Abstract
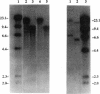
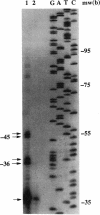
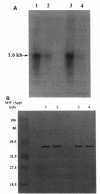
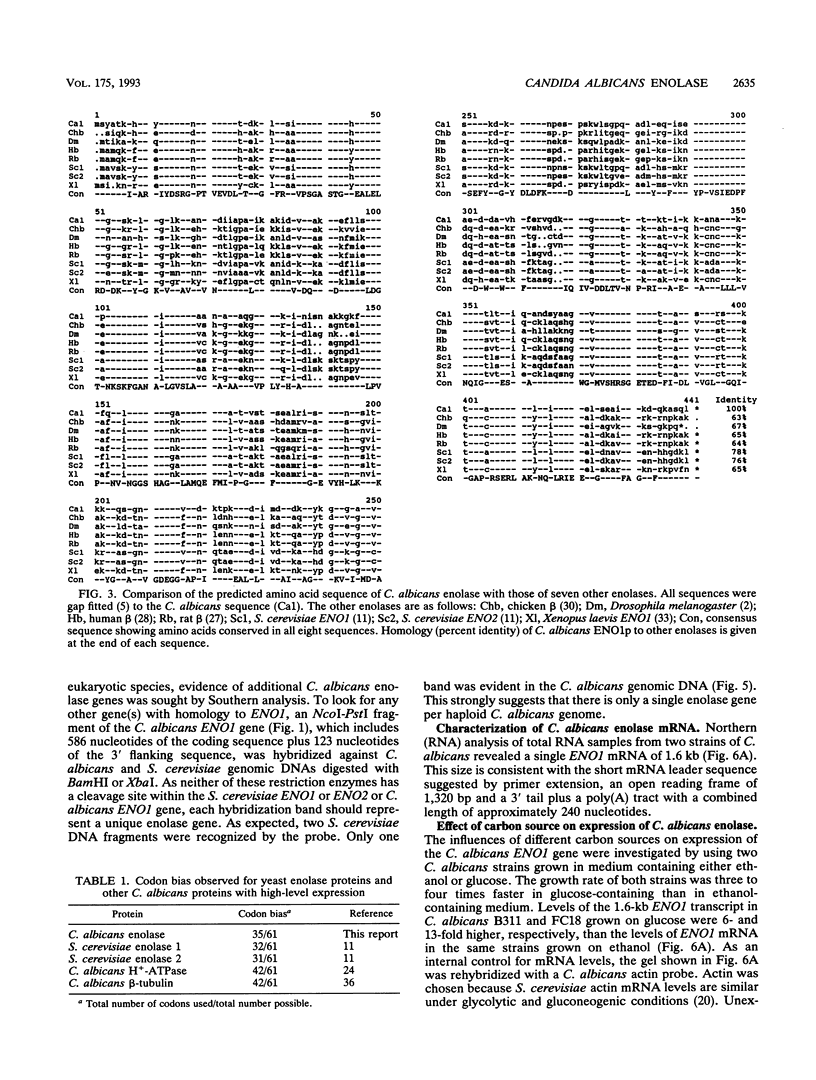
A DNA clone containing the putative Candida albicans enolase gene (ENO1) was isolated from a genomic DNA library. The sequenced insert contained a continuous open reading frame of 1,320 bp. The predicted 440-amino-acid protein is 78 and 76% identical, respectively, to Saccharomyces cerevisiae enolase proteins 1 and 2. Only one enolase gene could be detected in C. albicans genomic DNA by Southern analysis with a homologous probe. Northern (RNA) analysis detected a single, abundant C. albicans ENO1 transcript of approximately 1,600 nucleotides. When cells were grown on glucose, levels of ENO1 mRNA were markedly increased by comparison with ENO1 mRNA levels in cells grown on ethanol, a gluconeogenic carbon source. In contrast to this glucose-mediated transcriptional induction, the carbon source had no dramatic effect on the levels of enolase protein or enzyme activity in the C. albicans strains tested. These results suggest that posttranscriptional mechanisms are responsible for modulating expression of the C. albicans enolase gene.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. G., Corces V. G. The nucleotide sequence of a Drosophila melanogaster enolase gene. Nucleic Acids Res. 1990 Jan 11;18(1):191–191. doi: 10.1093/nar/18.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle P. K., Holland J. P., Willett C. E., Innis M. A., Holland M. J. Multiple factors bind the upstream activation sites of the yeast enolase genes ENO1 and ENO2: ABFI protein, like repressor activator protein RAP1, binds cis-acting sequences which modulate repression or activation of transcription. Mol Cell Biol. 1990 Sep;10(9):4872–4885. doi: 10.1128/mcb.10.9.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K. J., Hill W. B. Studies on the pink, adenine-deficient strains of Candida albicans. I. Cultural and morphological characteristics. Sabouraudia. 1970 May;8(1):48–59. [PubMed] [Google Scholar]
- Clifton D., Fraenkel D. G. The gcr (glycolysis regulation) mutation of Saccharomyces cerevisiae. J Biol Chem. 1981 Dec 25;256(24):13074–13078. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fling M. E., Kopf J., Tamarkin A., Gorman J. A., Smith H. A., Koltin Y. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol Gen Genet. 1991 Jun;227(2):318–329. doi: 10.1007/BF00259685. [DOI] [PubMed] [Google Scholar]
- Franklyn K. M., Warmington J. R., Ott A. K., Ashman R. B. An immunodominant antigen of Candida albicans shows homology to the enzyme enolase. Immunol Cell Biol. 1990 Jun;68(Pt 3):173–178. doi: 10.1038/icb.1990.24. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Kelly J. D., Cohen E. H. Transcription terminates in yeast distal to a control sequence. Cell. 1983 Jun;33(2):607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- Holland J. P., Labieniec L., Swimmer C., Holland M. J. Homologous nucleotide sequences at the 5' termini of messenger RNAs synthesized from the yeast enolase and glyceraldehyde-3-phosphate dehydrogenase gene families. The primary structure of a third yeast glyceraldehyde-3-phosphate dehydrogenase gene. J Biol Chem. 1983 Apr 25;258(8):5291–5299. [PubMed] [Google Scholar]
- Holland M. J., Holland J. P., Thill G. P., Jackson K. A. The primary structures of two yeast enolase genes. Homology between the 5' noncoding flanking regions of yeast enolase and glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1981 Feb 10;256(3):1385–1395. [PubMed] [Google Scholar]
- Kawaguchi Y., Honda H., Taniguchi-Morimura J., Iwasaki S. The codon CUG is read as serine in an asporogenic yeast Candida cylindracea. Nature. 1989 Sep 14;341(6238):164–166. doi: 10.1038/341164a0. [DOI] [PubMed] [Google Scholar]
- Kief D. R., Warner J. R. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Nov;1(11):1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft R., Tardiff J., Krauter K. S., Leinwand L. A. Using mini-prep plasmid DNA for sequencing double stranded templates with Sequenase. Biotechniques. 1988 Jun;6(6):544-6, 549. [PubMed] [Google Scholar]
- Lebioda L., Stec B. Crystal structure of enolase indicates that enolase and pyruvate kinase evolved from a common ancestor. Nature. 1988 Jun 16;333(6174):683–686. doi: 10.1038/333683a0. [DOI] [PubMed] [Google Scholar]
- Lee K. L., Buckley H. R., Campbell C. C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida Albicans. Sabouraudia. 1975 Jul;13(2):148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- Mason A. B., Brandt M. E., Buckley H. R. Enolase activity associated with a C. albicans cytoplasmic antigen. Yeast. 1989 Apr;5(Spec No):S231–S239. [PubMed] [Google Scholar]
- Matrisian L. M., Rautmann G., Magun B. E., Breathnach R. Epidermal growth factor or serum stimulation of rat fibroblasts induces an elevation in mRNA levels for lactate dehydrogenase and other glycolytic enzymes. Nucleic Acids Res. 1985 Feb 11;13(3):711–726. doi: 10.1093/nar/13.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer S. A., Dieckmann C. L. The yeast CBP1 gene produces two differentially regulated transcripts by alternative 3'-end formation. Mol Cell Biol. 1989 Oct;9(10):4161–4169. doi: 10.1128/mcb.9.10.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister L., Holland M. J. Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1985 Dec 5;260(28):15019–15027. [PubMed] [Google Scholar]
- McAlister L., Holland M. J. Targeted deletion of a yeast enolase structural gene. Identification and isolation of yeast enolase isozymes. J Biol Chem. 1982 Jun 25;257(12):7181–7188. [PubMed] [Google Scholar]
- McNeil J. B. Functional characterization of a pyrimidine-rich element in the 5'-noncoding region of the yeast iso-1-cytochrome c gene. Mol Cell Biol. 1988 Mar;8(3):1045–1054. doi: 10.1128/mcb.8.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk B. C., Kurtz M. B., Marrinan J. A., Perlin D. S. Cloning and characterization of the plasma membrane H(+)-ATPase from Candida albicans. J Bacteriol. 1991 Nov;173(21):6826–6836. doi: 10.1128/jb.173.21.6826-6836.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P. A., Bettany A. J., Brown J. P. Expression of a yeast glycolytic gene is subject to dosage limitation. Gene. 1990 Apr 30;89(1):85–92. doi: 10.1016/0378-1119(90)90209-a. [DOI] [PubMed] [Google Scholar]
- Moore P. A., Sagliocco F. A., Wood R. M., Brown A. J. Yeast glycolytic mRNAs are differentially regulated. Mol Cell Biol. 1991 Oct;11(10):5330–5337. doi: 10.1128/mcb.11.10.5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima Y., Mitsui H., Takayama Y., Kushiya E., Sakimura K., Takahashi Y. cDNA cloning and nucleotide sequence of rat muscle-specific enolase (beta beta enolase). FEBS Lett. 1989 Jan 2;242(2):425–430. doi: 10.1016/0014-5793(89)80515-0. [DOI] [PubMed] [Google Scholar]
- Peshavaria M., Hinks L. J., Day I. N. Structure of human muscle (beta) enolase mRNA and protein deduced from a genomic clone. Nucleic Acids Res. 1989 Nov 11;17(21):8862–8862. doi: 10.1093/nar/17.21.8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard J. J., Pelle R., Schonne E., Dworkin-Rastl E., Dworkin M. B. Tumorigenic Xenopus cells express several maternal and early embryonic mRNAs. Exp Cell Res. 1986 Nov;167(1):157–165. doi: 10.1016/0014-4827(86)90213-2. [DOI] [PubMed] [Google Scholar]
- Russell G. A., Dunbar B., Fothergill-Gilmore L. A. The complete amino acid sequence of chicken skeletal-muscle enolase. Biochem J. 1986 May 15;236(1):115–126. doi: 10.1042/bj2360115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M., Colthurst D. R., Wills N., McLaughlin C. S., Tuite M. F. Efficient translation of the UAG termination codon in Candida species. Curr Genet. 1990 Jun;17(6):487–491. doi: 10.1007/BF00313076. [DOI] [PubMed] [Google Scholar]
- Segil N., Shrutkowski A., Dworkin M. B., Dworkin-Rastl E. Enolase isoenzymes in adult and developing Xenopus laevis and characterization of a cloned enolase sequence. Biochem J. 1988 Apr 1;251(1):31–39. doi: 10.1042/bj2510031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. A., Allaudeen H. S., Whitman M. H., Koltin Y., Gorman J. A. Isolation and characterization of a beta-tubulin gene from Candida albicans. Gene. 1988;63(1):53–63. doi: 10.1016/0378-1119(88)90545-8. [DOI] [PubMed] [Google Scholar]
- Stec B., Lebioda L. Refined structure of yeast apo-enolase at 2.25 A resolution. J Mol Biol. 1990 Jan 5;211(1):235–248. doi: 10.1016/0022-2836(90)90023-F. [DOI] [PubMed] [Google Scholar]
- Strockbine N. A., Largen M. T., Zweibel S. M., Buckley H. R. Identification and molecular weight characterization of antigens from Candida albicans that are recognized by human sera. Infect Immun. 1984 Feb;43(2):715–721. doi: 10.1128/iai.43.2.715-721.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao S. G., Brunk C. F., Pearlman R. E. Hybridization of nucleic acids directly in agarose gels. Anal Biochem. 1983 Jun;131(2):365–372. doi: 10.1016/0003-2697(83)90185-9. [DOI] [PubMed] [Google Scholar]
- WESTHEAD E. W., MCLAIN G. A PURIFICATION OF BREWERS' AND BAKERS' YEAST ENOLASE YIELDING A SINGLE ACTIVE COMPONENT. J Biol Chem. 1964 Aug;239:2464–2468. [PubMed] [Google Scholar]
- Whelan W. L., Partridge R. M., Magee P. T. Heterozygosity and segregation in Candida albicans. Mol Gen Genet. 1980;180(1):107–113. doi: 10.1007/BF00267358. [DOI] [PubMed] [Google Scholar]