Abstract
Background
The low concentration and highly hydrophobic nature of proteins in lipid raft samples present significant challenges for the sensitive and accurate proteomic analyses of lipid raft proteins. Elimination of highly enriched lipids and interfering substances from raft samples is generally required before mass spectrometric analyses can be performed, but these procedures often lead to excessive protein loss and increased sample variability. For accurate analyses of the raft proteome, simplified protocols are needed to avoid excessive sample handling and purification steps.
Results
We have devised a simple protocol using a 'tube-gel' protein digestion that, when combined with mass spectrometry, can be used to obtain comprehensive and reproducible identification and quantitation of the lipid raft proteome prepared from neonatal mouse brain. Lipid rafts (detergent-resistant membranes using Triton X-100 extraction) prepared from neonatal mouse brain were directly incorporated into a polyacrylamide tube-gel matrix without prior protein separation. After in-gel digestion of proteins, nanospray LC-MS/MS was used to analyze the extracted peptides, and the resulting spectra were searched to identify the proteins present in the sample. Using the standard 'label-free' proteomics approach, the total number of MS/MS spectra for the identified proteins was used to provide a measure of relative protein abundances. This approach was successfully applied to lipid rafts prepared from neonatal mouse brain. A total of 216 proteins were identified: 127 proteins (58.8%) were predicted to be membrane proteins, or membrane-associated proteins and 175 proteins (~80%) showed less than a 2-fold variation in the relative abundance in replicate samples.
Conclusion
The tube-gel protein digestion protocol coupled with nanospray LC-MS/MS (TubeGeLC-MS/MS) offers a simple and reproducible method for identifying and quantifying the changes of relative abundances in lipid raft proteins from neonatal mouse brain and could become a useful approach for studying lipid raft proteins from various tissues.
Background
Lipid rafts are cholesterol- and sphingolipid-enriched specialized structures present in biological membranes [1-5] that can be isolated by various techniques. A common method for the isolation of the rafts is to prepare detergent-resistant membranes (DRMs) by extraction with the nonionic detergent Triton X-100 at cold temperature. Recent interest in lipid rafts arises from observations that some membrane proteins appear to partition preferentially into raft domains, and may require this environment for their biological activity [4,5]. Many previous studies have utilized two-dimensional gel electrophoresis (2DE) for proteomic profiling, but this method is limited by its lower sensitivity and it is often inefficient when analyzing raft proteins. Mass spectrometry (MS) has become a powerful tool for the analysis of complex protein mixtures. Proteomics profiling of either protein mixtures fractionated by 1DE or unfractionated protein mixtures by protease digestion and LC-MS/MS analysis has become increasingly popular. Peptides are identified by searching the resulting MS/MS spectra against protein sequence databases and protein presence is inferred from peptide presence. This general approach is referred to as 'top-down' or 'shotgun' proteomics. Several studies utilizing 1D gel filtration or in-solution protein digestion, combined with stable isotope labeling or label-free LC-MS/MS, have successfully profiled the protein composition and abundance in lipid rafts prepared from different biological sources [6-14]. However, quantitation of changes in the raft protein abundance under various experimental circumstances remains a major challenge. A number of technical factors are critical for analytical reliability, such as sample quality, reproducibility of the raft preparations, quality of the chromatography system, and the performance of the mass spectrophotometer. The most pressing problems for lipid raft proteomic investigations are those involving sample preparation and handling. Lipid raft samples prepared by different methods are composed of highly enriched lipids and low concentrations of hydrophobic proteins. Raft preparations also contain many non-proteinaceous substances including exogenous reagents, such as salts, buffers and detergents employed for sample preparation. These highly enriched lipids and non-protein components, or contaminants can often interfere with proteome analysis and their removal is a critical step before any proteome analysis can be performed. Although the low protein concentrations in raft samples do not present a limitation for analysis, methods used for removing lipids and other interfering substances from raft samples can lead to excessive protein loss. Thus, the process of lipid raft preparation suitable for mass spectrometry is a major factor in the variability of data obtained by these powerful proteomic techniques.
For accurate analyses of the raft proteome, a robust protocol avoids excessive purification steps, each of which lead to additional protein losses, is desirable. To avoid protein loss during sample preparation for mass spectrometry, a 'tube-gel' protein digestion protocol was adopted in which the lipid raft samples were directly incorporated into a polyacrylamide tube-gel without electrophoresis [15]. Detergents, lipids and other possible LC-MS/MS interfering materials in the raft samples are eliminated from the gel matrix while proteins are retained in the gel matrix. After the in-gel digestion of proteins, automated nanospray liquid chromatography tandem mass spectrometry (nanospray LC-MS/MS) is used to analyze the extracted peptides for protein identification. This protocol was used to analyze the protein profile of lipid rafts prepared from neonatal mouse brain. Neonatal mouse brain was chosen because there have been few proteomic studies of lipid rafts from neonatal brain [16-21]. Neonatal brain disorders are an important cause of mortality and morbidity contributing to the development of autism, cholesterol biosynthesis disorders, and a myriad of learning and developmental neurological and cognitive disabilities [22-27]. Developmental membrane defects have been postulated as one of the pathophysiological processes in these neonatal brain disorders. Additionally, the higher sterol content in brain tissue presents an additional challenge in preparing lipid raft samples for nanospray LC-MS/MS analysis.
Starting with limited amounts of frozen brain tissue, a total of 216 raft proteins were identified. Among the identified proteins, 127 (58.8%) were predicted to be plasma membrane (PM) or PM associated proteins including a number of authentic raft and/or GPI and lipid anchoring proteins, receptors, channel proteins, synaptic proteins, kinases, heterotrimeric G protein subunits, and some novel membrane proteins important for neurodevelopment. The major brain raft proteins, reported in previous investigations [8,18,19,28], were also identified as high abundance raft proteins in the present study. An advantage of this method is that it allows for raft proteins to be digested directly, dramatically reducing variations due to sample preparation prior to mass spectrometry. In this study, the standard 'label-free' proteomics approach in which total MS/MS spectral count is utilized to quantify the relative abundance of the identified proteins was used [29]. The results showed that the variations of relative abundance in ~80% of the identified proteins in replicate samples were less than 2-fold, suggesting that the method is highly reproducible. This approach offers a simple and reproducible protocol for identifying and quantifying changes in the relative abundance of the lipid raft proteins from neonatal mouse brain and could become a useful method for studying lipid raft proteins from various tissues.
Results and Discussion
Characterization of lipid rafts by sucrose density gradient ultracentrifugation
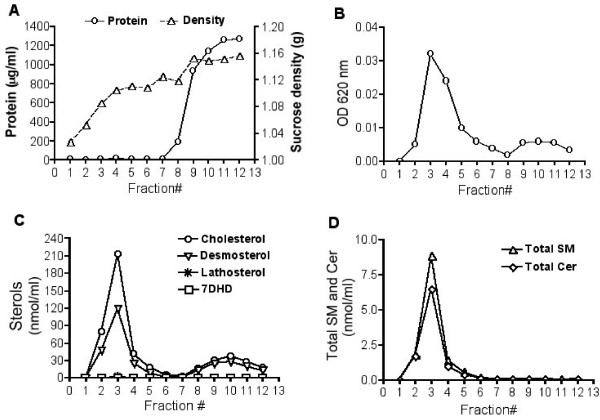
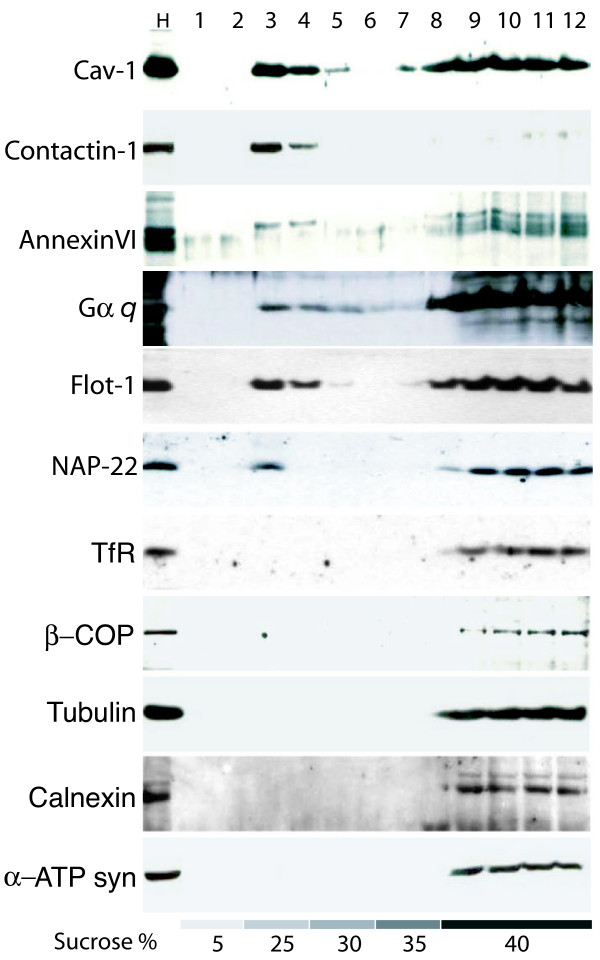
Biochemical isolation of lipid raft membranes by gradient ultracentrifugation, as well as their subsequent analysis, is a useful and simple method to determine if membrane components are located in raft microdomains. The distributions of total protein, sucrose density, and contents of sterols, sphingomyelin (SM) and ceramide (Cer), as well as the lighting-scattering properties at 620 nm, for each of the sucrose gradient fractions are summarized in Figure 1. Buoyant low density fractions 2–4 (DRMs/rafts) had the greatest light-scattering properties at 620 nm, consistent with a high content of lipids, but the non-raft fractions 8–11 had little or no absorbance. Conversely, most of the recovered proteins were present in the non-raft fractions and the total protein in the lipid raft fractions was too low to allow for accurate measurement by conventional methods. The lipid raft fractions 2~4 were highly enriched in sterols (a mixture composed of ~60% cholesterol, ~40% desmosterol, and trace amounts of other sterol precursors such as 7-dehydrodesmosterol and lathosterol), SM, and Cer compared to plasma membranes. Further characterization of the known raft and the non-raft marker proteins in the sucrose gradients was performed by immunoblotting (Figure 2). Known raft proteins, such as caveolin-1 (cav-1), flotillin-1 (flot-1), contactin-1 (Cntn-1), annexin -VI (Anx-VI, Anx6A), GTP-binding protein αq (Gαq), and NAP-22, were present in the low-density fractions (fractions 2~4). Various accepted non-raft markers, such as β-COP (a Golgi marker), transferrin receptor (TfR) (a non-raft membrane marker protein), α-tubulin (a cytoskeletal protein), calnexin (an ER resident membrane protein), and ATP synthase (a mitochondrial protein), were only present in the high-density fractions. Collectively, these results reflect the typical biochemical profiles of lipid rafts from brain tissue [8,30,31].
Figure 1.
Biochemical characterization of sucrose density gradient fractions of neonatal mouse brain. Panel A shows the distributions of protein and sucrose density in membrane fractions from sucrose gradients of neonatal brain. Panel B shows the light-scattering properties of each fraction by absorbance at 620 nm. The buoyant low density fractions 2~4 showed the greatest light-scattering properties at 620 nm, consistent with a high content of lipids. Panel C shows the distribution of sterols (cholesterol, desmosterol, lathosterol, and 7-dehydrodesmosterol (7DHD)) in each fraction from sucrose gradients of neonatal mouse brain and Panel D shows total sphingomyelin (SM) and ceramide (Cer).
Figure 2.
Localization of known raft and non-raft marker proteins in sucrose gradients. Post-nuclear homogenates (PNH) from neonatal brain tissues were extracted using 1% of Triton X-100 (TX) and fractionated in 5–40% discontinuous sucrose-density gradient as described in Methods. Twelve fractions of each 1.0 ml were collected from the top to bottom. Twenty μg of PNH protein (H) and equal 30 μl of each fraction of gradient were subjected to immunoblotting with antibodies against indicated proteins.
Analysis of core raft proteome in neonatal mouse brain by TubeGeLC-MS/MS
The core protein composition of lipid rafts from neonatal mouse brain was determined by using a tube-gel protein digestion coupled with nanospray LC-MS/MS (TubeGeLC-MS/MS) analyses. The major benefit of this modification is that the raft proteins (usually in limiting quantities) are digested in a tube-gel matrix without fractionation and purification. Thus, sample losses are minimized compared to in-gel digestion based on SDS-PAGE. Moreover, inclusion of detergents (Triton X-100 and SDS. See Methods) can facilitate the effective solubilization and denaturation of hydrophobic lipid raft membrane proteins [32,33]. After proteins are incorporated into the tube-gel, the detergents, lipids and other interfering substances can be efficiently eliminated by extensive washing with acetonitrile prior to protein enzymatic digestion and subsequent nanospray LC-MS/MS analysis, without any significant loss of the proteins that are trapped in the gel matrix [15]. This tube-gel approach has been successfully employed for high throughout mass spectrometric analysis of membrane proteins [15].
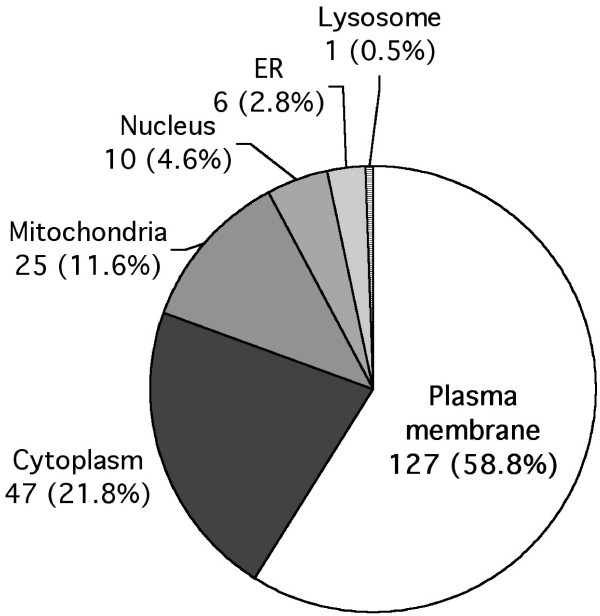
Two biological replicates (raft preparations from two neonatal brains), each with two MS technical replicates, were analyzed by TubeGeLC-MS/MS method. The peptides and corresponding proteins that were commonly identified in two biological samples (total 4 replicates) were considered as confident protein identifications. All identified proteins were then searched using UniProtKB/Swiss-Prot Release 52.3, TMHMM 2.0, and PubMed, to obtain information about their subcellular localization. The presence of predicted or verified transmembrane domains, glycosylphosphatidylinositol (GPI)-anchors and the lipid consensus sequences for myristoylation, pamitolyation, geranylgeranylation, farnesylation, and prenylation was used to classify proteins as either a membrane protein, or a membrane-associated protein [31]. Identified proteins were also analyzed by UniProtKB for the predicted presence of these motifs in order to provide an additional criterion for the evaluation. The overall experimental results for lipid raft core proteome of neonatal mouse brain thus characterized are shown in Figure 3. The complete lists of identified proteins, categorized as plasma membrane (PM) or PM-associated proteins and non-PM proteins, are shown in Tables 1 and 2, respectively.
Figure 3.
Cellular localization of identified proteins in lipid rafts from neonatal mouse brain. Cellular localization was annotated based on Gene Ontology (GO) terms and the PubMed literature database. The number of proteins and their percentage of the total identified proteins associated with each cellular location are indicated.
Table 1.
Plasma membrane associated proteins identified in the lipid rafts of neonatal mouse brain
| Acc. No. | Protein name | Pepa | % covb | MW (KDa) | PTMc | Spectral countd | SDe |
| P12960 | Contactin-1 (Neural cell surface protein F3) | 26 | 35.03 | 113.4 | GPI | 174.0 | 4.0 |
| Q91XV3 | Neuronal axonal membrane protein NAP-22 | 8 | 66.52 | 22.1 | Lipid anchor | 125.0 | 0.6 |
| P59216 | G-protein G(o), alpha subunit 1 | 7 | 25.28 | 40.1 | Lipid anchor | 101.0 | 2.0 |
| P14824 | Annexin A6 | 21 | 41.43 | 75.8 | 99.0 | 22.0 | |
| Q8BLK3 | Limbic system-associated membrane protein | 5 | 17.94 | 38.0 | GPI | 66.0 | 1.0 |
| Q80Z24 | Neuronal growth regulator 1 (Neurotractin) | 4 | 14.99 | 37.9 | GPI | 56.5 | 2.5 |
| Q61330 | Contactin-2 | 13 | 20.69 | 113.2 | GPI | 44.5 | 5.5 |
| P38401 | G-protein G(i), alpha-1 subunit | 5 | 19.03 | 40.4 | Lipid anchor | 42.5 | 10.5 |
| O08917 | Flotillin-1 | 8 | 29.04 | 47.5 | 40.0 | 0.0 | |
| Q8BFZ9 | SPFH domain-containing protein 2 | 3 | 11.30 | 38.9 | 38.0 | 1.2 | |
| Q9Z2S9 | Flotillin-2 (Reggie-1) (REG-1) | 6 | 22.22 | 41.7 | 36.5 | 1.5 | |
| P13595 | Neural cell adhesion molecule 1, 180 kDa isoform | 6 | 9.43 | 119.4 | 32.5 | 10.5 | |
| P38402 | G-protein G(i), alpha-2 subunit | 2 | 8.78 | 40.5 | Lipid anchor | 30.0 | 4.0 |
| Q9Z1G4 | Vacuolar proton pump subunit 1 | 7 | 9.43 | 96.5 | 28.5 | 0.5 | |
| Q62188 | Dihydropyrimidinase-related protein 3 (DRP-3) | 7 | 20.39 | 61.9 | 21.5 | 0.5 | |
| P27600 | G-protein alpha-12 subunit (G alpha-12) | 2 | 8.49 | 44.0 | Lipid anchor | 20.5 | 5.5 |
| O70443 | G-protein G(z), alpha subunit (G(x) alpha chain) | 4 | 17.85 | 40.9 | 19.5 | 1.5 | |
| Q99PJ0 | Neurotrimin | 3 | 11.95 | 38.0 | GPI | 17.0 | 1.0 |
| Q9WTR5 | Cadherin-13 | 4 | 9.26 | 78.3 | GPI | 17.0 | 2.0 |
| P21278 | G-protein alpha-11 subunit | 3 | 9.78 | 42.0 | 15.0 | 0.6 | |
| P51150 | Ras-related protein Rab-7 | 3 | 19.90 | 23.5 | Lipid anchor | 14.5 | 0.5 |
| Q8VDN2 | Sodium/potassium-transporting ATPase alpha-1 chain | 5 | 8.61 | 113.0 | 13.5 | 0.5 | |
| P32736 | Opioid-binding protein/cell adhesion molecule | 3 | 13.08 | 38.1 | GPI | 13.0 | 2.0 |
| P21279 | G-protein G(q), alpha subunit | 3 | 13.64 | 41.5 | Lipid anchor | 13.0 | 0.0 |
| P01831 | Thy-1 membrane glycoprotein precursor (Thy-1) | 3 | 26.71 | 18.1 | GPI | 12.5 | 0.5 |
| P59729 | Ras and Rab interactor 3 | 2 | 3.14 | 107.3 | 12.0 | 2.0 | |
| Q6TMK6 | G-protein G(I)/G(S)/G(T) beta subunit | 2 | 8.81 | 37.4 | 12.0 | 4.0 | |
| P06837 | Neuromodulin (Axonal membrane protein GAP-43 | 2 | 18.14 | 23.6 | Lipid anchor | 11.5 | 0.5 |
| Q6PIE5 | Sodium/potassium-transporting ATPase alpha-2 chain | 3 | 10.71 | 112.2 | 11.0 | 1.0 | |
| P62821 | Ras-related protein Rab-1A | 2 | 16.26 | 22.7 | Lipid anchor | 11.0 | 1.0 |
| P53994 | Ras-related protein Rab-2A | 3 | 20.38 | 23.5 | Lipid anchor | 10.5 | 0.5 |
| P38403 | G-protein G(k), alpha subunit | 3 | 13.64 | 40.6 | Lipid anchor | 10.0 | 2.0 |
| Q8BKV1 | Glypican-2 precursor | 2 | 5.88 | 63.3 | GPI | 10.0 | 2.0 |
| Q68FD5 | Clathrin heavy chain | 5 | 4.06 | 191.6 | 10.0 | 3.0 | |
| Q91X78 | SPFH domain-containing protein 1 | 6 | 24.48 | 38.9 | 9.5 | 2.5 | |
| P39688 | Proto-oncogene tyrosine-protein kinase Fyn | 2 | 3.57 | 59.9 | Lipid anchor | 9.5 | 1.5 |
| Q61735 | Integrin-associated protein (IAP) | 2 | 4.64 | 33.1 | 9.5 | 0.5 | |
| P97792 | Coxsackievirus and adenovirus receptor homolog | 2 | 6.04 | 39.9 | Lipid anchor | 9.5 | 1.5 |
| Q9WUC3 | Lymphocyte antigen Ly-6H precursor | 2 | 12.32 | 14.67 | GPI | 9.5 | 1.5 |
| Q6PIC6 | Sodium/potassium-transporting ATPase alpha-3 chain | 3 | 4.45 | 111.7 | 9.0 | 1.0 | |
| Q8BMT4 | Leucine-rich repeat-containing protein 33 | 2 | 2.75 | 77.1 | 9.0 | 1.0 | |
| P48036 | Annexin A5 | 5 | 21.07 | 35.8 | 9.0 | 1.0 | |
| P17182 | Alpha-enolase | 3 | 11.57 | 47.1 | 9.0 | 0.6 | |
| O08532 | L-type calcium channel subunit delta | 2 | 2.75 | 124.6 | 8.5 | 2.5 | |
| P63044 | Vesicle-associated membrane protein 2 (VAMP-2) | 3 | 35.09 | 12.7 | 8.0 | 0.0 | |
| P54227 | Stathmin (Phosphoprotein p19) | 2 | 14.97 | 17.3 | 8.0 | 1.0 | |
| P61027 | Ras-related protein Rab-10 | 3 | 16.58 | 22.5 | Lipid anchor | 8.0 | 1.0 |
| Q9QZF2 | Glypican-1 precursor | 3 | 8.99 | 61.4 | GPI | 8.0 | 2.0 |
| Q3U1F9 | Transmembrane phosphoprotein Cbp | 2 | 7.48 | 46.5 | Lipid anchor | 7.0 | 0.6 |
| Q69Z26 | Contactin-4 | 3 | 2.15 | 117.5 | GPI | 7.0 | 1.0 |
| Q9R0N7 | Synaptotagmin-7 (Synaptotagmin VII) (SytVII) | 2 | 7.71 | 45.5 | 6.5 | 0.5 | |
| Q9Z0P4 | Paralemmin | 3 | 10.73 | 41.6 | Lipid anchor | 6.5 | 1.5 |
| O35454 | Chloride channel protein 6 (ClC-6) | 4 | 6.56 | 97.0 | 6.5 | 1.5 | |
| P51863 | Vacuolar ATP synthase subunit d | 2 | 6.86 | 40.3 | 6.0 | 0.6 | |
| P62814 | Vacuolar ATP synthase subunit B, brain isoform | 3 | 8.04 | 56.6 | 6.0 | 1.0 | |
| Q9ER00 | Syntaxin-12 | 2 | 11.72 | 31.19 | 6.0 | 2.0 | |
| Q7SIG6 | Development and differentiation-enhancing factor 2 | 2 | 2.84 | 106.8 | 6.0 | 1.0 | |
| Q6PHN9 | Ras-related protein Rab-35 | 2 | 5.50 | 23.0 | Lipid anchor | 5.5 | 0.5 |
| P63213 | G-protein G(I)/G(S)/G(O) gamma-2 subunit | 2 | 8.78 | 37.4 | 5.5 | 0.5 | |
| P14094 | Sodium/potassium-transporting ATPase beta-1 chain | 2 | 8.25 | 35.2 | 5.0 | 2.0 | |
| Q6PCX7 | Repulsive guidance molecule A | 2 | 6.62 | 50.0 | GPI | 5.0 | 2.0 |
| P70296 | Phosphatidylethanolamine-binding protein (PEBP) | 3 | 28.65 | 20.7 | 5.0 | 3.0 | |
| P59108 | Copine-2 (Copine II) | 3 | 6.76 | 61.0 | 5.0 | 1.7 | |
| Q9WV55 | VAMP-associated protein A | 2 | 4.98 | 27.3 | 4.5 | 0.5 | |
| P46096 | Synaptotagmin-1 | 2 | 5.71 | 47.4 | Lipid anchor | 4.5 | 1.5 |
| Q9D1G1 | Ras-related protein Rab-1B | 2 | 16.50 | 22.2 | Lipid anchor | 4.5 | 0.5 |
| P07356 | Annexin A2 (Annexin II) | 3 | 13.95 | 38.5 | 4.5 | 1.5 | |
| Q99KR6 | RING finger protein 34 | 2 | 4.53 | 42.0 | 4.0 | 1.0 | |
| Q91V41 | Ras-related protein Rab-14 | 2 | 12.62 | 23.9 | Lipid anchor | 4.0 | 1.2 |
| P10852 | 4F2 cell-surface antigen heavy chain | 2 | 5.14 | 58.3 | 4.0 | 0.6 | |
| P35279 | Ras-related protein Rab-6A (Rab-6) | 2 | 5.34 | 23.6 | Lipid anchor | 3.5 | 1.5 |
| Q9CQD1 | Ras-related protein Rab-5A | 2 | 10.28 | 23.6 | Lipid anchor | 3.5 | 0.5 |
| O35963 | Ras-related protein Rab-33B | 2 | 4.82 | 25.8 | Lipid anchor | 3.5 | 0.5 |
| Q8K386 | Ras-related protein Rab-15 | 2 | 5.21 | 24.3 | Lipid anchor | 3.5 | 1.5 |
| P60764 | Ras-related C3 botulinum toxin substrate 3 | 2 | 7.33 | 21.4 | Lipid anchor | 3.5 | 0.5 |
| Q9QXL2 | Kinesin family member 21A | 2 | 1.14 | 186.53 | 3.5 | 0.5 | |
| Q8R4A8 | G-protein G(s), alpha subunit | 3 | 13.64 | 45.7 | Lipid anchor | 3.5 | 1.5 |
| Q9JMB8 | Contactin-6 | 3 | 3.21 | 113.8 | GPI | 3.5 | 2.5 |
| Q65CL1 | Alpha-3 catenin (Alpha T-catenin) | 1 | 1.79 | 99.8 | 3.5 | 1.5 | |
| Q60547 | Synaptonemal complex protein 3 | 3 | 6.01 | 27.1 | 3.0 | 0.6 | |
| Q9DAS9 | G-protein G(I)/G(S)/G(O) gamma-12 subunit | 4 | 22.86 | 78.7 | Lipid anchor | 3.0 | 1.0 |
| P97449 | Aminopeptidase N (Membrane protein p161) | 1 | 1.87 | 109.7 | 3.0 | 1.0 | |
| Q9JHS3 | Late endosomal/lysosomal Mp1-interacting protein | 2 | 14.52 | 13.48 | 3.0 | 1.0 | |
| P31324 | Prkar2b | 1 | 4.11 | 46.04 | 3.0 | 1.5 | |
| P61264 | Syntaxin-1B2 (Syntaxin 1B) | 2 | 8.01 | 33.3 | 2.5 | 1.5 | |
| P35278 | Ras-related protein Rab-5C | 2 | 6.51 | 23.4 | Lipid anchor | 2.5 | 0.5 |
| P63011 | Ras-related protein Rab-3A | 2 | 8.68 | 25.0 | Lipid anchor | 2.5 | 0.5 |
| P97855 | Ras-GTPase-activating protein binding protein 1 | 1 | 3.02 | 51.8 | 2.5 | 0.5 | |
| Q9CYH2 | Protein C10orf58 homolog | 2 | 5.99 | 24.4 | 2.5 | 0.5 | |
| P11505 | Plasma membrane calcium-transporting ATPase 1 | 1 | 1.43 | 138.7 | 2.5 | 0.5 | |
| P05480 | Neuronal proto-oncogene tyrosine-protein kinase Src | 2 | 5.38 | 60.6 | Lipid anchor | 2.5 | 0.5 |
| Q07310 | Neurexin-3-alpha | 1 | 4.11 | 174.0 | 2.5 | 0.5 | |
| O35136 | Neural cell adhesion molecule 2 | 1 | 1.56 | 93.2 | GPI | 2.5 | 1.5 |
| O89051 | Integral membrane protein 2B | 2 | 6.04 | 30.3 | 2.5 | 0.5 | |
| O08842 | GDNF family receptor alpha-2 | 1 | 3.68 | 51.6 | GPI | 2.5 | 0.5 |
| O08545 | Ephrin-A3 precursor | 2 | 8.60 | 21.2 | GPI | 2.5 | 0.5 |
| Q9R1T7 | Inducible T-cell co-stimulator (CD278 antigen) | 2 | 8.54 | 225.30 | 2.5 | 0.5 | |
| P60879 | Synaptosomal-associated protein 25 | 2 | 6.83 | 23.3 | Lipid anchor | 2.0 | 1.0 |
| P80236 | Ras-related C3 botulinum toxin substrate 1 | 3 | 18.42 | 8.8 | 2.0 | 1.0 | |
| P68404 | Protein kinase C beta type | 1 | 2.38 | 76.9 | 2.0 | 0.6 | |
| Q04690 | Neurofibromin | 2 | 3.81 | 319.6 | 2.0 | 0.6 | |
| Q60437 | Insulin receptor substrate p53 | 2 | 6.15 | 57.64 | 2.0 | 0.6 | |
| Q61411 | GTPase HRas | 2 | 12.77 | 21.3 | Lipid anchor | 2.0 | 0.6 |
| P51655 | Glypican-4 precursor (K-glypican) | 2 | 4.86 | 62.6 | GPI | 2.0 | 1.0 |
| P23818 | Glutamate receptor 1 (GluR-1) | 1 | 2.21 | 101.57 | 2.0 | 0.6 | |
| Q8VBX4 | C-type lectin domain family 4 member K | 1 | 4.24 | 37.6 | 2.0 | 0.6 | |
| Q8JZW4 | Copine-5 (Copine V) | 1 | 2.53 | 65.6 | 2.0 | 1.0 | |
| Q8BLR2 | Copine-4 (Copine IV) | 1 | 2.70 | 62.4 | 2.0 | 0.6 | |
| Q02013 | Aquaporin-1 | 1 | 7.49 | 28.66 | 2.0 | 1.2 | |
| Q07076 | Annexin A7 | 1 | 3.46 | 49.9 | 2.0 | 1.7 | |
| P97429 | Annexin A4 (Annexin IV) | 3 | 10.09 | 35.9 | 2.0 | 0.6 | |
| Q9DBE8 | Alpha-1,3-mannosyltransferase ALG2 | 1 | 3.86 | 47.4 | 2.0 | 1.0 | |
| P84078 | ADP-ribosylation factor 1 | 2 | 6.15 | 20.6 | 2.0 | 1.0 | |
| Q6QIY3 | Sensory neuron sodium channel | 1 | 3.09 | 220.6 | 2.0 | 0.6 | |
| P49817 | Caveolin-1 | 1 | 7.91 | 20.54 | 2.0 | 0.6 | |
| P18708 | Vesicle-fusing ATPase | 2 | 2.69 | 82.54 | 1.5 | 0.5 | |
| O70439 | Syntaxin-7 | 2 | 5.41 | 29.8 | 1.5 | 0.5 | |
| P16546 | Spectrin alpha chain, brain | 1 | 1.24 | 274.7 | 1.5 | 0.5 | |
| Q9QZB0 | Regulator of G-protein signaling 17 | 1 | 6.22 | 24.3 | 1.5 | 0.5 | |
| Q9JIR4 | Regulating synaptic membrane exocytosis protein 1 | 2 | 6.07 | 179.7 | 1.5 | 0.5 | |
| Q05909 | Receptor-type tyrosine-protein phosphatase gamma | 2 | 9.72 | 161.2 | 1.5 | 0.5 | |
| P46638 | Ras-related protein Rab-11B | 2 | 9.72 | 24.5 | Lipid anchor | 1.5 | 0.5 |
| P05696 | Protein kinase C alpha type(PKC-alpha) | 1 | 2.09 | 76.8 | 1.5 | 0.5 | |
| O35764 | Neuronal pentraxin receptor | 1 | 2.84 | 52.37 | 1.5 | 0.5 | |
| Q8CGK7 | G-protein G(olf), alpha subunit | 6 | 20.74 | 44.3 | 1.5 | 0.5 | |
| P50153 | G-protein G(I)/G(S)/G(O) gamma-4 subunit | 2 | 9.35 | 84.1 | Lipid anchor | 1.5 | 0.5 |
| Q99KJ8 | Dynactin subunit 2 | 1 | 4.75 | 44.0 | 1.5 | 0.5 |
aPep: peptide counts; b % cov: protein coverage%; c PTM: posttranslational lipid modification, GPI and lipid anchor: myristoylation, pamitolyation, geranylgeranylation, farnesylation, and prenylation; d Spectral count: total MS/MS spectral counts. Number represents mean value of 4 replicates; c standard deviation of spectral counts in 4 replicates.
Table 2.
Non-PM proteins identified in the lipid rafts of neonatal mouse brain
| Acc. No. | Protein name | Pepa | % Covb | MW (KDa) | Loc.c | Spectral countd | SDe |
| P69893 | Tubulin beta-1 chain | 12 | 38.60 | 49.67 | cyto | 262.0 | 14.0 |
| P68361 | Tubulin alpha-1 chain | 9 | 30.44 | 50.15 | cyto | 175.0 | 20.0 |
| Q71FK5 | Actin, cytoplasmic 1 (Beta-actin) | 5 | 21.39 | 41.74 | cyto | 102.5 | 4.5 |
| Q03265 | ATP synthase alpha chain | 10 | 26.63 | 59.75 | mc | 63.0 | 4.0 |
| P56480 | ATP synthase beta chain | 9 | 20.83 | 56.30 | mc | 51.5 | 5.5 |
| P04104 | Keratin, type II cytoskeletal 1 | 3 | 5.59 | 65.1 | cyto | 37.0 | 4.0 |
| P62629 | Elongation factor 1-alpha 1 | 3 | 9.33 | 50.11 | cyto | 36.0 | 5.0 |
| P19378 | Heat shock cognate 71 kDa protein | 4 | 10.39 | 70.8 | cyto | 33.0 | 1.0 |
| Q922U2 | Keratin, type II cytoskeletal 5 | 2 | 4.15 | 61.8 | cyto | 26.5 | 6.5 |
| P14873 | Microtubule-associated protein 1B (MAP 1B) | 7 | 4.34 | 270.41 | cyto | 25.5 | 3.5 |
| Q6IFZ6 | Keratin, type II cytoskeletal 1b | 2 | 4.20 | 61.4 | cyto | 25.5 | 3.5 |
| P97427 | Dihydropyrimidinase-related protein 1 | 4 | 8.93 | 62.17 | cyto | 25.0 | 0.0 |
| Q62188 | Dihydropyrimidinase-related protein 3 | 7 | 20.39 | 61.94 | cyto | 21.5 | 0.5 |
| P14733 | Lamin-B1 | 5 | 12.80 | 66.66 | nuc | 19.0 | 3.0 |
| P68372 | Tubulin beta-2C chain | 2 | 6.76 | 49.83 | cyto | 18.5 | 1.5 |
| Q6IG00 | Keratin, type II cytoskeletal 4 | 2 | 1.68 | 57.7 | cyto | 18.0 | 1.0 |
| Q60932 | Voltage-dependent anion-selective channel protein 1 | 7 | 38.31 | 32.35 | mc | 16.5 | 3.5 |
| P48962 | ADP/ATP translocase 1 | 3 | 11.15 | 32.77 | mc | 15.0 | 2.0 |
| Q922F4 | Tubulin beta-6 chain | 2 | 3.81 | 50.09 | cyto | 14.5 | 0.5 |
| Q04447 | Creatine kinase B-type | 3 | 12.63 | 42.71 | cyto | 13.5 | 2.5 |
| P67778 | Prohibitin | 4 | 21.03 | 29.82 | mc | 12.5 | 1.5 |
| Q10758 | Keratin, type II cytoskeletal 8 | 2 | 3.33 | 53.9 | cyto | 12.5 | 0.5 |
| Q61696 | Heat shock 70 kDa protein 1A | 2 | 5.78 | 70.08 | cyto | 12.0 | 1.0 |
| O08553 | Dihydropyrimidinase-related protein 2 | 5 | 13.84 | 62.17 | cyto | 11.5 | 2.5 |
| P46633 | Heat shock protein HSP 90-alpha (HSP 86) | 2 | 3.56 | 84.72 | cyto | 10.5 | 2.5 |
| P63101 | 14-3-3 protein zeta/delta | 2 | 12.30 | 27.77 | cyto | 10.5 | 0.5 |
| Q60930 | Voltage-dependent anion-selective channel protein 2 | 3 | 14.29 | 31.73 | mc | 10.0 | 0.0 |
| Q9ERD7 | Tubulin beta-3 chain | 2 | 9.35 | 50.42 | cyto | 10.0 | 1.0 |
| P50672 | Cytochrome c oxidase subunit 2 | 2 | 11.50 | 25.82 | mc | 9.0 | 0.0 |
| Q9DCT2 | NADH-ubiquinone oxidoreductase 30 kDa subunit | 2 | 6.35 | 34.00 | mc | 8.5 | 0.5 |
| P18760 | Cofilin-1 (Cofilin, non-muscle isoform) | 2 | 15.24 | 18.43 | nuc | 8.5 | 1.5 |
| P07823 | 78 kDa glucose-regulated protein | 5 | 11.18 | 72.38 | er | 8.5 | 1.5 |
| P11497 | Acetyl-CoA carboxylase 1 | 1 | 4.24 | 37.62 | cyto | 8.0 | 1.0 |
| Q60931 | Voltage-dependent anion-selective channel protein 3 | 3 | 13.83 | 30.75 | mc | 7.5 | 0.5 |
| P09445 | Elongation factor 2 | 2 | 2.92 | 95.27 | cyto | 7.5 | 0.5 |
| P62977 | Ubiquitin | 1 | 21.33 | 8.57 | cyto | 7.0 | 1.0 |
| P19783 | Cytochrome c oxidase subunit IV isoform 1 | 1 | 7.14 | 19.53 | mc | 7.0 | 2.0 |
| P14152 | Malate dehydrogenase | 1 | 3.61 | 36.35 | mc | 6.5 | 0.5 |
| P11499 | Heat shock protein HSP 90-beta | 2 | 3.88 | 83.20 | cyto | 6.5 | 0.5 |
| P12787 | Cytochrome c oxidase polypeptide Va | 1 | 10.42 | 16.03 | mc | 6.5 | 0.5 |
| P31253 | Ubiquitin-activating enzyme E1 X | 2 | 7.35 | 50.99 | cyto | 6.0 | 3.0 |
| P35564 | Calnexin | 2 | 4.92 | 67.28 | er | 6.0 | 1.0 |
| Q91VD9 | NADH-ubiquinone oxidoreductase 75 kDa subunit | 2 | 3.99 | 79.75 | mc | 5.5 | 2.5 |
| P56135 | ATP synthase f chain, mitochondrial | 2 | 26.74 | 10.21 | mc | 5.5 | 1.5 |
| P51881 | ADP/ATP translocase 2 | 2 | 8.11 | 32.80 | mc | 5.5 | 0.5 |
| Q8R429 | SR Ca(2+)-ATPase 1 | 2 | 3.12 | 109.43 | er | 5.0 | 1.0 |
| Q9DB20 | ATP synthase O subunit | 2 | 9.91 | 23.36 | mc | 5.0 | 1.0 |
| Q8R429 | Calcium pump 1 (SERCA1) | 2 | 3.12 | 109.43 | er | 5.0 | 1.0 |
| Q91V61 | Sideroflexin-3 | 1 | 4.06 | 35.41 | mc | 4.5 | 0.5 |
| P03995 | Glial fibrillary acidic protein, astrocyte (GFAP) | 1 | 2.56 | 49.92 | cyto | 4.5 | 1.5 |
| Q9CQV8 | 14-3-3 protein beta/alpha | 1 | 5.43 | 21.22 | cyto | 4.5 | 2.5 |
| P68368 | Tubulin alpha-4 chain | 2 | 3.11 | 50.14 | cyto | 3.5 | 0.5 |
| P62962 | Profilin-1 (Profilin I) | 2 | 21.74 | 14.83 | cyto | 3.5 | 0.5 |
| Q8QZT1 | Acetyl-CoA acetyltransferase | 2 | 7.09 | 44.82 | mc | 3.5 | 0.5 |
| P62962 | Profilin-1 | 2 | 21.74 | 14.82 | cyto | 3.5 | 0.5 |
| Q02053 | Ubiquitin-activating enzyme E1 1 | 3 | 4.82 | 117.81 | cyto | 3.0 | 1.0 |
| P42932 | T-complex protein 1 subunit theta | 3 | 6.59 | 59.43 | cyto | 3.0 | 2.0 |
| O35129 | Prohibitin-2 | 4 | 21.03 | 29.82 | mc | 3.0 | 1.0 |
| P31324 | Prkar2b | 1 | 4.11 | 46.04 | cyto | 3.0 | 0.0 |
| P20357 | Microtubule-associated protein 2 (MAP 2) | 2 | 1.20 | 198.98 | cyto | 3.0 | 0.0 |
| P34926 | Microtubule-associated protein 1A (MAP 1A) | 2 | 2.36 | 299.53 | cyto | 3.0 | 0.0 |
| P52480 | Pyruvate kinase isozyme M2 | 4 | 12.48 | 57.76 | mc | 3.0 | 2.0 |
| P63209 | S-phase kinase-associated protein 1A | 1 | 9.32 | 18.53 | cyto | 3.0 | 1.0 |
| P60879 | Synaptonemal complex protein 3 | 3 | 6.01 | 27.1 | nuc | 3.0 | 1.0 |
| O88809 | Neuronal migration protein doublecortin | 1 | 3.56 | 40.61 | cyto | 2.5 | 0.5 |
| P17156 | Heat shock-related 70 kDa protein 2 | 2 | 3.01 | 69.74 | cyto | 2.5 | 1.5 |
| Q9EQF6 | Dihydropyrimidinase-related protein 5 | 1 | 3.20 | 61.52 | cyto | 2.5 | 0.5 |
| Q8BH59 | Calcium-binding mitochondrial carrier protein Aralar1 | 2 | 4.59 | 74.57 | mc | 2.5 | 0.5 |
| P48670 | Vimentin | 1 | 4.25 | 51.85 | cyto | 2.5 | 1.5 |
| P80315 | T-complex protein 1 subunit delta | 1 | 2.98 | 57.94 | cyto | 2.0 | 0.0 |
| P11984 | T-complex protein 1 subunit alpha A | 1 | 4.14 | 60.34 | cyto | 2.0 | 0.0 |
| Q9JKK8 | Serine-protein kinase ATR | 1 | 2.36 | 84.26 | nuc | 2.0 | 0.0 |
| Q04899 | Serine/threonine-protein kinase PCTAIRE-3 | 1 | 3.78 | 51.85 | nuc | 2.0 | 1.0 |
| Q99PT1 | Rho GDP-dissociation inhibitor 1 (Rho GDI 1) | 1 | 7.88 | 23.41 | er | 2.0 | 0.0 |
| Q61879 | Myosin-10 | 1 | 3.09 | 49.59 | cyto | 2.0 | 0.0 |
| P24638 | Lysosomal acid phosphatase | 1 | 2.13 | 48.51 | lysosome | 2.0 | 1.0 |
| O70251 | Elongation factor 1-beta | 2 | 12.56 | 24.56 | cyto | 2.0 | 0.0 |
| Q9CPQ8 | ATP synthase g chain, mitochondrial | 1 | 18.63 | 11.43 | mc | 2.0 | 0.0 |
| O35627 | Orphan nuclear receptor NR1I3 | 1 | 2.52 | 40.89 | nuc | 2.0 | 1.0 |
| P53026 | 60S ribosomal protein L10a | 1 | 6.98 | 24.78 | nuc | 2.0 | 1.0 |
| P97524 | Very-long-chain acyl-CoA synthetase | 1 | 2.58 | 70.69 | er | 1.5 | 0.5 |
| Q01853 | Transitional endoplasmic reticulum ATPase | 2 | 3.11 | 89.18 | cyto | 1.5 | 0.5 |
| Q99JR1 | Sideroflexin-1 | 1 | 5.63 | 35.52 | mc | 1.5 | 0.5 |
| Q62627 | PRKC apoptosis WT1 regulator protein | 1 | 4.53 | 35.87 | nuc | 1.5 | 0.5 |
| Q9DCS9 | NADH-ubiquinone oxidoreductase PDSW subunit | 1 | 10.92 | 20.89 | mc | 1.5 | 0.5 |
| P08249 | Malate dehydrogenase | 2 | 9.79 | 35.60 | mc | 1.5 | 0.5 |
| Q8BGU5 | Cyclin fold protein 1 | 1 | 5.00 | 39.39 | nuc | 1.5 | 0.5 |
| Q8CEE6 | PAS-kinase (PASKIN) | 1 | 1.16 | 151.27 | cyto | 1.5 | 0.5 |
| P35980 | 60S ribosomal protein L18 | 1 | 6.99 | 21.5 | nuc | 1.5 | 0.5 |
aPep: peptide counts; b % cov: protein coverage%; c Loc.: subcellular localization. mc. mitochondria, cyto. cytoplasm, er. endoplasmic reticulum; eSpectral count: total MS/MS spectral counts. Number represents mean value of 4 replicates; c standard deviation of spectral counts in 4 replicates.
The identified core proteome in the lipid rafts of neonatal mouse brain covered a wide range of sizes (8.0~319.6 kDa). Up to 216 non-redundant proteins were identified from 100 μl of a lipid raft fraction by TubeGeLC-MS/MS, 75% of these proteins were identified by at least two peptide matches and 25% of those identified were based upon a single peptide match. Although protein identifications based upon a single peptide match may be problematic, this does not necessarily imply a potential false identification [34]. For example, caveolin-1 (cav-1) was detected in raft samples from neonatal mouse brain by immunoblots, but was represented in each of the 4 replicates identified by mass spectrometry by a single peptide match. Using the Gene Ontology (GO) classifications and PubMed database searches, 127 (58.8%) of the proteins identified were PM or PM-associated proteins, with 18 (14.2%) having a GPI-anchoring site and 34 (26.8%) with other lipid-anchoring sites as described above. Many of the PM proteins identified were reported previously as being lipid raft proteins by conventional biochemical procedures. Typical raft marker proteins, such as caveolin-1, flotillin-1 and -2, Fyn and Src, were identified in the lipid raft preparations from neonatal mouse brain. Functional categories revealed that the identified PM proteins cover a broad range of neural functions involving neurodevelopment. Several proteins are known to function as part of the neurotransmitter release and re-uptake machinery; 3 syntaxin (Stx) proteins, Stx1A, Stx1B, Stx7; synaptosomal-associated protein 25 (Snap25); synaptotagmin (Syt) proteins, Syt1 and Sty7; vesicle-associated membrane proteins (Vamp), Vamp1 and Vamp2; regulating synaptic membrane exocytosis protein 1 (RIM1); and the glutamate receptor (GluR1) were all present in the raft fractions. Relatively large numbers of guanine nucleotide-binding protein (G protein) isoforms and Ras subfamily of GTPases were identified; G(s)α, G(i)α1, G(i)α2, G(o)α, G(k)α3, G(olf)α, G(q)α, G(z)α, Gα11, Gα12, G(I)/G(S)/G(T)β1, G(I)/G(S)/G(O)γ2, G(I)/G(S)/G(O)γ4, G(I)/G(S)/G(O)γ12 and Rab1A, Rab1B, Rab2A, Rab3A, Rab5A, Rab5C, Rab10, Rab11B, Rab14, Rab15, Rab33B, Rab35, p21Rac1, p21Rac3, Rab GDIα, RIN3, and G3BP. These proteins have been implicated in a variety of developmental processes in neonatal brain, including signal transduction, neurotransmitter release, and membrane trafficking [2]. Another important group of proteins in the neonatal brain-raft proteome comprises the cell adhesion/recognition molecules for cell-cell communication. Twenty-four such proteins were identified; contactin1 (Cntn1), Cntn2, Cntn4, and Cntn6, neurotrimin, Thy1, neurotractin (Kilon protein), Nap22, Gap43, paralemmin, desmocollin-2, neurexin-3α, Ncam2, Ncam180, dynactin, glypican (Gpc)1, Gpc 2, Gpc4, limbic system-associated membrane protein (LSAMP), transmembrane phosphoprotein Cbp, neurofibromin, opioid-binding cell adhesion molecule (Obcam), Alpha-3 catenin, and cadherin-13. These cell surface communication proteins are known to participate in the formation of neuronal networks in the brain during development, specifically axon growth, synapse formation, and fasciculation [35-38]. Several transporters and non-receptor type channel proteins were also identified; Na(+)/K(+) ATPase (ATP1A1, ATP1A2 and ATP1A3), small conductance calcium-activated potassium channel protein 3 (Kcnn3), plasma membrane calcium-transporting ATPase 1, Slc3a2, GDNF family receptor alpha-2, integrin-associated protein, integral membrane protein 2B, sensory neuron sodium channel, L-type calcium channel subunit delta, aquaporin-1, and chloride channel protein 6 (Clc-6). Calcium and phospholipid binding proteins cupine (Cnpe)2, Cnpe4, Cnpe591, annexin (Anx)2A, Anx4A, Anx5A, Anx6A, and Anx7A were also identified. A number of proteins of unknown function were also identified, such as receptor-type tyrosine-protein phosphatase gamma, protein C10orf58 homolog, Coxsackie's virus and adenovirus receptor homologs. As expected, since these raft proteins are from neonatal mouse brain, myelin proteins (present in adult brain tissue) such as myelin basic protein (MBP), myelin proteolipid protein (PLP), oligodendrocyte-myelin glycoprotein (Omg), and 2',3'-cyclic-nucleotide 3'-phosphodiesterase (CNPase), were not represented. Functional annotation and grouping of the major neonate-brain raft proteome will provide a basis for determining the potential targets of lipid raft disorganization in mouse models of neonatal brain disorders.
As reported in most raft proteomic studies [8,10,11,14,19,39-43], non-PM proteins were also found in the raft samples in the present study (Fig. 2 and Table 2). Eighty-nine of the 216 (41.2%) identified proteins from neonatal mouse brain rafts were predicted to be non-PM proteins by their GO terms. They are comprised of 47 cytoplasmic proteins including 20 cellular structural proteins (such as tubulins, actins, keratins, and microtubule-associated proteins), 25 mitochondrial proteins, 10 nuclear proteins, 6 ER proteins, and 1 lysosomal protein. Proteins from other subcellular compartments such as endosome and Golgi apparatus were poorly represented. The presence of subcellular membrane and cytoplasmic proteins in lipid raft fractions have been discussed in several proteomic studies [1,8,11,43-46]. One possibility is the contamination of non-plasma membrane proteins during gradient purification. The position of membrane particles in the density gradient ultracentrifugation is determined mainly by the ratio of its lipid and protein contents; different ratios of lipids to proteins for the various intracellular membrane particles could lead them to have different buoyant properties in density gradients. In this context, any method used for preparing cell membrane 'lipid rafts' is likely to generate a fraction containing membranes from a number of sub-cellular membranes, but not necessarily one enriched specifically in plasma membrane lipid rafts [8]. Certain subcellular proteins highly enriched in raft samples may be structurally involved and play critical roles in cell membrane lipid raft organization. For example, the cellular structural proteins such as tubulins, actins, keratins, and microtubular proteins, are highly enriched in lipid raft samples including brain-rafts as shown in this study and many other reports [7,8,13,28]. These cytoskeletal proteins not only contribute to the structural organization of cytoplasm but also play important roles in regulating the topography of the plasma membrane and trafficking and in modulating the localization of lipid raft proteins in eukaryotic cells [47,48]. Additionally, many proteins could have multiple cellular localizations regulated by multiple mechanisms. For example, cytoplasmic microtubule-associated proteins and 14-3-3 proteins, histones, and mitochondrial ATP synthases and voltage-dependent anion-selective channel 1 (VDAC1), have also been identified in cell plasma membranes [44,49-52]. Thus, enrichment of certain non-PM proteins in lipid rafts (DRMs) may represent a true observation of protein localization in different biological conditions and not necessarily be due to cross-contamination acquired during purification.
Compilation of proteins into abundance lists
All proteins identified as PM protein or non-PM proteins in lipid rafts of neonatal mouse brain by TubeGeLC/MS/MS are compiled in Tables 1 and 2, respectively, and were sorted by their relative abundance calculated from the MS/MS spectral counts. Mass spectrometry of proteins and peptides is not quantitative, therefore, it is difficult to assess the abundance of a particular protein from the MS data per se. However, recent studies with label-free LC-MS/MS shotgun proteomics [29,53-57] revealed a relationship between protein abundance and sampling statistics, such as sequence coverage, peptide count, and spectral count. The use of sampling statistics is a promising method for measuring the relative protein abundance and detecting differentially expressed proteins. In general, the greater the amount of protein, the greater the MS signal intensity, number of spectral counts, sequenced peptides/sequence coverage, total ion current (TIC), and total Xcorr or scores that combine these values. Label-free proteomics has emerged as an alternative to stable isotope labeling for protein quantitation. The MS/MS spectral count, which compares the number of MS/MS spectra assigned to each protein, was selected for relative protein abundance in this study. Although this method has a tendency to overestimate the abundance of large proteins because they yield more peptides and therefore more spectral counts than the smaller proteins, the results indicate that this may not be a fundamental problem [29]. In the current study, contactin-1 (113.4 kDa) had a MS/MS spectral count of 174, but the sodium/potassium-transporting ATPase alpha-3 chain, a protein of almost identical size (111.7 kDa), had a spectral count of 9 (Table 1). It is reasonable to assume that the former protein is much more abundant than the latter. When plotting MS/MS spectral count versus protein size for all proteins identified (data not shown), both the maximum spectral count distribution was highest for proteins with a size distribution of 20~50 kDa. Therefore, the bias that may be potentially caused by size towards larger proteins may not be overly large, when using MS/MS spectral counts as a measure of abundance [29]. About 50% of proteins were identified with fewer than 5 total spectral counts, presumably due to their relatively low abundances. A total of 11 identified proteins with > 40 spectral counts were arbitrarily categorized as the most abundant proteins in the lipid rafts from neonatal mouse brain. These include contactin-1, NAP-22, Gα(o), annexin-A6, Lsamp, neurotractin, contactin-2, Gα(i), and flotillin-1, as well as intracellular structural and mitochondrial proteins such as tubulins, actins, and ATP synthases. Proteins with total spectral counts from 5 to 40 were arbitrarily categorized as medium abundance proteins. The relative abundances agree well with published data [7,18,19,30,39,40,42,43,45,49,58-71] and support our contention that the TubeGeLC-MS/MS approach provides a fair representation of the protein composition of the lipid rafts from neonatal mouse brain. The spectral count data for each identified protein provides proteome-wide semi-quantitative information on the relative abundance of lipid raft proteins.
Comparison of protein identifications between GeLC-MS/MS and TubeGeLC-MS/MS
In-gel digestion can be efficiently employed after protein mixtures are resolved by SDS-PAGE or directly polymerized into a 'tube-gel' without electrophoresis [7,15]. Both of these in gel-based protein digestion protocols give clean LC-MS/MS baselines as interfering substances, such as detergents, salts and lipids, can be effectively removed during washing steps. To compare the GeLC-MS/MS versus the TubeGeLC-MS/MS, four separate experiments were conducted using 100 μl of sucrose-gradient isolated rafts that were subjected to a 1D SDS-PAGE combined with nanospray LC-MS/MS spectrometry (GeLC-MS/MS) modified by an established protocol [72], as described in Methods. The results for the peptides and corresponding proteins that were identified in a minimum of 2 of 4 independent experiments were used for comparative analyses. The comparison showed that about 200 proteins could also be identified by GeLC-MS/MS approach, with similar protein identifications, especially for high and medium abundance proteins, as compared to TubeGeLC-MS/MS (data not shown). However, the reproducibility of protein identified by GeLC-MS/MS was less than TubeGeLC-MS/MS (see below).
Reproducibility of raft proteome characterization by TubeGeLC-MS/MS
To test the reproducibility of proteins identified, 3 raft samples from 3 separate neonatal brains prepared by identical methods at the same time were processed by both TubeGeLC-MS/MS and GeLC-MS/MS protocols, and the resulting protein identifications for within technique variations compared. There was a 68.8 ± 6.5% (SD) concordance in the proteins identified by TubeGeLC-MS/MS protocol among the 3 raft samples. As expected, the high abundance proteins showed a higher reproducibility of identification. The non-concordant proteins of ~30% may reflect some false identification because 55% of the non-concordant proteins had single or two peptide identifications. In addition, lipid raft isolations per se have a degree of variability. The results from the GeLC-MS/MS protocol yielded 45 ± 11% of concordance for within technique protein identifications.
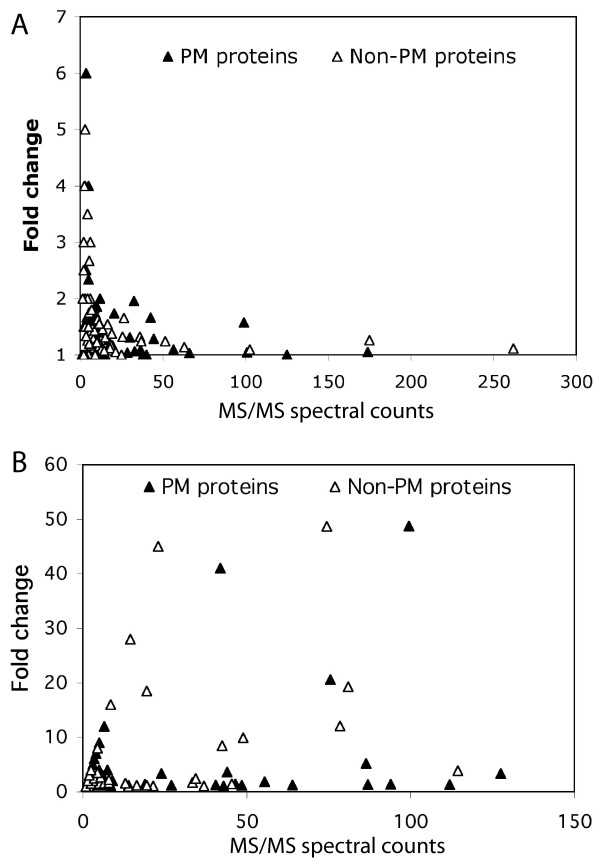
A MS/MS spectral-count method was employed as a semi-quantitative measure for comparing proteins in different samples. Variability in protein abundance, calculated as MS/MS spectral counts, between the brain raft samples from two separate animals was evaluated and compared between the two approaches. The ratio of the spectral count per protein between these two samples was presented as fold-change and plotted against the average of the spectral count of the two samples. With the TubeGeLC-MS/MS method the fold-change was less than 2 for ~80% of the identified proteins; the higher the abundance, the lower of fold changes as shown in Figure 4A. However, greater variations for low abundance proteins were evident, indicating that the sensitivity of quantifying changes for low abundance proteins was generally lower. The fold-change results of the same samples analyzed by the GeLC/MS/MS protocol are shown in Figure 4B; greater variations were evident for both high and low abundant proteins. These results suggested that there was larger experimental variation associated with 1D gel protein separation and extraction from the gel slices prior protein digestion and mass spectrometry using the GeLC/MS/MS method. One of the explanations is that lipid associated proteins and other hydrophobic proteins may not fully enter the gel lanes in the GeLC/MS/MS protocol, causing variations in quantitative analyses. Employing the TubeGeLC-MS/MS approach, despite the experimental variation in isolating the lipid rafts, the protein composition from replicate samples was less variable, indicating that this simple change in sample handling results in more reproducible results.
Figure 4.
Variations of quantification by spectral counting. The variations in spectral counts for each protein were compared between the TubeGeLC/MS/MS and the GeLC/MS/MS protocols. The ratio of the spectral count per protein between two samples is presented as fold-change and plotted against the average of the spectral count of two samples. Panel A shows the results from the TubeGeLC/MS/MS method and panel B from the GeLC/MS/MS protocol.
Conclusion
We have successfully combined a 'tube-gel' protein digestion protocol with nanospray LC-MS/MS analysis to carry out a high throughput proteomic mapping of lipid raft proteins isolated from neonatal mouse brain. Characterization of analytically difficult lipid raft proteins was simplified by this method. The MS/MS spectral count information from mass spectrometric analyses allowed for the label-free quantitation of relative protein abundances of more than 200 raft proteins from a single sample. The major advantage of this protocol is that the raft proteins are directly digested in a gel matrix without fractionation and purification, thus dramatically minimizing variation in protein yields due to losses during sample manipulation prior to mass spectrometry. With careful isolation of rafts, this protocol should allow for a reproducible quantitation of relative protein abundance in lipid rafts. This methodology should allow the investigation of the role of these specialized membranes under various biological conditions.
Methods
Reagents and antibodies
Sources for antibodies were as follows: caveolin-1 (Cav-1), contactin-1 (Cntn-1), annexin-VI (Anx VI, Anx6A), GTP-binding protein αq (Gαq), NAP-22, calnexin, α-tubulin from Santa Cruz Biotechnology, CA USA; flotillin-1 (flot-1): BD Transduction Laboratories, CA USA; mouse monoclonal antibody against β-COP: Sigma-Aldrich, MO USA; and mouse anti-human transferrin receptor antibody: Zymed Laboratories, CA USA. Trypsin Gold (MS grade) was obtained from Promega, WI, USA. All other reagents were from ThermoFisher Scientific, MA, USA.
Preparation of raft-enriched detergent-resistant membranes from neonatal mouse brain
All animal experiments were performed with the approval of the Institutional Animal Care and Research Advisory Committee at the Clement Zablocki Veterans Medical Center. Neonatal mice (postnatal day 1, C57Bl/6J, Jackson Laboratories) were sacrificed by decapitation. Details of protocols used to prepare the raft-enriched detergent-resistant membranes have been described previously [30,73]. Briefly, frozen brains from neonatal mice, 50~60 mg of wet brain tissue, were homogenized in an ice-cold lysis buffer containing 5% glycerol in buffer A (50 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 0.15 M NaCl, 20 mM NaF, 1 mM Na3VO4, 5 mM β-mercaptoethanol, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM PMSF), using a tissue homogenizer (PRO Scientific Inc., Oxford, CT USA) by three pulses of 10 seconds each, followed by 20 strokes of a Dounce homogenizer, pestle A. Tissue debris and nuclei were removed by centrifugation at 1,000 g for 5 minutes and the pellet was re-extracted. The protein concentration of the post-nuclear homogenates (PNH) was measured using Protein Reagent (Bio-Rad, CA USA), adjusted to 2 mg/ml and 2 ml of the homogenates extracted with 1% Triton X-100 (TX) on ice for 30 min. The samples were mixed with an equal volume of ice-cold 80% (w/v) sucrose in buffer A, and then overlaid with 2.0 ml each of 35, 30, 25, and 5% (w/v) sucrose (all in buffer A). The sucrose gradient was centrifuged at 36,000 rpm in a Sorval 90 ultracentrifuge using a TH-641 rotor for 15 hr at 4°C. After ultracentrifugation, TX-resistant lipid rafts appeared as an insoluble white light-scattering band at the interface between the 5% and 25% sucrose layer. Twelve 1.0 ml fractions were collected from the top to bottom, with fractions 2–4 containing the rafts (density range of 1.055~1.115 g/ml). Non-raft fractions 8–11 were collected in the density range 1.130~1.180 g/ml.
Tube-gel protein digestion
A Tube-Gel digestion method has been successfully used for high throughput analysis of membrane proteins and proven to be compatible with detergents in protein samples [15]. In these experiments, fraction 3 of the sucrose gradient was used as the lipid raft fraction. The raft fraction was directly incorporated into a polyacrylamide gel matrix as follows: 100 μl of the raft solution, 25 μl of acrylamide solution (40%, 29:1), 1.0 μl of 10% SDS, 0.5 μl of 10% ammonium persulfate, and 0.1 μl of TEMED were mixed in a 0.5 ml Eppendorf tube. The co-polymerization reaction was carried out for 30 min at room temperature. Post-polymerization, no liquid was extruded from the tube-gel, indicating that all of the materials were trapped in the gel matrix. The gel block was removed, cut into small pieces, and washed five times with 50% acetonitrile (v/v) in 25 mM ammonium bicarbonate for 15 min, using sonication and agitation. The gel pieces were dried using a SpeedVac, subjected to in-gel digestion using 100 μl of 10 ng/μl trypsin dissolved in 25 mM AMBIC and incubated at 37°C overnight. Peptides were then extracted from the gel using 500 μl of 0.1% formic acid in MS-grade water followed by 2 extractions with the same volume of 0.1% formic acid in 70% acetonitrile. Corresponding fractions were combined and dried using a SpeedVac. The dried samples were resuspended in 6 M guanidine-hydrochloride and 5 mM potassium phosphate, pH 6.0, purified using C-18 zip-tips from Millipore Corp., and subjected to nanospray LC-MS/MS analysis. This protocol is referred to as TubeGeLC-MS/MS.
Alternatively, lipid raft proteins were digested using an established protocol with some minor modification by 1-D electrophoresis coupled with nanospray LC-MS/MS (GeLC-MS/MS) [72]. Rather than conventional SDS-PAGE separation and multiple LC-MS/MS analyses, proteins in 100 μl of raft fractions, were first separated on 6% SDS-PAGE gels, long enough for the protein mixtures to penetrate the separation gel and then stained with silver. The stained areas of the gel containing the complex mixture of proteins were excised, digested with trypsin and applied to the nanospray LC-MS/MS to analyze raft proteome as described above.
Nanospray LC-MS/MS spectrometry and data Analysis
Automated nanospray liquid chromatography tandem mass spectrometry (nanospray LC-MS/MS) was performed using an LTQ-LC/MS from ThermoFisher Scientific. Peptide mixtures were separated using a C18 reverse phase column (0.75-Å internal diameter at a flow rate of 1 μl/min) in line with the mass spectrometer. The mobile phases consisted of 0.1% formic acid containing 5% acetonitrile (A) and 0.1% formic acid in 95% acetonitrile (B), respectively. A 260-min linear gradient was typically used.
The MS data obtained were searched using the SEQUEST algorithm against the UniProt Rodent database v49.1. The search was limited only to tryptic peptides, and identifications were filtered from the search results using the Epitomize program [74]. Epitomize reads all the SEQUEST.out files in a directory, filters the files based on user-defined levels of Xcorr, and outputs the proteins identified. The Xcorr versus charge state filter used was set to Xcorr values of 1.8, 2.3 and 3.0 for charge states +1, +2 and +3, respectively. These filter values are similar to others previously reported for SEQUEST analyses [75]. Protein hits that passed the filter were annotated using the generic Gene Ontology (GO) slim. All proteins were identified by two or more peptides, and those identified with single peptide were included in the analysis if identified in two or more scans. Finally, the peptides listed were manually verified for correct identification by comparing the experimental spectra with the theoretical band ion spectra. Quantitative analyses were done using the open-source software program ZoomQuant, which provides a validation and a quantization platform for protein mass spectrometry [74,76].
Biochemical analysis of lipids in gradient fractions
Sterol composition in each of the 12 fractions was quantitatively determined by gas chromatography/mass spectroscopy (GC/MS). An aliquot of ethanol containing the internal standard 5α-cholestane (25 μg) was added to each sample tube, and samples were hydrolyzed at 50°C in ethanol containing 1 M NaOH for 1 hour. Sterols were extracted in hexane (final volume 30 ml), dried under nitrogen, and derivatized with HMDS-TMCS. GC-MS analysis was performed using a Focus DSQ system (ThermoFisher Scientific). The trimethylsilyl-derived sterols were separated on a TR-35MS capillary column (35 m × 0.25 mm internal diameter × 0.25 μm film) with helium as the carrier gas at the rate of 1.8 ml/min. The temperature program was 150°C for 1 minute, followed by increases of 20°C/min up to 310°C, which was then held for 6 minutes. The injector was operated in the splitless mode at 250°C. Standard curves were generated by MS analysis of various amounts of each sterol. The contents of sphingomyelin (SM), ceramide (Cer) in each of the 12 fractions was quantitatively determined by LC/ESI/MS/MS on a Thermo Finnigan TSQ 7000 triple quadrupole mass spectrometer, operating in a Multiple Reaction Monitoring (MRM) positive ionization mode, as described previously [77].
SDS/PAGE and immunoblots
A 30 μl aliquot of each fraction from the sucrose gradient was analyzed by SDS/PAGE on 10 or 12% (w/v) acrylamide gels. Separated proteins were transferred to nitrocellulose membranes for immunoblotting analyses. Membranes were blocked in 5% (w/v) non-fat milk in TBS-Tween [0.05% (w/v) Tween 20 in 10 mM Tris/100 mM NaCl, pH 7.5], and then incubated with the primary antibodies of choice. Membranes were subsequently incubated with HRP-conjugated second antibodies, and specific interactions were revealed using the ECL® (Enhanced Chemiluminescence) detection system (Amersham, CA, USA).
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
HY and BW conceived of the study and designed the experiments. HY, ML, and GST carried out the experiments. HY, BW, BH and SBP analyzed the data and prepared the manuscript. SBP supervised and coordinated the project and HY and SBP obtained funding for this project. HY and SBP wrote the manuscript, and all authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Yanhong Cai for her technical services and assistance with the animal husbandry. The authors thank the members of the Protein Core of Medical College of Wisconsin for the proteomic Service. We also thank Dr. Michael Olivier for critically reading this manuscript. This research was supported by Biomedical Research Grant RG-11311-M from the American Lung Association (HY) and by PHS grant HL68660 from the National Heart, Blood and Lung Institute, NIH (SBP). This work also was supported in part by the NHLBI Proteomics Center contract NIH-N01 HV-28182.
Contributor Information
Hongwei Yu, Email: hyu@mcw.edu.
Bassam Wakim, Email: bwakim@mcw.edu.
Man Li, Email: manli@mcw.edu.
Brian Halligan, Email: halligan@mcw.edu.
G Stephen Tint, Email: tintgs@umdnj.edu.
Shailendra B Patel, Email: sbpatel@mcw.edu.
References
- Brown DA. Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology (Bethesda) 2006;21:430–439. doi: 10.1152/physiol.00032.2006. [DOI] [PubMed] [Google Scholar]
- Allen JA, Halverson-Tamboli RA, Rasenick MM. Lipid raft microdomains and neurotransmitter signalling. Nat Rev Neurosci. 2007;8:128–140. doi: 10.1038/nrn2059. [DOI] [PubMed] [Google Scholar]
- Pike LJ. Lipid rafts: heterogeneity on the high seas. Biochem J. 2004;378:281–292. doi: 10.1042/BJ20031672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Ehehalt R. Cholesterol, lipid rafts, and disease. J Clin Invest. 2002;110:597–603. doi: 10.1172/JCI200216390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Vaz WL. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- Blonder J, Hale ML, Lucas DA, Schaefer CF, Yu LR, Conrads TP, Issaq HJ, Stiles BG, Veenstra TD. Proteomic analysis of detergent-resistant membrane rafts. Electrophoresis. 2004;25:1307–1318. doi: 10.1002/elps.200405891. [DOI] [PubMed] [Google Scholar]
- Martosella J, Zolotarjova N, Liu H, Moyer SC, Perkins PD, Boyes BE. High recovery HPLC separation of lipid rafts for membrane proteome analysis. J Proteome Res. 2006;5:1301–1312. doi: 10.1021/pr060051g. [DOI] [PubMed] [Google Scholar]
- Magee AI, Parmryd I. Detergent-resistant membranes and the protein composition of lipid rafts. Genome Biol. 2003;4:234. doi: 10.1186/gb-2003-4-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLellan DL, Steen H, Adam RM, Garlick M, Zurakowski D, Gygi SP, Freeman MR, Solomon KR. A quantitative proteomic analysis of growth factor-induced compositional changes in lipid rafts of human smooth muscle cells. Proteomics. 2005;5:4733–4742. doi: 10.1002/pmic.200500044. [DOI] [PubMed] [Google Scholar]
- Elortza F, Nuhse TS, Foster LJ, Stensballe A, Peck SC, Jensen ON. Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol Cell Proteomics. 2003;2:1261–1270. doi: 10.1074/mcp.M300079-MCP200. [DOI] [PubMed] [Google Scholar]
- Foster LJ, De Hoog CL, Mann M. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc Natl Acad Sci U S A. 2003;100:5813–5818. doi: 10.1073/pnas.0631608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Wollscheid B, Watts JD, Scheer B, Aebersold R, DeFranco AL. Quantitative proteomic analysis of B cell lipid rafts reveals that ezrin regulates antigen receptor-mediated lipid raft dynamics. Nat Immunol. 2006;7:625–633. doi: 10.1038/ni1337. [DOI] [PubMed] [Google Scholar]
- Jia JY, Lamer S, Schumann M, Schmidt MR, Krause E, Haucke V. Quantitative proteomics analysis of detergent-resistant membranes from chemical synapses: evidence for cholesterol as spatial organizer of synaptic vesicle cycling. Mol Cell Proteomics. 2006;5:2060–2071. doi: 10.1074/mcp.M600161-MCP200. [DOI] [PubMed] [Google Scholar]
- Sprenger RR, Horrevoets AJ. Proteomic study of caveolae and rafts isolated from human endothelial cells. Methods Mol Biol. 2007;357:199–213. doi: 10.1385/1-59745-214-9:199. [DOI] [PubMed] [Google Scholar]
- Lu X, Zhu H. Tube-gel digestion: a novel proteomic approach for high throughput analysis of membrane proteins. Mol Cell Proteomics. 2005;4:1948–1958. doi: 10.1074/mcp.M500138-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishmael JE, Safic M, Amparan D, Vogel WK, Pham T, Marley K, Filtz TM, Maier CS. Nonmuscle myosins II-B and Va are components of detergent-resistant membrane skeletons derived from mouse forebrain. Brain Res. 2007;1143:46–59. doi: 10.1016/j.brainres.2007.01.061. [DOI] [PubMed] [Google Scholar]
- Kim KB, Lee JW, Lee CS, Kim BW, Choo HJ, Jung SY, Chi SG, Yoon YS, Yoon G, Ko YG. Oxidation-reduction respiratory chains and ATP synthase complex are localized in detergent-resistant lipid rafts. Proteomics. 2006;6:2444–2453. doi: 10.1002/pmic.200500574. [DOI] [PubMed] [Google Scholar]
- Kisby GE, Standley M, Park T, Olivas A, Fei S, Jacob T, Reddy A, Lu X, Pattee P, Nagalla SR. Proteomic analysis of the genotoxicant methylazoxymethanol (MAM)-induced changes in the developing cerebellum. J Proteome Res. 2006;5:2656–2665. doi: 10.1021/pr060126g. [DOI] [PubMed] [Google Scholar]
- Sheikh AM, Barrett C, Villamizar N, Alzate O, Miller S, Shelburne J, Lodge A, Lawson J, Jaggers J. Proteomics of cerebral injury in a neonatal model of cardiopulmonary bypass with deep hypothermic circulatory arrest. J Thorac Cardiovasc Surg. 2006;132:820–828. doi: 10.1016/j.jtcvs.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Spitzer AR, Chace D. Mass spectrometry in neonatal medicine and clinical diagnosis--the [corrected] potential use of mass spectrometry in neonatal brain [corrected] monitoring. Clin Perinatol. 2006;33:729–44, viii. doi: 10.1016/j.clp.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Yang ZJ, Appleby VJ, Coyle B, Chan WI, Tahmaseb M, Wigmore PM, Scotting PJ. Novel strategy to study gene expression and function in developing cerebellar granule cells. J Neurosci Methods. 2004;132:149–160. doi: 10.1016/j.jneumeth.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Kovacs WJ, Shackelford JE, Tape KN, Richards MJ, Faust PL, Fliesler SJ, Krisans SK. Disturbed cholesterol homeostasis in a peroxisome-deficient PEX2 knockout mouse model. Mol Cell Biol. 2004;24:1–13. doi: 10.1128/MCB.24.1.1-13.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edison R, Muenke M. The interplay of genetic and environmental factors in craniofacial morphogenesis: holoprosencephaly and the role of cholesterol. Congenit Anom (Kyoto) 2003;43:1–21. doi: 10.1111/j.1741-4520.2003.tb01022.x. [DOI] [PubMed] [Google Scholar]
- Nissenkorn A, Michelson M, Ben-Zeev B, Lerman-Sagie T. Inborn errors of metabolism: a cause of abnormal brain development. Neurology. 2001;56:1265–1272. doi: 10.1212/wnl.56.10.1265. [DOI] [PubMed] [Google Scholar]
- FitzPatrick DR, Keeling JW, Evans MJ, Kan AE, Bell JE, Porteous ME, Mills K, Winter RM, Clayton PT. Clinical phenotype of desmosterolosis. Am J Med Genet. 1998;75:145–152. doi: 10.1002/(SICI)1096-8628(19980113)75:2<145::AID-AJMG5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Opitz JM, de la Cruz F. Cholesterol metabolism in the RSH/Smith-Lemli-Opitz syndrome: summary of an NICHD conference. Am J Med Genet. 1994;50:326–338. doi: 10.1002/ajmg.1320500406. [DOI] [PubMed] [Google Scholar]
- Powers JM, Tummons RC, Moser AB, Moser HW, Huff DS, Kelley RI. Neuronal lipidosis and neuroaxonal dystrophy in cerebro-hepato-renal (Zellweger) syndrome. Acta Neuropathol (Berl) 1987;73:333–343. doi: 10.1007/BF00688256. [DOI] [PubMed] [Google Scholar]
- Li N, Shaw AR, Zhang N, Mak A, Li L. Lipid raft proteomics: analysis of in-solution digest of sodium dodecyl sulfate-solubilized lipid raft proteins by liquid chromatography-matrix-assisted laser desorption/ionization tandem mass spectrometry. Proteomics. 2004;4:3156–3166. doi: 10.1002/pmic.200400832. [DOI] [PubMed] [Google Scholar]
- Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Arnaud L, Cooper JA. Lipid-dependent recruitment of neuronal Src to lipid rafts in the brain. J Biol Chem. 2003;278:40806–40814. doi: 10.1074/jbc.M306440200. [DOI] [PubMed] [Google Scholar]
- Melkonian KA, Ostermeyer AG, Chen JZ, Roth MG, Brown DA. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J Biol Chem. 1999;274:3910–3917. doi: 10.1074/jbc.274.6.3910. [DOI] [PubMed] [Google Scholar]
- Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: un amour impossible? Electrophoresis. 2000;21:1054–1070. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1054::AID-ELPS1054>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Blonder J, Chan KC, Issaq HJ, Veenstra TD. Identification of membrane proteins from mammalian cell/tissue using methanol-facilitated solubilization and tryptic digestion coupled with 2D-LC-MS/MS. Nat Protoc. 2006;1:2784–2790. doi: 10.1038/nprot.2006.359. [DOI] [PubMed] [Google Scholar]
- States DJ, Omenn GS, Blackwell TW, Fermin D, Eng J, Speicher DW, Hanash SM. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat Biotechnol. 2006;24:333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- Elmariah SB, Hughes EG, Oh EJ, Balice-Gordon RJ. Neurotrophin signaling among neurons and glia during formation of tripartite synapses. Neuron Glia Biol. 2005;1:1–11. doi: 10.1017/S1740925X05000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foty RA, Steinberg MS. Cadherin-mediated cell-cell adhesion and tissue segregation in relation to malignancy. Int J Dev Biol. 2004;48:397–409. doi: 10.1387/ijdb.041810rf. [DOI] [PubMed] [Google Scholar]
- Scheiffele P. Cell-cell signaling during synapse formation in the CNS. Annu Rev Neurosci. 2003;26:485–508. doi: 10.1146/annurev.neuro.26.043002.094940. [DOI] [PubMed] [Google Scholar]
- Fetissov SO, Bergstrom U, Johansen JE, Hokfelt T, Schalling M, Ranscht B. Alterations of arcuate nucleus neuropeptidergic development in contactin-deficient mice: comparison with anorexia and food-deprived mice. Eur J Neurosci. 2005;22:3217–3228. doi: 10.1111/j.1460-9568.2005.04513.x. [DOI] [PubMed] [Google Scholar]
- Yang JW, Rodrigo R, Felipo V, Lubec G. Proteome analysis of primary neurons and astrocytes from rat cerebellum. J Proteome Res. 2005;4:768–788. doi: 10.1021/pr049774v. [DOI] [PubMed] [Google Scholar]
- Dremina ES, Sharov VS, Schoneich C. Protein tyrosine nitration in rat brain is associated with raft proteins, flotillin-1 and alpha-tubulin: effect of biological aging. J Neurochem. 2005;93:1262–1271. doi: 10.1111/j.1471-4159.2005.03115.x. [DOI] [PubMed] [Google Scholar]
- Head BP, Patel HH, Roth DM, Murray F, Swaney JS, Niesman IR, Farquhar MG, Insel PA. Microtubules and actin microfilaments regulate lipid raft/caveolae localization of adenylyl cyclase signaling components. J Biol Chem. 2006;281:26391–26399. doi: 10.1074/jbc.M602577200. [DOI] [PubMed] [Google Scholar]
- Chen S, Bawa D, Besshoh S, Gurd JW, Brown IR. Association of heat shock proteins and neuronal membrane components with lipid rafts from the rat brain. J Neurosci Res. 2005;81:522–529. doi: 10.1002/jnr.20575. [DOI] [PubMed] [Google Scholar]
- Say YH, Hooper NM. Contamination of nuclear fractions with plasma membrane lipid rafts. Proteomics. 2007;7:1059–1064. doi: 10.1002/pmic.200600849. [DOI] [PubMed] [Google Scholar]
- Bae TJ, Kim MS, Kim JW, Kim BW, Choo HJ, Lee JW, Kim KB, Lee CS, Kim JH, Chang SY, Kang CY, Lee SW, Ko YG. Lipid raft proteome reveals ATP synthase complex in the cell surface. Proteomics. 2004;4:3536–3548. doi: 10.1002/pmic.200400952. [DOI] [PubMed] [Google Scholar]
- Igbavboa U, Eckert GP, Malo TM, Studniski AE, Johnson LN, Yamamoto N, Kobayashi M, Fujita SC, Appel TR, Muller WE, Wood WG, Yanagisawa K. Murine synaptosomal lipid raft protein and lipid composition are altered by expression of human apoE 3 and 4 and by increasing age. J Neurol Sci. 2005;229-230:225–232. doi: 10.1016/j.jns.2004.11.037. [DOI] [PubMed] [Google Scholar]
- Thouvenot E, Lafon-Cazal M, Demettre E, Jouin P, Bockaert J, Marin P. The proteomic analysis of mouse choroid plexus secretome reveals a high protein secretion capacity of choroidal epithelial cells. Proteomics. 2006;6:5941–5952. doi: 10.1002/pmic.200600096. [DOI] [PubMed] [Google Scholar]
- Pelkmans L, Puntener D, Helenius A. Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science. 2002;296:535–539. doi: 10.1126/science.1069784. [DOI] [PubMed] [Google Scholar]
- Chang L, Goldman RD. Intermediate filaments mediate cytoskeletal crosstalk. Nat Rev Mol Cell Biol. 2004;5:601–613. doi: 10.1038/nrm1438. [DOI] [PubMed] [Google Scholar]
- Franzen R, Tanner SL, Dashiell SM, Rottkamp CA, Hammer JA, Quarles RH. Microtubule-associated protein 1B: a neuronal binding partner for myelin-associated glycoprotein. J Cell Biol. 2001;155:893–898. doi: 10.1083/jcb.200108137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Watson K, Edwards RJ, Shaunak S, Parmelee DC, Sarraf C, Gooderham NJ, Davies DS. Extra-nuclear location of histones in activated human peripheral blood lymphocytes and cultured T-cells. Biochem Pharmacol. 1995;50:299–309. doi: 10.1016/0006-2952(95)00142-M. [DOI] [PubMed] [Google Scholar]
- Lawen A, Ly JD, Lane DJ, Zarschler K, Messina A, De Pinto V. Voltage-dependent anion-selective channel 1 (VDAC1)--a mitochondrial protein, rediscovered as a novel enzyme in the plasma membrane. Int J Biochem Cell Biol. 2005;37:277–282. doi: 10.1016/j.biocel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Andreev VP, Li L, Cao L, Gu Y, Rejtar T, Wu SL, Karger BL. A New Algorithm Using Cross-Assignment for Label-Free Quantitation with LC-LTQ-FT MS. J Proteome Res. 2007 doi: 10.1021/pr0606880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienkoop S, Larrainzar E, Niemann M, Gonzalez EM, Lehmann U, Weckwerth W. Stable isotope-free quantitative shotgun proteomics combined with sample pattern recognition for rapid diagnostics. J Sep Sci. 2006;29:2793–2801. doi: 10.1002/jssc.200600290. [DOI] [PubMed] [Google Scholar]
- Le Bihan T, Goh T, Stewart. Salter AM, Bukhman YV, Dharsee M, Ewing R, Wisniewski JR. Differential analysis of membrane proteins in mouse fore- and hindbrain using a label-free approach. J Proteome Res. 2006;5:2701–2710. doi: 10.1021/pr060190y. [DOI] [PubMed] [Google Scholar]
- Wang G, Wu WW, Zeng W, Chou CL, Shen RF. Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: Reproducibility, linearity, and application with complex proteomes. J Proteome Res. 2006;5:1214–1223. doi: 10.1021/pr050406g. [DOI] [PubMed] [Google Scholar]
- Ru QC, Zhu LA, Silberman J, Shriver CD. Label-free semiquantitative peptide feature profiling of human breast cancer and breast disease sera via two-dimensional liquid chromatography-mass spectrometry. Mol Cell Proteomics. 2006;5:1095–1104. doi: 10.1074/mcp.M500387-MCP200. [DOI] [PubMed] [Google Scholar]
- Cui XY, Hu QD, Tekaya M, Shimoda Y, Ang BT, Nie DY, Sun L, Hu WP, Karsak M, Duka T, Takeda Y, Ou LY, Dawe GS, Yu FG, Ahmed S, Jin LH, Schachner M, Watanabe K, Arsenijevic Y, Xiao ZC. NB-3/Notch1 pathway via Deltex1 promotes neural progenitor cell differentiation into oligodendrocytes. J Biol Chem. 2004;279:25858–25865. doi: 10.1074/jbc.M313505200. [DOI] [PubMed] [Google Scholar]
- Brady ST, Lasek RJ. Nerve-specific enolase and creatine phosphokinase in axonal transport: soluble proteins and the axoplasmic matrix. Cell. 1981;23:515–523. doi: 10.1016/0092-8674(81)90147-1. [DOI] [PubMed] [Google Scholar]
- Deininger SO, Rajendran L, Lottspeich F, Przybylski M, Illges H, Stuermer CA, Reuter A. Identification of teleost Thy-1 and association with the microdomain/lipid raft reggie proteins in regenerating CNS axons. Mol Cell Neurosci. 2003;22:544–554. doi: 10.1016/S1044-7431(03)00028-9. [DOI] [PubMed] [Google Scholar]
- Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, Takeda Y, Chia W, Sankar N, Ng YK, Ling EA, Maciag T, Small D, Trifonova R, Kopan R, Okano H, Nakafuku M, Chiba S, Hirai H, Aster JC, Schachner M, Pallen CJ, Watanabe K, Xiao ZC. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell. 2003;115:163–175. doi: 10.1016/S0092-8674(03)00810-9. [DOI] [PubMed] [Google Scholar]
- Guirland C, Suzuki S, Kojima M, Lu B, Zheng JQ. Lipid rafts mediate chemotropic guidance of nerve growth cones. Neuron. 2004;42:51–62. doi: 10.1016/S0896-6273(04)00157-6. [DOI] [PubMed] [Google Scholar]
- Kashihara M, Miyata S, Kumanogoh H, Funatsu N, Matsunaga W, Kiyohara T, Sokawa Y, Maekawa S. Changes in the localization of NAP-22, a calmodulin binding membrane protein, during the development of neuronal polarity. Neurosci Res. 2000;37:315–325. doi: 10.1016/S0168-0102(00)00132-2. [DOI] [PubMed] [Google Scholar]
- Leshchyns'ka I, Sytnyk V, Morrow JS, Schachner M. Neural cell adhesion molecule (NCAM) association with PKCbeta2 via betaI spectrin is implicated in NCAM-mediated neurite outgrowth. J Cell Biol. 2003;161:625–639. doi: 10.1083/jcb.200303020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, Morii H, Kumanogoh H, Sano M, Naruse Y, Sokawa Y, Mori N. Localization of neuronal growth-associated, microtubule-destabilizing factor SCG10 in brain-derived raft membrane microdomains. J Biochem (Tokyo) 2001;129:691–697. doi: 10.1093/oxfordjournals.jbchem.a002908. [DOI] [PubMed] [Google Scholar]
- Miyata S, Funatsu N, Matsunaga W, Kiyohara T, Sokawa Y, Maekawa S. Expression of the IgLON cell adhesion molecules Kilon and OBCAM in hypothalamic magnocellular neurons. J Comp Neurol. 2000;424:74–85. doi: 10.1002/1096-9861(20000814)424:1<74::AID-CNE6>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Roussel G, Nussbaum F, Schoentgen F, Jolles P, Nussbaum JL. Immunological investigation of a 21-kilodalton cytosolic basic protein in rat brain. Dev Neurosci. 1988;10:65–74. doi: 10.1159/000111957. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Bonnet A, Erb M, Erne B, Bartsch U, Kern F, Mantei N, Sherman D, Suter U. The raft-associated protein MAL is required for maintenance of proper axon--glia interactions in the central nervous system. J Cell Biol. 2004;166:731–742. doi: 10.1083/jcb.200406092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicht K, Buttner A, Siedler F, Scheffer B, Zill P, Eisenmenger W, Ackenheil M, Bondy B. Comparative proteomic analysis with postmortem prefrontal cortex tissues of suicide victims versus controls. J Psychiatr Res. 2007;41:493–501. doi: 10.1016/j.jpsychires.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M, Nakamura K, Taga T. Roles of lipid rafts in integrin-dependent adhesion and gp130 signalling pathway in mouse embryonic neural precursor cells. Genes Cells. 2004;9:801–809. doi: 10.1111/j.1365-2443.2004.00764.x. [DOI] [PubMed] [Google Scholar]
- Yang JW, Suder P, Silberring J, Lubec G. Proteome analysis of mouse primary astrocytes. Neurochem Int. 2005;47:159–172. doi: 10.1016/j.neuint.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Schirle M, Heurtier MA, Kuster B. Profiling core proteomes of human cell lines by one-dimensional PAGE and liquid chromatography-tandem mass spectrometry. Mol Cell Proteomics. 2003;2:1297–1305. doi: 10.1074/mcp.M300087-MCP200. [DOI] [PubMed] [Google Scholar]
- Yu H, Li M, Tint GS, Chen J, Xu G, Patel SB. Selective reconstitution of liver cholesterol biosynthesis promotes lung maturation but does not prevent neonatal lethality in Dhcr7 null mice. BMC Dev Biol. 2007;7:27. doi: 10.1186/1471-213X-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks WA, Halligan BD, Slyper RY, Twigger SN, Greene AS, Olivier M. Simultaneous quantification and identification using 18O labeling with an ion trap mass spectrometer and the analysis software application "ZoomQuant". J Am Soc Mass Spectrom. 2005;16:916–925. doi: 10.1016/j.jasms.2005.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza SP, Halligan BD, Greene AS, Olivier M. An improved method for the analysis of membrane proteins by mass spectrometry. Physiol Genomics. 2007 doi: 10.1152/physiolgenomics.00279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan BD, Slyper RY, Twigger SN, Hicks W, Olivier M, Greene AS. ZoomQuant: an application for the quantitation of stable isotope labeled peptides. J Am Soc Mass Spectrom. 2005;16:302–306. doi: 10.1016/j.jasms.2004.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39:82–91. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]