Abstract
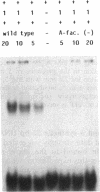
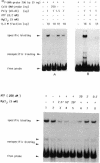
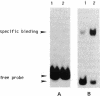
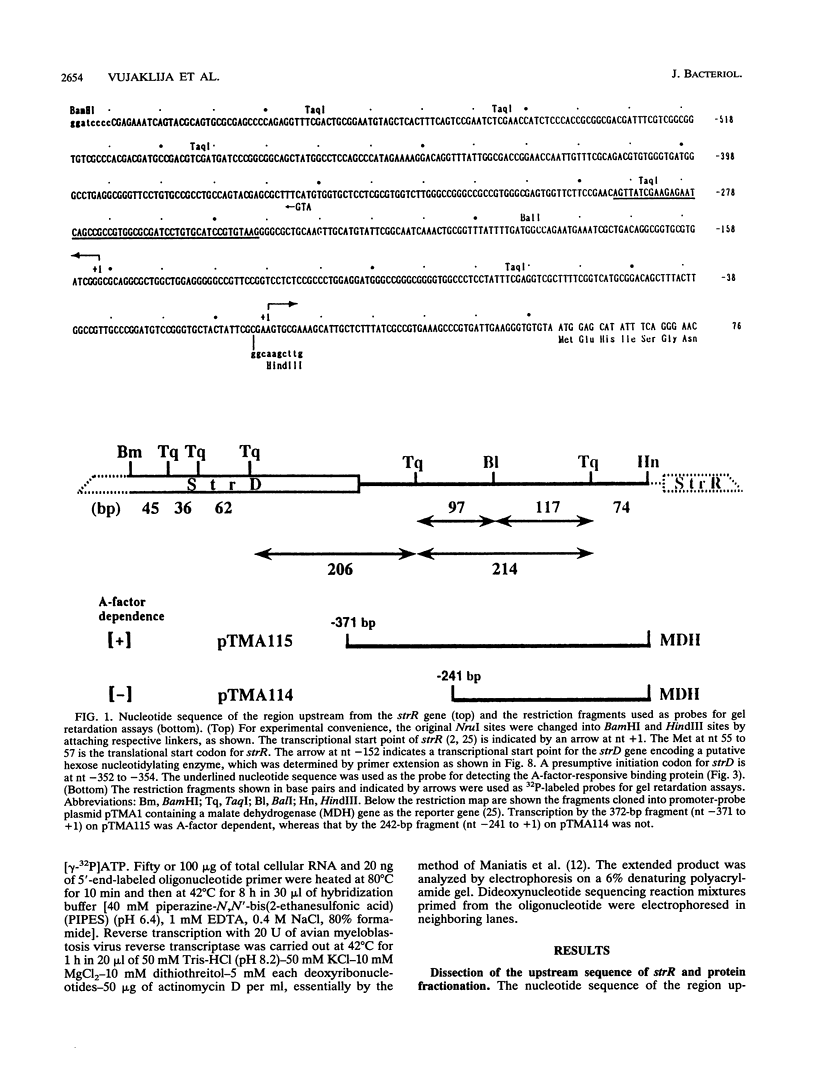
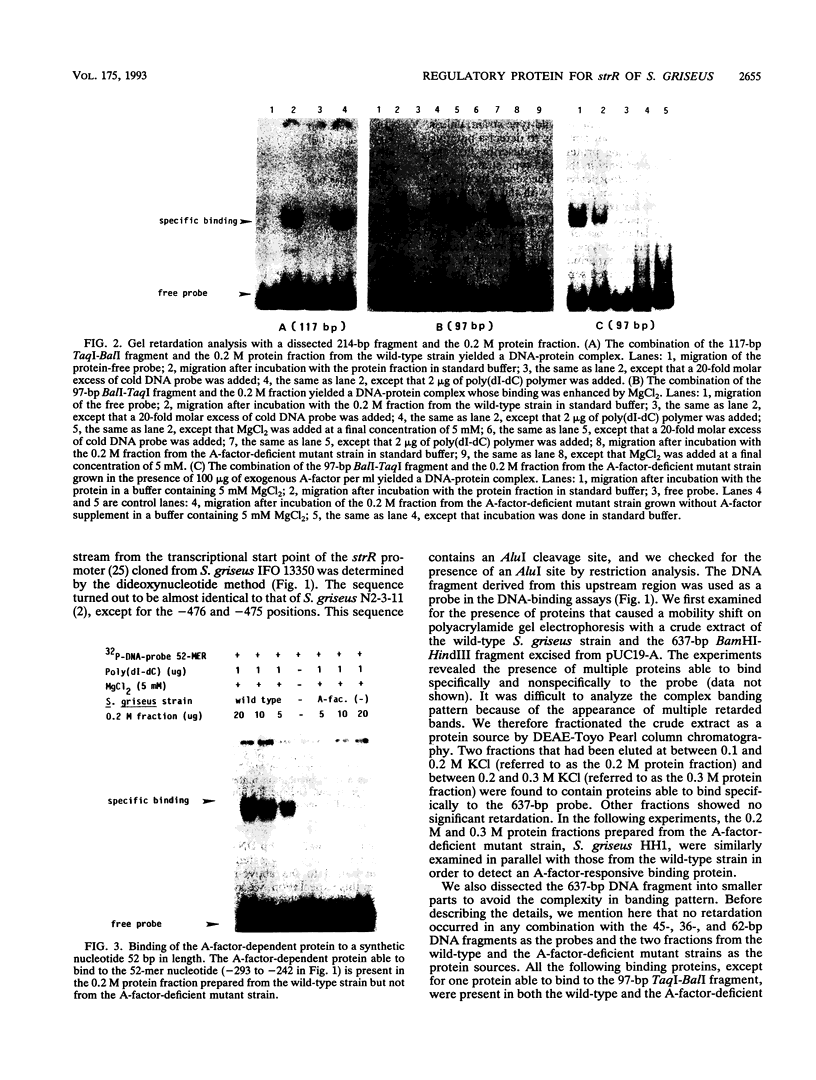
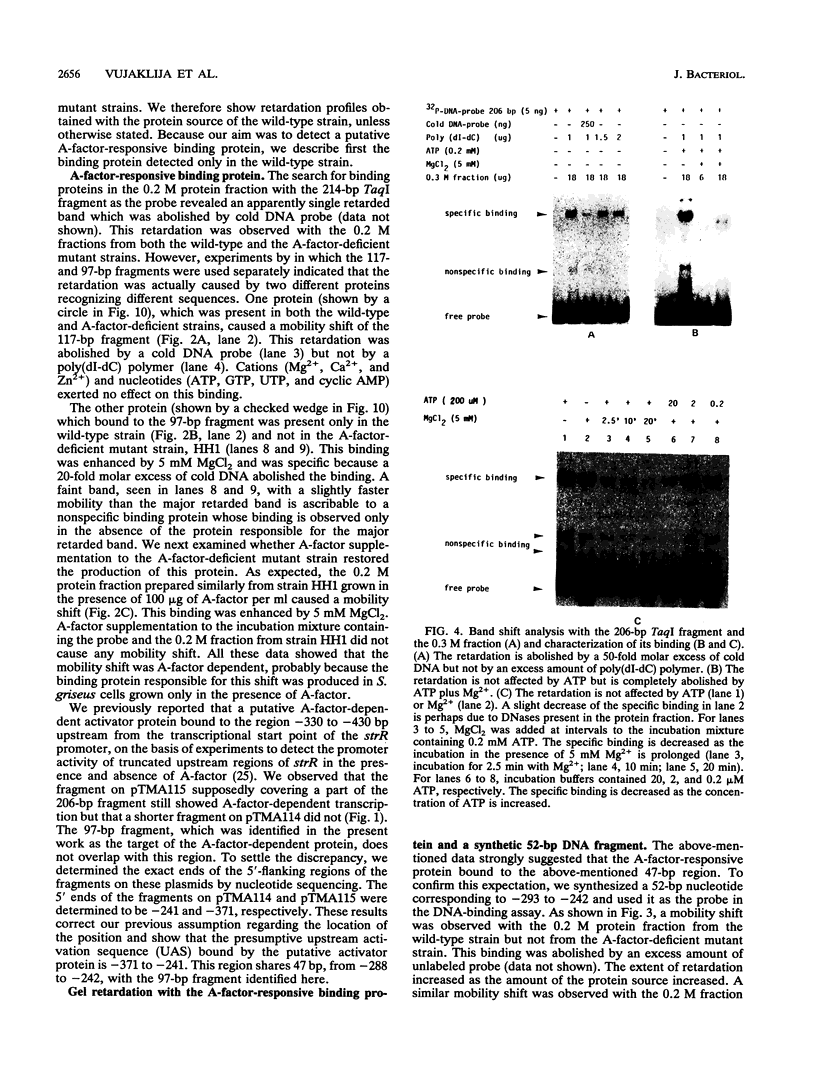
DNA-binding assays using mobility shift polyacrylamide gel electrophoresis revealed the presence of a protein that specifically bound to a restriction fragment -288 to -191 bp upstream from the transcriptional start point of strR, a regulatory gene for streptomycin biosynthesis in Streptomyces griseus. The binding site corresponded to an upstream activation sequence predicted from the results of in vivo promoter assays. The binding was greatly enhanced by 5 mM Mg2+. This binding was detected with the protein source only from the wild-type strain and not from an A-factor-deficient mutant strain. The exogenous supplementation of A-factor to the A-factor-deficient mutant strain caused the appearance of the protein in the DNA-binding assay. A synthetic nucleotide 52 bp in length (region from -293 to -242), which was synthesized on the basis of data obtained from both retardation assays with dissected DNA fragments and in vivo promoter assays, was retarded by the A-factor-dependent protein. In addition to this A-factor-dependent protein, at least three proteins with different recognition site affinities capable of binding to the upstream region of the strR promoter were detected. The binding of one of these proteins to both sides of the upstream activation sequence bound by the A-factor-dependent protein was completely abolished in the presence of ATP and Mg2+ in the incubation mixture. The region bound by these proteins showed anomalous electrophoretic mobility, like that of a bent DNA molecule, which is probably caused by the presence of many blocks consisting of A and T. The region bound by these proteins was found to be transcribed in the orientation opposite to that of strR.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Distler J., Ebert A., Mansouri K., Pissowotzki K., Stockmann M., Piepersberg W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 1987 Oct 12;15(19):8041–8056. doi: 10.1093/nar/15.19.8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler J., Mansouri K., Mayer G., Stockmann M., Piepersberg W. Streptomycin biosynthesis and its regulation in Streptomycetes. Gene. 1992 Jun 15;115(1-2):105–111. doi: 10.1016/0378-1119(92)90547-3. [DOI] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Hara O., Beppu T. Mutants blocked in streptomycin production in Streptomyces griseus - the role of A-factor. J Antibiot (Tokyo) 1982 Mar;35(3):349–358. doi: 10.7164/antibiotics.35.349. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Beppu T. Autoregulatory factors and communication in actinomycetes. Annu Rev Microbiol. 1992;46:377–398. doi: 10.1146/annurev.mi.46.100192.002113. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Beppu T. Regulation of secondary metabolism and cell differentiation in Streptomyces: A-factor as a microbial hormone and the AfsR protein as a component of a two-component regulatory system. Gene. 1992 Jun 15;115(1-2):167–172. doi: 10.1016/0378-1119(92)90555-4. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Kumada Y., Beppu T. Unstable genetic determinant of A-factor biosynthesis in streptomycin-producing organisms: cloning and characterization. J Bacteriol. 1984 May;158(2):481–487. doi: 10.1128/jb.158.2.481-487.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokhlov A. S., Tovarova I. I., Borisova L. N., Pliner S. A., Shevchenko L. N., Kornitskaia E. Ia, Ivkina N. S., Rapoport I. A. A-faktor, obespechivaiushchii biosintez streptomitsina mutantnym shtammom Actinomyces streptomycini. Dokl Akad Nauk SSSR. 1967 Nov-Dec;177(1):232–235. [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Linial M., Shlomai J. Sequence-directed bent DNA helix is the specific binding site for Crithidia fasciculata nicking enzyme. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8205–8209. doi: 10.1073/pnas.84.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Effron P. N., Goodman T. C., Singleton C. K., Wells R. D., Wartell R. M., Englund P. T. Physical characterization of a kinetoplast DNA fragment with unusual properties. J Biol Chem. 1984 Jul 25;259(14):8974–8979. [PubMed] [Google Scholar]
- Miyake K., Horinouchi S., Yoshida M., Chiba N., Mori K., Nogawa N., Morikawa N., Beppu T. Detection and properties of A-factor-binding protein from Streptomyces griseus. J Bacteriol. 1989 Aug;171(8):4298–4302. doi: 10.1128/jb.171.8.4298-4302.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake K., Kuzuyama T., Horinouchi S., Beppu T. The A-factor-binding protein of Streptomyces griseus negatively controls streptomycin production and sporulation. J Bacteriol. 1990 Jun;172(6):3003–3008. doi: 10.1128/jb.172.6.3003-3008.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T. Static bend of DNA helix at the activator recognition site of the ompF promoter in Escherichia coli. Gene. 1987;54(1):57–64. doi: 10.1016/0378-1119(87)90347-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Buchman A. R., Davis R. W. Bent DNA at a yeast autonomously replicating sequence. Nature. 1986 Nov 6;324(6092):87–89. doi: 10.1038/324087a0. [DOI] [PubMed] [Google Scholar]
- Strohl W. R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992 Mar 11;20(5):961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. DNA structure. Curves with a function. Nature. 1989 Sep 21;341(6239):184–185. doi: 10.1038/341184a0. [DOI] [PubMed] [Google Scholar]
- Ueki N., Kinashi H., Mizuno T. Primary characterization of curved DNA segments cloned from Streptomyces DNA with extremely high G/C-contents. Agric Biol Chem. 1991 Jan;55(1):173–180. [PubMed] [Google Scholar]
- Vujaklija D., Ueda K., Hong S. K., Beppu T., Horinouchi S. Identification of an A-factor-dependent promoter in the streptomycin biosynthetic gene cluster of Streptomyces griseus. Mol Gen Genet. 1991 Sep;229(1):119–128. doi: 10.1007/BF00264220. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]