Abstract
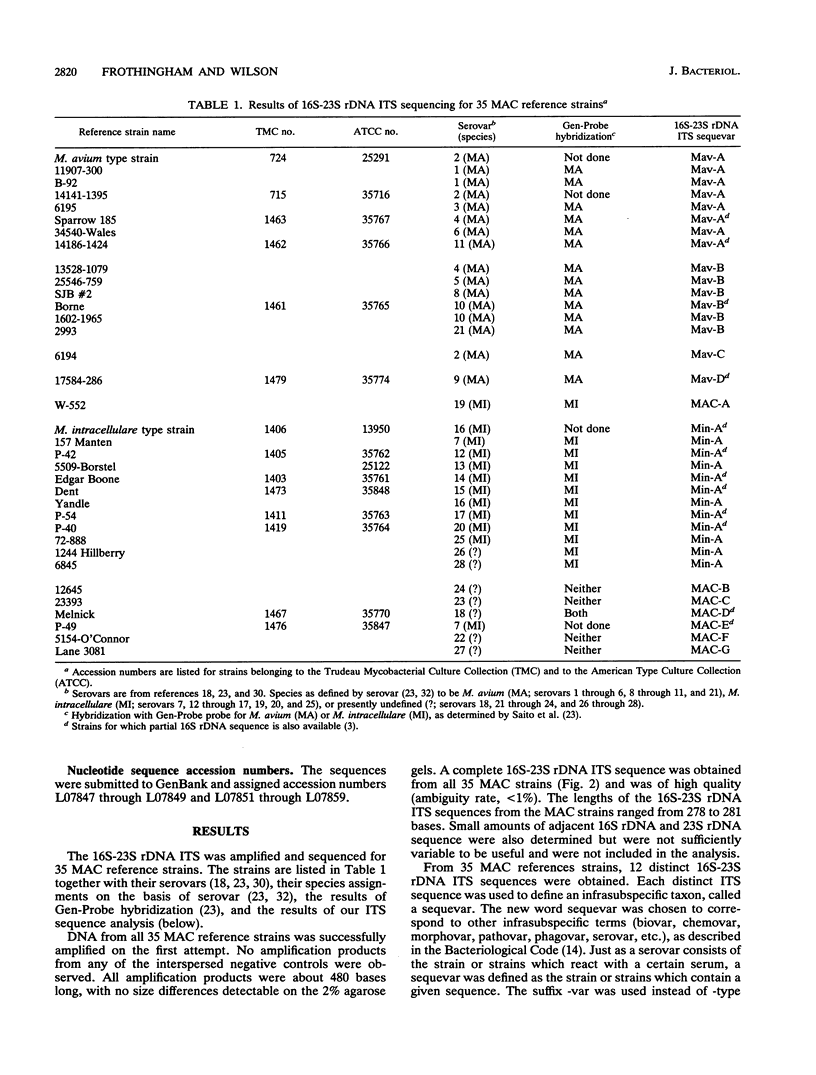
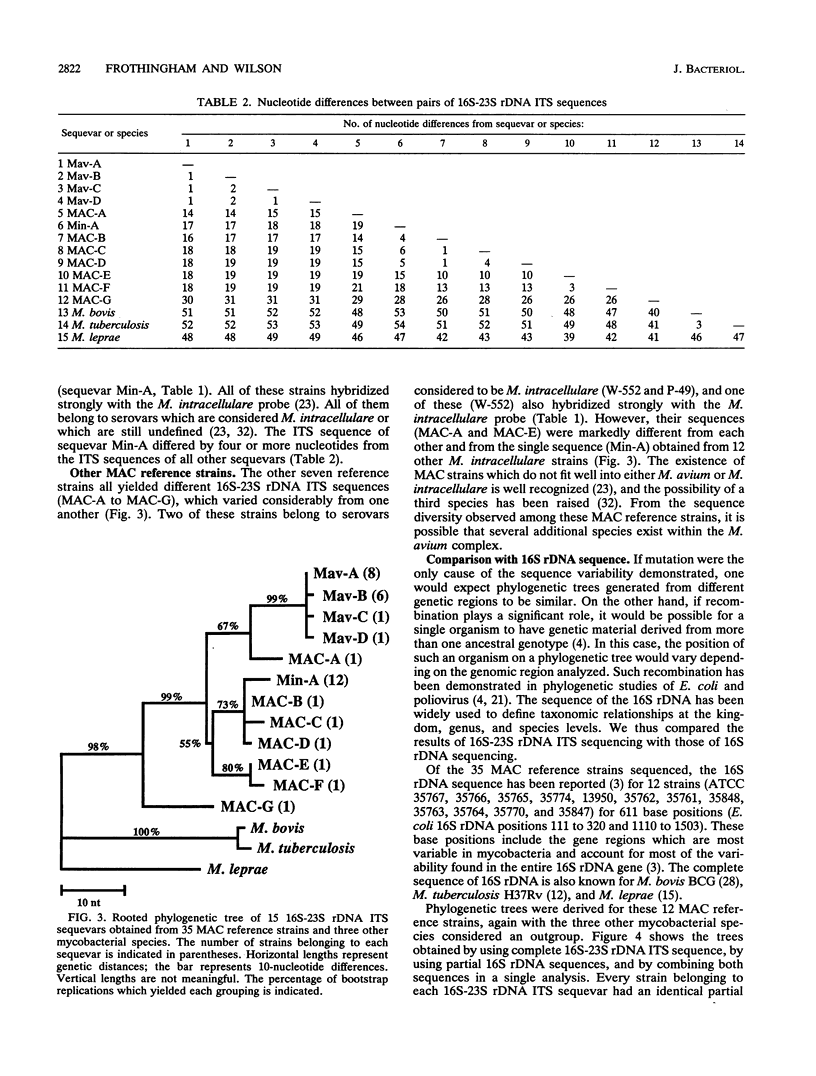
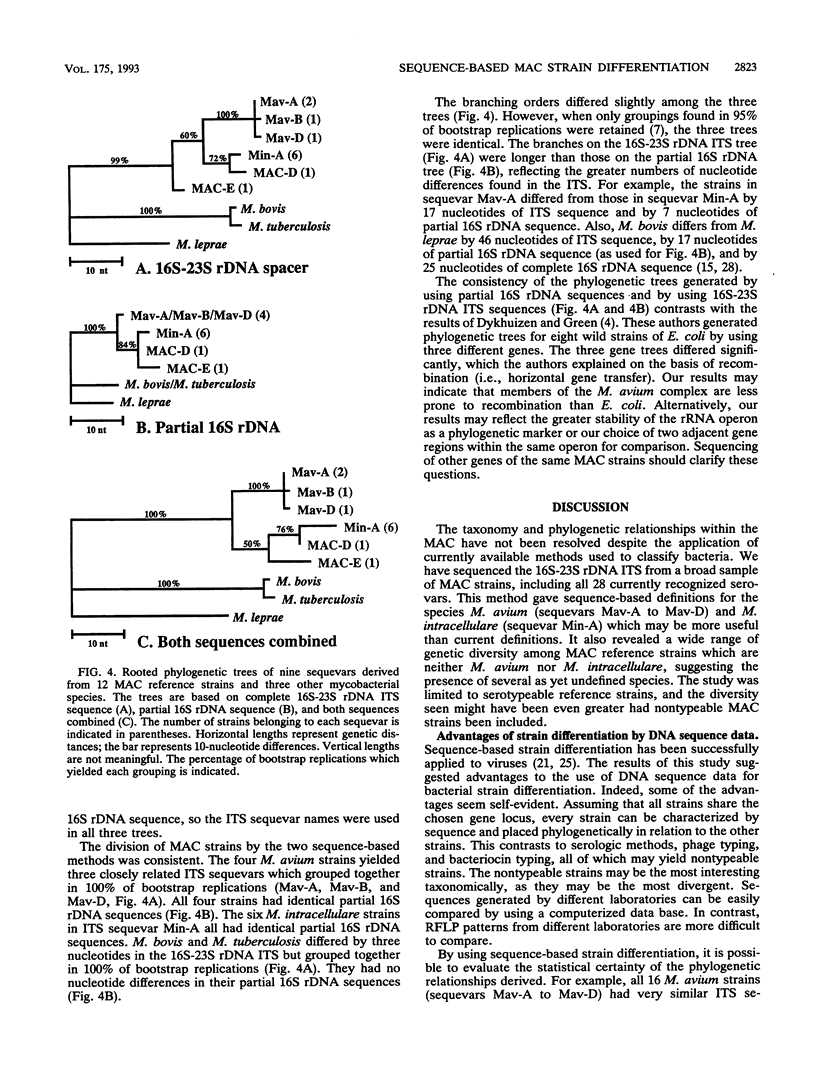
The complete 16S-23S rDNA internal transcribed spacer (ITS) was sequenced in 35 reference strains of the Mycobacterium avium complex. Twelve distinct ITS sequences were obtained, each of which defined a "sequevar"; a sequevar consists of the strain or strains which have a particular sequence. ITS sequences were identified which corresponded to M. avium (16 strains, four ITS sequevars) and Mycobacterium intracellulare (12 strains, one ITS sequevars). The other seven M. avium complex strains had ITS sequences which varied greatly from those of M. avium and M. intracellulare and from each other. The 16S-23S rDNA ITS was much more variable than 16S rDNA, which is widely used for genus and species identification. Phylogenetic trees based on the ITS were compatible with those based on 16S rDNA but were more detailed and had longer branches. The results of ITS sequencing were consistent with the results of hybridization with M. avium and M. intracellulare probes (Gen-Probe) for 30 of 31 strains tested. Serologic testing correlated poorly with ITS sequencing. Strains with the same sequence were different serovars, and those of the same serovar had different sequences. Sequencing of the 16S-23S rDNA ITS should be useful for species and strain differentiation for a wide variety of bacteria and should be applicable to studies of epidemiology, diagnosis, virulence, and taxonomy.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barry T., Colleran G., Glennon M., Dunican L. K., Gannon F. The 16s/23s ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1991 Aug;1(1):51–56. doi: 10.1101/gr.1.1.51. [DOI] [PubMed] [Google Scholar]
- Bercovier H., Kafri O., Sela S. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem Biophys Res Commun. 1986 May 14;136(3):1136–1141. doi: 10.1016/0006-291x(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Böddinghaus B., Wolters J., Heikens W., Böttger E. C. Phylogenetic analysis and identification of different serovars of Mycobacterium intracellulare at the molecular level. FEMS Microbiol Lett. 1990 Jul;58(2):197–203. doi: 10.1111/j.1574-6968.1990.tb13978.x. [DOI] [PubMed] [Google Scholar]
- Dykhuizen D. E., Green L. Recombination in Escherichia coli and the definition of biological species. J Bacteriol. 1991 Nov;173(22):7257–7268. doi: 10.1128/jb.173.22.7257-7268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein B. I. New molecular techniques for microbial epidemiology and the diagnosis of infectious diseases. J Infect Dis. 1990 Apr;161(4):595–602. doi: 10.1093/infdis/161.4.595. [DOI] [PubMed] [Google Scholar]
- Falkinham J. O., 3rd, Parker B. C., Gruft H. Epidemiology of infection by nontuberculous mycobacteria. I. Geographic distribution in the eastern United States. Am Rev Respir Dis. 1980 Jun;121(6):931–937. doi: 10.1164/arrd.1980.121.6.931. [DOI] [PubMed] [Google Scholar]
- Frothingham R., Allen R. L., Wilson K. H. Rapid 16S ribosomal DNA sequencing from a single colony without DNA extraction or purification. Biotechniques. 1991 Jul;11(1):40–44. [PubMed] [Google Scholar]
- Frothingham R., Blitchington R. B., Lee D. H., Greene R. C., Wilson K. H. UV absorption complicates PCR decontamination. Biotechniques. 1992 Aug;13(2):208–210. [PubMed] [Google Scholar]
- Grange J. M., Yates M. D., Boughton E. The avian tubercle bacillus and its relatives. J Appl Bacteriol. 1990 May;68(5):411–431. doi: 10.1111/j.1365-2672.1990.tb02892.x. [DOI] [PubMed] [Google Scholar]
- Hampson S. J., Portaels F., Thompson J., Green E. P., Moss M. T., Hermon-Taylor J., McFadden J. J. DNA probes demonstrate a single highly conserved strain of Mycobacterium avium infecting AIDS patients. Lancet. 1989 Jan 14;1(8629):65–68. doi: 10.1016/s0140-6736(89)91427-x. [DOI] [PubMed] [Google Scholar]
- Kempsell K. E., Ji Y. E., Estrada I. C., Colston M. J., Cox R. A. The nucleotide sequence of the promoter, 16S rRNA and spacer region of the ribosomal RNA operon of Mycobacterium tuberculosis and comparison with Mycobacterium leprae precursor rRNA. J Gen Microbiol. 1992 Aug;138(Pt 8):1717–1727. doi: 10.1099/00221287-138-8-1717. [DOI] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesack W., Pitulle C., Sela S., Stackebrandt E. Nucleotide sequence of the 16S rRNA from Mycobacterium leprae. Nucleic Acids Res. 1990 Sep 25;18(18):5558–5558. doi: 10.1093/nar/18.18.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesack W., Sela S., Bercovier H., Pitulle C., Stackebrandt E. Complete nucleotide sequence of the Mycobacterium leprae 23 S and 5 S rRNA genes plus flanking regions and their potential in designing diagnostic oligonucleotide probes. FEBS Lett. 1991 Apr 9;281(1-2):114–118. doi: 10.1016/0014-5793(91)80372-a. [DOI] [PubMed] [Google Scholar]
- McFadden J. J., Butcher P. D., Thompson J., Chiodini R., Hermon-Taylor J. The use of DNA probes identifying restriction-fragment-length polymorphisms to examine the Mycobacterium avium complex. Mol Microbiol. 1987 Nov;1(3):283–291. doi: 10.1111/j.1365-2958.1987.tb01934.x. [DOI] [PubMed] [Google Scholar]
- Pitulle C., Dorsch M., Kazda J., Wolters J., Stackebrandt E. Phylogeny of rapidly growing members of the genus Mycobacterium. Int J Syst Bacteriol. 1992 Jul;42(3):337–343. doi: 10.1099/00207713-42-3-337. [DOI] [PubMed] [Google Scholar]
- Regnery R. L., Spruill C. L., Plikaytis B. D. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J Bacteriol. 1991 Mar;173(5):1576–1589. doi: 10.1128/jb.173.5.1576-1589.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico-Hesse R., Pallansch M. A., Nottay B. K., Kew O. M. Geographic distribution of wild poliovirus type 1 genotypes. Virology. 1987 Oct;160(2):311–322. doi: 10.1016/0042-6822(87)90001-8. [DOI] [PubMed] [Google Scholar]
- Rogall T., Wolters J., Flohr T., Böttger E. C. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int J Syst Bacteriol. 1990 Oct;40(4):323–330. doi: 10.1099/00207713-40-4-323. [DOI] [PubMed] [Google Scholar]
- Saito H., Tomioka H., Sato K., Tasaka H., Dawson D. J. Identification of various serovar strains of Mycobacterium avium complex by using DNA probes specific for Mycobacterium avium and Mycobacterium intracellulare. J Clin Microbiol. 1990 Aug;28(8):1694–1697. doi: 10.1128/jcm.28.8.1694-1697.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet P., Normand P., Moiroud A., Bardin R. Identification of Frankia strains in nodules by hybridization of polymerase chain reaction products with strain-specific oligonucleotide probes. Arch Microbiol. 1990;153(3):235–240. doi: 10.1007/BF00249074. [DOI] [PubMed] [Google Scholar]
- Smith J. S., Orciari L. A., Yager P. A., Seidel H. D., Warner C. K. Epidemiologic and historical relationships among 87 rabies virus isolates as determined by limited sequence analysis. J Infect Dis. 1992 Aug;166(2):296–307. doi: 10.1093/infdis/166.2.296. [DOI] [PubMed] [Google Scholar]
- Stahl D. A., Urbance J. W. The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria. J Bacteriol. 1990 Jan;172(1):116–124. doi: 10.1128/jb.172.1.116-124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoflet E. S., Koeberl D. D., Sarkar G., Sommer S. S. Genomic amplification with transcript sequencing. Science. 1988 Jan 29;239(4839):491–494. doi: 10.1126/science.3340835. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Nagata A., Ono Y., Yamada T. Complete nucleotide sequence of the 16S rRNA gene of Mycobacterium bovis BCG. J Bacteriol. 1988 Jun;170(6):2886–2889. doi: 10.1128/jb.170.6.2886-2889.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasem C. F., McCarthy C. M., Murray L. W. Multilocus enzyme electrophoresis analysis of the Mycobacterium avium complex and other mycobacteria. J Clin Microbiol. 1991 Feb;29(2):264–271. doi: 10.1128/jcm.29.2.264-271.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne L. G., Sramek H. A. Agents of newly recognized or infrequently encountered mycobacterial diseases. Clin Microbiol Rev. 1992 Jan;5(1):1–25. doi: 10.1128/cmr.5.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. H., Blitchington R. B., Greene R. C. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990 Sep;28(9):1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakrus M. A., Good R. C. Geographic distribution, frequency, and specimen source of Mycobacterium avium complex serotypes isolated from patients with acquired immunodeficiency syndrome. J Clin Microbiol. 1990 May;28(5):926–929. doi: 10.1128/jcm.28.5.926-929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]



