets of genes is critical in determining cell fate. Importantly, these transcription patterns are propagated to progeny by epigenetic mechanisms that ensure the maintenance of cellular identity. Gene expression of developmental-associated genes is largely regulated by multimeric protein complexes that function to activate or repress transcription, both of which must operate in concert for proper differentiation. The best studied of these are regulatory complexes that contain the Polycomb group (PcG) or Trithorax (TRX) proteins [1]. The PcG proteins, which were first discovered in Drosophila, function to specify positional identity by creating a repressive chromatin structure at homeotic (Hox) genes resulting in their transcriptional silencing [2,3]. Currently there are three known PcG complexes: PRC1, PRC2 and the newly characterized PhoRC [4]. The main component of the PhoRC complex is Pleiohomeotic (Pho), a sequence-specific DNA binding protein that targets Polycomb response elements (PREs) in the genome [5]. Drosophila Pho was recently shown to heterodimerize with a novel PcG protein, known as SFMBT; which is required for Hox gene silencing [6].
The mammalian version of SFMBT was first cloned seven years ago, however, little is currently known about its biological function [7]. The translated protein contains four tandem MBT domains and a conserved protein-interacting SAM domain, which was first identified in the PcG gene Scm and is also found in both ph and l(3)mbt [8]. The MBT domain is evolutionarily restricted to metazoan lineages, is invariably found in tandem arrays of two to four repeats and proteins harboring MBT domains, such as Drosophila Sfmbt (dSfmbt), human SCML2 and L(3)MBT, have been linked to PcG silencing, although their function in these pathways remains elusive [6,9,10]. Outside of these observations, little else is known about the mammalian homologs of SFMBT.
To gain further insights into the biological significance of human SFMBT (hSFMBT), we investigated the structure and function of this protein. Consistent with its role as a putative PcG protein, hSFMBT specifically partitions to the nucleus and is a potent repressor of transcription. We discovered that hSFMBT strongly interacts with the nuclear matrix and that it also selectively binds histones H3 and H4, both in vitro and in vivo. Binding occurs at the N-terminal tail suggesting that hSFMBT functions to sequester transcriptionally inert chromatin at the nuclear periphery. Interestingly, we discovered that all four of the MBT repeats of hSFMBT were sufficient and necessary for nuclear matrix attachment, transcriptional repression and histone binding. In addition, all four MBT domains were required for repressor activity indicating that the higher-order structure formed by the four MBT repeats is essential for biological function. This is consistent With the structural characterization of the MBT repeats in human SCML2 and L(3)MBT, where the MBT domains fold cooperatively through interdigitation to form unique higher-order structures [9,11]. Lastly, we found that hSFMBT is preferentially expressed in certain cell types suggesting that it may be an important regulator of transcriptional programs during developmental and differentiation processes.
2. Materials and methods
2.1. Cell culture
HeLa, HEK-293 and K562 cells (ATCC) were cultured as previously described [12].
2.2 Plasmids
The human full length SFMBT cDNA clone (MGC:3342 IMAGE:3029598), 4xMBT (aa 20−453), ΔMBT3,4 (aa 20−235), ΔMBT1,4 (aa 125−351), ΔMBT1,2 (aa 239−453), ΔMBT4 (aa 20−351) and ΔMBT1 (aa 125−453) were produced by PCR amplification and inserted in-frame in the pCMX-Gal4-DBD plasmid [13], the pGEX-4T-1 plasmid (GE Healthcare) or pEGFP-C1 plasmid (Clontech).
2.3 Microscopy
Microscopy of HeLa cells was performed as previously described [12].
2.4 Antibodies
A synthetic peptide corresponding to amino acids 767−780 of human SFMBT (NP_057413) was used to immunize rabbits under the Standard Protocol (Zymed Laboratories). A 1:1000 dilution of sera was used for Western analysis and peptide competition experiments, as previously described [12]. The GFP antibody (ABCAM) was used 1:10,000, HP1β (Chemicon) was used 1:5000, RNAP II (Covance) was used 1:100,000, Gal4-DBD (Santa Cruz) was used 1:5000, H3 (ABCAM) and H4 (Upstate) were used 1:5000, Ubc9 (Santa Cruz) was used 1:5000, monoclonal FLAG (SIGMA) was used 1:10,000 and the GST antibody (Upstate) was used 1:500 for Western analysis. For immunoprecipitations, 20 μl of the Gal4-DBD antibody or 20 μl M2 FLAG beads (SIGMA) were used.
2.5 Transfections and reporter assays
HEK-293 cells were plated in 6-well plates (4x105 cells/ml) and co-transfected 24 hours later using Lipofectamine with Plus reagent (Invitrogen) and 100 ng of each Gal4-DBD fusion plasmid, 1 μg of reporter vector (pGK1-luc [14] or pSV40-luc [15]), 5 ng of pRL (Promega) and the pUC19 plasmid to bring the total DNA amount to 2 μg, according to the manufacture's instructions. Cell lysates were collected 48 hours post-transfection and luciferase activity was measured using Luciferase Assay Substrate (Promega) and a TopCount NXT microplate reader (Packard Bioscience). Quantitative measurements were obtained by normalizing to renilla luciferase activity. Standard error bars were generated by performing all experiments in triplicate.
2.6 Chromatin fractionation
Isolation of the S1, S2 and P fractions were performed as previously described with minor modifications [16]. Nuclei from 2x106 cells were isolated and resuspended in 300 μl nuclear buffer (20 mM Tris-HCl, pH 7.5, 70 mM NaCl, 20 mM KCl, 5 mM MgCl2, 3 mM CaCl2, and protease inhibitors), split equally into 4 aliquots and incubated with 6 units of micrococcal nuclease (Fermentas) at 37°C for 1min, 4min and 16min; digestion was terminated by adding EDTA and EGTA to a final concentration of 5 mM. Samples were centrifuged at 18,000 x g for 30”. The supernatant (S1) was collected and placed on ice while the nuclear pellet was lysed in 2 mM EDTA for 15 min at 4°C. The supernatant (S2) was collected following centrifugation and the pellet (P) was resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM EDTA, 0.5 mM SDS). Western analysis of the fractions was performed with the indicated antibodies.
2.7 GST pull-downs
pGEX-4T-1, pGEX-4T-1-SFMBT and pGEX-4T-1−4xMBT were transformed into BL21 E. coli (Novagen). Expression of recombinant GST fusion proteins was induced using 0.5 mM IPTG (Calbiochem) for 3 hours at 37°C. Proteins were purified using glutathione-sepharose 4B beads (GE Healthcare) and eluted in 10 mM reduced glutathione, 50 mM Tris pH 8.0, according to the manufacturer's protocol, dialyzed in PBS, quantitated and stored in 1 mM DTT and 1 mM PMSF at 4°C. Equal molar (100 pmol) amounts of each GST fusion protein was incubated with acid extracted histones [17], myelin basic protein or bovine serum albumin (EMD) in 400 μl PBS with 1% Triton X-100 (PBS-T) for 1 hour at 4°C prior to the addition of a 25 μl of pre-equilibrated glutathione-sepharose 4B beads. Following a 30' incubation at 4°C, beads were washed thoroughly with PBS-T and bound proteins were eluted in 40 μl elution buffer. One-eighth of the eluted material was used for Western analysis.
2.8 Immunoprecipitations
HEK-293 cells were collected 24 hours post-transfection, washed in PBS, resuspended in 300 μl lysis buffer (50 mM Tris pH 7.0, 150 mM NaCl, 0.5 mM DTT, 1% Triton X-100, protease inhibitors) and rotated for 30' at 4°C. Lysates were clarified by centrifugation at 18,000 x g for 1'. The supernatant was dialyzed in PBS for 1 hour at 4°C and incubated with either 20 μl Gal4-DBD antibody or 20 μl M2 FLAG-conjugated beads overnight at 4°C. Beads were washed with PBS, the bound material was eluted by boiling in SDS loading buffer and fractionated by SDS-PAGE prior to Western analysis.
3. Results
3.1 SFMBT is a conserved Polycomb-like protein
In order to gain insights into the possible biological functions of hSFMBT, we compared its amino acid composition and structure to other metazoan MBT-containing proteins. The human, mouse and rat SFMBT proteins are structurally similar, where each contains four N-terminal tandem MBT repeats and a SAM domain near the C-terminus. While Drosophila SFMBT (dSfmbt) retains the C-terminal SAM domain, its four tandem MBT repeats are located towards the C-terminal and dSfmbt contains a zinc finger motif which is lacking in mammals (not shown). Despite these structural differences, a sequence alignment of the MBT domains of dSfmbt, hSFMBT, and hL(3)MBT reveals a substantial degree of homology within their respective repeats (Fig. 1). A pairwise alignment of the individual repeats from the fly and human SFMBT proteins revealed that there is 48% sequence homology and 34% sequence identity between the two. This high degree of conservation strongly suggests that hSFMBT functions as a Polycomb-group protein, similar to dSfmbt [6].
Fig. 1. The MBT domains of dSfmbt, hSFMBT and hL(3)MBT are conserved.
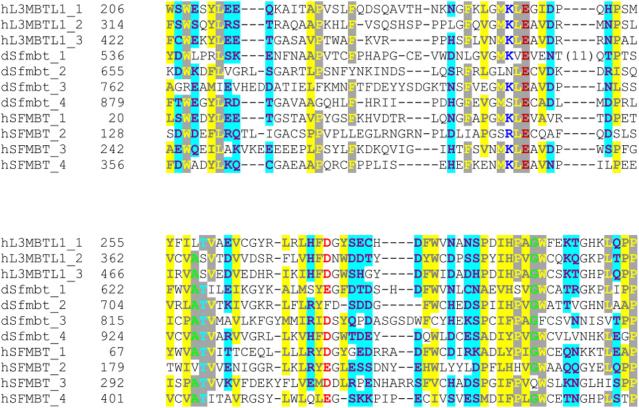
Sequences comprising the MBT repeats of each protein were obtained from the SMART Domain Database [40]. Sequences were aligned using ClustalX [41] and rendered using CHROMA [42]. Residues sharing sequence identity within the MBT repeats are denoted with a gray background, while conserved hydrophobic and hydrophilic residues are illustrated with yellow and cyan backgrounds, respectively. Conserved acid residues are red. The MBT repeats of fly and human SFMBT are 34% identical and 48% homologous.
3.2 hSFMBT is a cell type-specific nuclear protein
Since hSFMBT is a putative PcG protein, we hypothesized that it would function as a nuclear protein. To determine this, a GFP plasmid fused in-frame to full length wild type hSFMBT was created and transiently transfected into HeLa cells. Fluorescence microscopy of the cells revealed that GFP-SFMBT was enriched within nuclei as demonstrated by its co-localization with nuclear DAPI staining (Fig. 2A). In contrast, cells transfected with a GFP construct lacking an insert was evenly dispersed throughout the HeLa cells. The transfected HeLa cells were also fractionated into nuclear and cytoplasmic components and Western analysis of the fractions using a GFP antibody confirmed that the GFP-SFMBT fusion protein was specifically enriched within the nuclear compartment (Fig. 2B).
Fig. 2. Human SFMBT is a cell type-specific nuclear protein.

(A) Fluorescent microscopy of HeLa cells expressing GFP-SFMBT (top) or GFP-null (bottom) fusion proteins. GFP-SFMBT (green) specifically co-localizes with nuclear DAPI staining (blue) whereas GFP-null is dispersed evenly through the cell. (B) Western analysis of the nuclear (N) and cytoplasmic (C) fractions of HeLa cells expressing GFP-null or GFP-SFMBT fusion proteins. GFP-SFMBT selectively partitions to the nuclear compartment compared to GFP-null. (C) Western analysis of whole cell lysates from several common cell lines using a novel hSFMBT-specific antibody indicates that hSFMBT is expressed in a cell type-specific manner. (D) Western analysis of the nuclear (N) and cytoplasmic (C) fractions of K562 cells using the hSFMBT antibody confirms that the endogenous protein partitions to the nucleus.
To verify that endogenous hSFMBT partitioned to the nucleus, a novel polyclonal SFMBT-specific antibody was created (Supplemental Fig. 1) and used in Western analysis of whole cell lysates from several commonly used human cell lines. Interestingly, hSFMBT was only detected in specific cell types, mainly those of hematological origin (Fig. 2C). While the highest levels of hSFMBT were detected in the erythroblastic K562 and myeloblastic HL-60 cells, hSFMBT was also detected in the B-cell lymphoblastic Daudi cells. In contrast, hSFMBT was barely detected in epithelial cell lines derived from uterine (HeLa), breast (MCF7) and kidney (HEK-293) tissues. Since the K562 cells expressed the highest levels of hSFMBT, they were fractionated into nuclear and cytoplasmic components for Western analysis with the SFMBT antibody. Similar to the GFP-SFMBT fusion protein, we found that endogenous hSFMBT was also specifically enriched within the nuclear compartment (Fig. 2D). Collectively, these findings demonstrate that hSFMBT is a cell type-specific nuclear protein.
3.3 hSFMBT strongly associates with the nuclear matrix
To further dissect the sub-nuclear localization of hSFMBT, nuclei were isolated from K562 cells, as depicted in Fig. 3A, and were partially digested with micrococcal nuclease (MNase) for 1, 4 or 16 minutes before isolating the various chromatin components by centrifugation [16,18]. DNA analysis demonstrates that the MNase-sensitive soluble S1 fraction is mainly composed of mono- and dinucleosomal sized DNA fragments, typically associated with euchromatin (Fig. 3B). In contrast, the insoluble S2 fraction is composed of MNase-resistant oligonucleosomes, typically associated with heterochromatin, as observed by the higher molecular weight laddering. The P fraction represents the nuclear material that remains bound to the nuclear matrix. With increased MNase digestion time, more of the S2 and P fractions become soluble and shift into the S1 and S2 fractions, respectively. Western analysis of these fractions using the hSFMBT antibody revealed that endogenous hSFMBT selectively and strongly associates with the nuclear matrix as it failed to shift from the P fraction even at extended MNase digestion times (Fig. 3C). Similarly, RNAP II was found to preferentially associate with the nuclear matrix, as previously reported [19]. In contrast, the beta isoform of heterochromatin protein 1 (HP1β) was enriched in the S2 fraction but was liberated to the S1 fraction upon extended digestion with MNase [20]. These findings demonstrate that hSFMBT strongly interacts with the nuclear matrix.
Fig. 3. The four tandem MBT repeats mediate the strong association of hSFMBT with the nuclear matrix.

(A) Flowchart of nuclear fractionation assay. Nuclei from the indicated cells were subjected to partial micrococcal nuclease (MNase) digestion for 1, 4 or 16 minutes. Following digestion, the soluble euchromatic fraction (S1), insoluble heterochromatic fraction (S2) and matrix-associated fraction (P) were collected for analysis. (B) DNA from each fraction and a 100 bp ladder (L) were electrophoresed on a 2% agarose gel. Bands corresponded to the expected sizes of mono-, di-, tri- and oligonucleosomes are indicated. (C) Equal volumes of each fraction were separated by SDS-PAGE followed by Western analysis for hSFMBT, RNA polymerase II (RNAP II) or heterochromatin protein 1 (HP1β). hSFMBT is found exclusively in the P fraction and remains bound to the nuclear matrix even after extensive digestion with MNase, similar to RNAP II. (D) Nuclear fractionation assay performed with HEK-293 cells transfected with either a Gal4-DBD-4xMBT (top) or Gal4-DBD-null (bottom) plasmid. Western analysis using a Gal4-DBD antibody indicates that the four MBT repeats are sufficient for nuclear matrix attachment.
Due to the evolutionary conservation of the four tandem MBT domains (Fig. 1), we hypothesized that the MBT repeats were sufficient for nuclear matrix attachment. To test this, a Gal4-DBD fusion construct with an hSFMBT truncation mutant containing only the four MBT repeats (4xMBT) was transfected into HEK-293 cells for analysis (Fig. 3A). Similar to the findings for endogenous hSFMBT, the Gal4-DBD-4xMBT fusion protein was preferentially bound to the nuclear matrix while the Gal4-DBD-null plasmid was ubiquitously distributed amongst the fractions (Fig. 3D). Therefore, these findings indicate that the four MBT repeats of hSFMBT are sufficient for nuclear matrix attachment.
3.4 The four MBT repeats of hSFMBT are sufficient for potent transcriptional repression
Since SFMBT associates with the nuclear matrix, similar to RNAP II, we hypothesized that SFMBT could function as a co-activator of transcription. To test this hypothesis, the pGK1-luc reporter construct containing five tandem repeats of the Gal4 Upstream Activating Sequence (5xUAS) followed by a TATA box and luciferase reporter gene was employed as previously described (Fig. 4A) [21]. HEK-293 cells were co-transfected with pGK1-luc and Gal4-DBD fusion constructs containing either the CARM1 co-activator as the positive control [22], full length hSFMBT or 4xMBT; a Gal4-DBD-null vector served as the negative control. In addition, cells were co-transfected with the pRL reporter vector to normalize for transfection efficiency [23]. As predicted, the CARM1 co-activator increased luciferase gene expression by 7-fold compared to control (Fig. 4A). In contrast, both Gal4-DBD-SFMBT and Gal4-DBD-4xMBT failed to activate transcription but, instead, greatly reduced even basal levels of luciferase expression when compared to the Gal4-DBD-null negative control. These findings demonstrate that hSFMBT does not act as a co-activator of transcription and, rather, suggests that hSFMBT may function as a transcriptional repressor protein.
Fig. 4. All four MBT repeats of hSFMBT are required for potent transcriptional repression.

(A) In co-transfected HEK-293 cells the Gal4-DBD fusion protein binds the Gal4 Upstream Activating Sequence (UAS) of the pGK1-luc reporter construct and will promote luciferase transcription if it functions as a co-activator, such as CARM1. Comparative quantitative measurements were made by normalizing to renilla activity and plotting the fold change in luciferase activation relative to the Gal4-DBD-null negative control. Both hSFMBT and 4xMBT squelch basal luciferase expression when compared to control. (B) In the repression assay, the Gal4-DBD fusion protein binds the Gal4 UAS of the pSV40-luc reporter construct, which constitutively drives expression of luciferase, and will decrease luciferase transcription if it functions as a repressor, such as SMRT. Comparative quantitative measurements were made as described above and the fold decrease in luciferase activity was plotted relative to the Gal4-DBD-null negative control. Both hSFMBT and 4xMBT directly repressed luciferase transcription since a non-targeted FLAG-SFMBT construct failed to induce repression. (C) Deletion of a single MBT repeat restores luciferase transcription indicating that all four MBT repeats of hSFMBT are required to induce repression.
To test this hypothesis, the pSV40-luc reporter construct containing five tandem repeats of the Gal4 UAS (5xUAS) followed by an SV40 promoter that constitutively activates robust transcription of a luciferase reporter gene was employed as previously described (Fig. 4B) [24]. HEK-293 cells were co-transfected with the pSV40-luc reporter and the Gal4-DBD fusion constructs generated above. The Gal4-DBD-SMRT repressor protein served as the positive control for repression [25] and the pRL reporter vector was also used to normalize for transfection efficiency. Consistent with previous reports, the SMRT repressor protein produced a 5-fold decrease in transcription of the luciferase gene compared to the negative control (Fig. 4B). Interestingly, both Gal4-DBD-SFMBT and Gal4-DBD-4xMBT produced a more than 7-fold and 9-fold decrease in luciferase gene expression, respectively. The failure of a FLAG-tagged full length SFMBT construct to reduce luciferase activity in these assays confirmed that the observed repressive effects of Gal4-DBD-SFMBT were not indirectly due to ectopically expressed hSFMBT. These data indicate that hSFMBT is a potent repressor of transcription and that the four MBT repeats of SFMBT are sufficient to induce repression.
3.5 All four MBT repeats of hSFMBT are required for repressor function
To further define which of the MBT repeats were required for the observed repressive effects of hSFMBT, truncation mutants of the Gal4-DBD-4xMBT fusion construct were created, as depicted in Fig. 4C, and used in the repression assays. Initial studies of constructs lacking the two MBT repeats closest to the N-terminal (ΔMBT1,2) or C-terminal or (ΔMBT3,4) both resulted in a complete loss of repression. Therefore, we speculated that the two central MBT repeats were required for repression, however, these two repeats (ΔMBT1,4) alone were not sufficient to restore repression suggesting that an additional MBT repeat flanking either the N-terminal (ΔMBT4) or C-terminal (ΔMBT1) was also required. Interestingly, the lack of a single N- or C-terminal MBT domain also resulted in a complete loss of repression. The lack of repression could not be attributed to differences in expression of the fusion proteins (Supplemental Fig. 2). Collectively, these findings indicate that all four MBT repeats of hSFMBT are required to induce potent transcriptional repression.
3.6 hSFMBT binding to histones H3 and H4 is mediated by the four MBT repeats
Recent structure studies suggest that the MBT domain closely resembles the chromodomain of HP1 and Pc [11,26], which selectively bind methylated histone H3 lysine 9 and 27, respectively [27-29]. Consistent with this, it was recently demonstrated that Drosophila SFMBT binds the mono- and dimethylated forms of histones H3 lysine 9 and H4 lysine 20 in vitro (5). To explore the possibility that human SFMBT could bind histones in vivo, the Gal4-DBD-4xMBT or Gal4-DBD-null constructs were transfected into HEK-293 cells and immunoprecipitated with a Gal4-DBD antibody (Fig. 5A). Western analysis of the bound material indicates that the four MBT repeats of SFMBT could specifically bind endogenous histones H3 and H4 with a higher preference for H3. This was a specific interaction as Gal4-DBD-4xMBT failed to immunoprecipitate several ubiquitously expressed proteins, including UBC9 [30].
Fig. 5. hSFMBT preferentially binds histones H3 and H4.
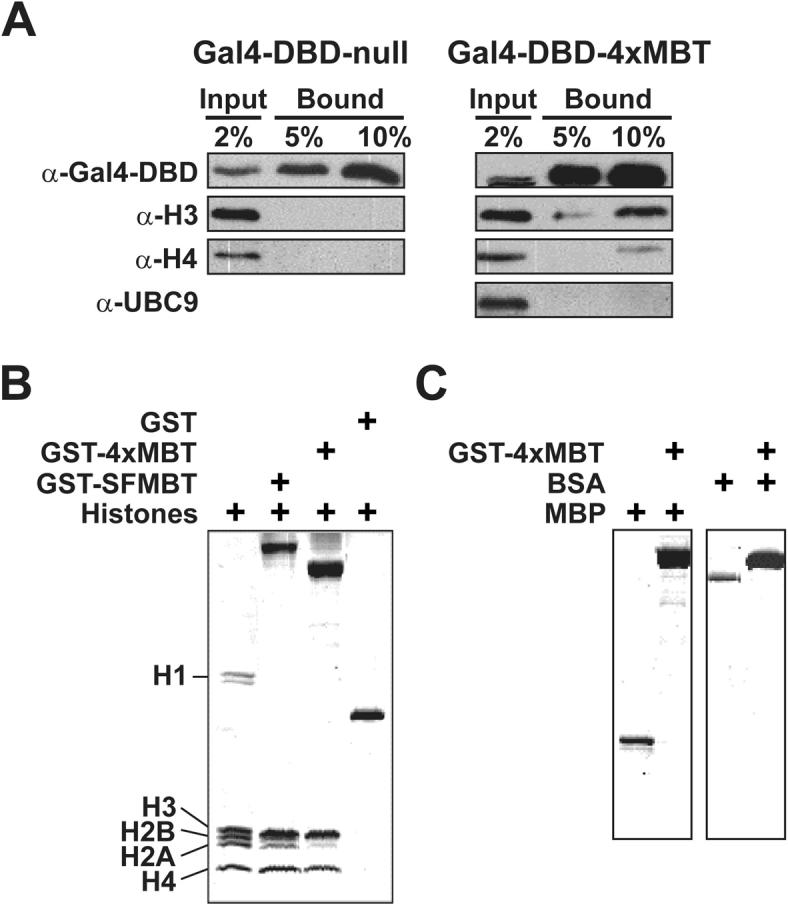
(A) Western analysis was performed with a general histone H3, H4 or UBC9 antibody on 2% of the input, 5% and 10% of the Gal4-DBD immunoprecipitated material from HEK-293 cells transfected with a Gal4-DBD-null or Gal4-DBD-4xMBT plasmid. The data demonstrate that the four tandem MBT repeats of hSFMBT (4xMBT) specifically immunoprecipitated histones H3>H4. (B) GST pull-down experiments with equal molar amounts of recombinant GST, GST-SFMBT or GST-4xMBT incubated with 50 μg of HEK-293 acid-extracted histones. Coomassie staining following SDS-PAGE demonstrates that the four MBT repeats of hSFMBT are sufficient for direct binding to histones H3 and H4. (C) GST pull-down experiments with recombinant GST-4xMBT and either myelin basic protein (MBP) or bovine serum albumin (BSA). GST-4xMBT failed to pull-down both MBP and BSA indicating a specific interaction between 4xMBT and histones H3 and H4.
To determine if hSFMBT directly binds histones, GST pull-down assays were performed using either purified recombinant GST, GST-SFMBT or GST-4xMBT and acid-extracted histones from the HEK-293 cells. While the negative control GST recombinant protein failed to pull-down histones, both GST-SFMBT and GST-4xMBT bound core histones with higher selectivity for H3 compared to H4, consistent with the in vivo findings (Fig. 5B). Both constructs also bound histones H2A and H2B, but to varying degrees and with far less affinity compared to H3 and H4. Histone H1 was not detected in any of the pull-down experiments. To confirm that the observed in vitro interaction was specific to histones, the experiments were repeated using GST-4xMBT with either myelin basic protein (MBP) or bovine serum albumin (BSA) – two highly positively charged proteins that mimic histones [31,32]. In both cases, GST-4xMBT failed to pull-down MBP or BSA (Fig. 5C). Collectively, these findings indicate that the four MBT repeats of hSFMBT directly and selectively bind histones H3 and H4 in vivo.
3.7 hSFMBT binds the N-terminal tail of histone H3
To determine the region of histone H3 responsible for interacting with the MBT repeats of hSFMBT, GST pull-down experiments were performed with purified recombinant GST or GST-4xMBT and either acid extracted histones or “tail-less” histones isolated from trypsinized HEK-293 oligonucleosomes [33]. Western analysis of the bound fractions using a general H3 antibody revealed that the four MBT repeats of hSFMBT failed to bind the H3 histone-fold region suggesting that the N-terminal tail is required for the interaction (Fig. 6A).
Fig. 6. The four MBT repeats of hSFMBT bind the N-terminal tail of histone H3.
(A) Acid-extracted histones or “tail-less” histones prepared from trypsinized nucleosomes were used as substrates for GST-null and GST-4xMBT pull-down experiments. Western analysis of the bound material using a general histone H3 antibody demonstrates that 4xMBT binding requires the N-terminal tail of H3. (B) HEK-293 cells were co-transfected with FLAG-SFMBT and either GST or GST-H3 1−41 and immunoprecipitated with FLAG conjugated beads. Western analysis using FLAG, GST, H3 or UBC9 antibodies on 4% of the input, 5% and 10% of the FLAG immunoprecipitated material indicates that FLAG-SFMBT specifically binds the H3 N-terminal tail. (C) Immunoprecipitations for GST were performed on the cell lysates in (B). Western analysis using GST, FLAG or UBC9 antibodies on 4% of the input, 5% and 10% of the GST immunoprecipitated material confirms that the first 41 amino acids of H3 bind hSFMBT.
To confirm this hypothesis, co-immunoprecipitations were performed in HEK-293 cells using the full length FLAG-SFMBT construct and an N-terminal GST fusion construct containing the first 41 amino acids of human H3 (H3 1−41) [34]. While Western analysis of the FLAG-SFMBT immunoprecipitated material demonstrated that GST alone did not interact with hSFMBT, the GST-H3 1−41 fusion protein did bind hSFMBT (Fig. 6B). Importantly, a FLAG-null immunoprecipitation failed to bind GST-H3 1−41 and the GST-H3 1−41 fusion protein did not bind the FLAG beads indicating a specific interaction (Supplemental Fig. 3). To confirm these observations, the experiment was repeated using the same lysates but immunoprecipitated with glutathione-sepharose beads instead of FLAG beads. Western analysis of the GST-H3 1−41 immunoprecipitated material verified binding to FLAG-SFMBT (Fig. 6C). This was a specific interaction as the GST-H3 1−41 failed to immunoprecipitate UBC9. Collectively, these finding demonstrate that four MBT repeats of hSFMBT specifically bind the N-terminal tail of histone H3.
4. Discussion
Although Drosophila SFMBT was recently found to be a PcG protein required for Hox gene repression, the biological function of human SFMBT remained unknown [6]. In this study we demonstrate for the first time that hSFMBT is a nuclear matrix-associated protein that acts as a potent repressor of transcription. Our novel findings indicate that hSFMBT binds the N-terminal tail of histone H3 suggesting that this interaction is required for targeting hSFMBT to specific chromatin regions destined for repression. The tissue-restricted expression of hSFMBT to certain cell lineages further suggests that it functions to repress sets of genes required for specific developmental and differentiation programs, similar to other PcG proteins. Due to its functional similarity to dSfmbt, it is likely that hSFMBT also plays a role in PcG-mediated gene repression.
The amino acid sequence comparison between human and fly SFMBT revealed a high degree of conservation in the four tandem MBT repeats, suggesting that this region plays a critical role in protein function. Consistent with this hypothesis, we found that the four tandem MBT repeats of hSFMBT were necessary and sufficient for nuclear matrix-association, histone binding and transcriptional repression. Importantly, the lack of any one of the four MBT domains resulted in the abolishment of repressor activity indicating that all four repeats are required to form a functional structural unit that is necessary for biological activity. These findings are consistent with the crystal structure of hL(3)MBT where each of its three MBT repeats formed tight globular modules that interdigitate to create a novel three-leaved propeller-like structure [26]. Based on these findings, it is likely that the four tandem MBT repeats of hSFMBT also create a novel propeller-like structure that is required for functional activity.
One possible role of this structure is to interact with other members of a putative multi-protein complex. This is likely since most PcG family members are part of larger multimeric chromatin-associated complexes that are required for long term silencing of developmental and oncogenic genes [35,36]. Another possibility is that the MBT domains bind to specific histone modifications. Based on its homology to the histone methyllysine-binding Chromo and Tudor domains, it was previously proposed that the MBT domain could, likewise, bind modified histone tails [37]. The ligand binding pockets of the Chromo and Tudor domains bind the methylated lysine via a hydrophobic cage created by conserved aromatic residues within the motif [28,38]. Homologous aromatic residues are found within the MBT domain suggesting its ability to bind methylated histones. Indeed, it was recently reported that the MBT repeats of hL(3)MBT bound dimethylated H4 lysine 20 and, importantly, that the four tandem MBT repeats of dSfmbt preferentially bound the mono- and dimethylated forms of H3 lysine 9 and H4 lysine 20 in vitro [6,39]. Therefore, it is possible that hSFMBT will also bind methylated histone tails; this will be determined in future experiments.
Acknowledgements
This work was supported by generous grants from the Robert E. and May R. Wright Foundation and the Donald E. & Delia B. Baxter Foundation. J.C.R. is a Pew Scholar in the Biomedical Sciences. R.C.T. is supported by National Institutes of Health (GM073839). The authors wish to thank Michael Stallcup (University of Southern California) for the pGK1-luc, Gal4-DBD and CARM1 plasmids; Mitchell Lazar (University of Pennsylvania) for the pSV40-luc, pCMX-Gal4-DBD and SMRT plasmids; Woojin An (University of Southern California) for the GST-H3 1−41 plasmid and members of the Rice Lab for helpful suggestions and comments.
Abbreviations
- SFMBT
Scm-related gene containing four mbt domains
- dSfmbt
Drosophila SFMBT
- hSFMBT
human SFMBT
- MBT
Malignant Brain Tumor
- SAM
Sterile Alpha Motif
- L(3)MBT
Lethal (3) Malignant Brain Tumor
- SCML2
Sex Comb on Midleg-Like 2
- PcG
Polycomb
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
REFERENCES
- 1.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–43. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 2.Bantignies F, Cavalli G. Cellular memory and dynamic regulation of polycomb group proteins. Curr Opin Cell Biol. 2006;18:275–83. doi: 10.1016/j.ceb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–70. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 5.Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–64. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 6.Klymenko T, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–22. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Usui H, Ichikawa T, Kobayashi K, Kumanishi T. Cloning of a novel murine gene Sfmbt, Scm-related gene containing four mbt domains, structurally belonging to the Polycomb group of genes. Gene. 2000;248:127–35. doi: 10.1016/s0378-1119(00)00131-1. [DOI] [PubMed] [Google Scholar]
- 8.Bornemann D, Miller E, Simon J. The Drosophila Polycomb group gene Sex comb on midleg (Scm) encodes a zinc finger protein with similarity to polyhomeotic protein. Development. 1996;122:1621–30. doi: 10.1242/dev.122.5.1621. [DOI] [PubMed] [Google Scholar]
- 9.Montini E, et al. Identification of SCML2, a second human gene homologous to the Drosophila sex comb on midleg (Scm): A new gene cluster on Xp22. Genomics. 1999;58:65–72. doi: 10.1006/geno.1999.5755. [DOI] [PubMed] [Google Scholar]
- 10.Wismar J, et al. The Drosophila melanogaster tumor suppressor gene lethal(3)malignant brain tumor encodes a proline-rich protein with a novel zinc finger. Mech Dev. 1995;53:141–54. doi: 10.1016/0925-4773(95)00431-9. [DOI] [PubMed] [Google Scholar]
- 11.Sathyamurthy A, Allen MD, Murzin AG, Bycroft M. Crystal structure of the malignant brain tumor (MBT) repeats in Sex Comb on Midleg-like 2 (SCML2) J Biol Chem. 2003;278:46968–73. doi: 10.1074/jbc.M306469200. [DOI] [PubMed] [Google Scholar]
- 12.Sims JK, Houston SI, Magazinnik T, Rice JC. A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J Biol Chem. 2006;281:12760–6. doi: 10.1074/jbc.M513462200. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Zamir I, Lazar MA. Differential recognition of liganded and unliganded thyroid hormone receptor by retinoid X receptor regulates transcriptional repression. Mol Cell Biol. 1997;17:6887–97. doi: 10.1128/mcb.17.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webb P, et al. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol Endocrinol. 1998;12:1605–18. doi: 10.1210/mend.12.10.0185. [DOI] [PubMed] [Google Scholar]
- 15.Harding HP, Lazar MA. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol Cell Biol. 1995;15:4791–802. doi: 10.1128/mcb.15.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rose SM, Garrard WT. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J Biol Chem. 1984;259:8534–44. [PubMed] [Google Scholar]
- 17.Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR- Set7 expression and its localization to mitotic chromosomes. Genes Dev. 2002;16:2225–30. doi: 10.1101/gad.1014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang SY, Garrard WT. Electrophoretic analyses of nucleosomes and other protein-DNA complexes. Methods Enzymol. 1989;170:116–42. doi: 10.1016/0076-6879(89)70044-6. [DOI] [PubMed] [Google Scholar]
- 19.Jackson DA, Cook PR. Transcription occurs at a nucleoskeleton. Embo J. 1985;4:919–25. doi: 10.1002/j.1460-2075.1985.tb03719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiru A, et al. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. Embo J. 2004;23:489–99. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J Biol Chem. 2006;281:8476–85. doi: 10.1074/jbc.M511093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–7. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 23.Rice JC, Massey-Brown KS, Futscher BW. Aberrant methylation of the BRCA1 CpG island promoter is associated with decreased BRCA1 mRNA in sporadic breast cancer cells. Oncogene. 1998;17:1807–12. doi: 10.1038/sj.onc.1202086. [DOI] [PubMed] [Google Scholar]
- 24.Ishizuka T, Lazar MA. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol Cell Biol. 2003;23:5122–31. doi: 10.1128/MCB.23.15.5122-5131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zamir I, Zhang J, Lazar MA. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997;11:835–46. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 26.Wang WK, Tereshko V, Boccuni P, MacGrogan D, Nimer SD, Patel DJ. Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets. Structure. 2003;11:775–89. doi: 10.1016/s0969-2126(03)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–81. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–3. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 29.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–8. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson VG, Rangasamy D. Intracellular targeting of proteins by sumoylation. Exp Cell Res. 2001;271:57–65. doi: 10.1006/excr.2001.5366. [DOI] [PubMed] [Google Scholar]
- 31.Farbiszewski R, Rzeczycki W. The digestion of basic proteins by extract of Guerin tumor lysosomes. Biochem Biophys Res Commun. 1975;65:280–5. doi: 10.1016/s0006-291x(75)80090-8. [DOI] [PubMed] [Google Scholar]
- 32.Moskaitis JE, Shriver LC, Campagnoni AT. The association of myelin basic protein with itself and other proteins. Neurochem Res. 1987;12:409–17. doi: 10.1007/BF00972291. [DOI] [PubMed] [Google Scholar]
- 33.Fletcher TM, Hansen JC. Core histone tail domains mediate oligonucleosome folding and nucleosomal DNA organization through distinct molecular mechanisms. J Biol Chem. 1995;270:25359–62. doi: 10.1074/jbc.270.43.25359. [DOI] [PubMed] [Google Scholar]
- 34.An W, Roeder RG. Direct association of p300 with unmodified H3 and H4 N termini modulates p300-dependent acetylation and transcription of nucleosomal templates. J Biol Chem. 2003;278:1504–10. doi: 10.1074/jbc.M209355200. [DOI] [PubMed] [Google Scholar]
- 35.Satijn DP, Otte AP. Polycomb group protein complexes: do different complexes regulate distinct target genes? Biochim Biophys Acta. 1999;1447:1–16. doi: 10.1016/s0167-4781(99)00130-x. [DOI] [PubMed] [Google Scholar]
- 36.Simon JA, Tamkun JW. Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr Opin Genet Dev. 2002;12:210–8. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- 37.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain 'Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 38.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–73. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–64. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodstadt L, Ponting CP. CHROMA: consensus-based colouring of multiple alignments for publication. Bioinformatics. 2001;17:845–6. doi: 10.1093/bioinformatics/17.9.845. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



