Abstract
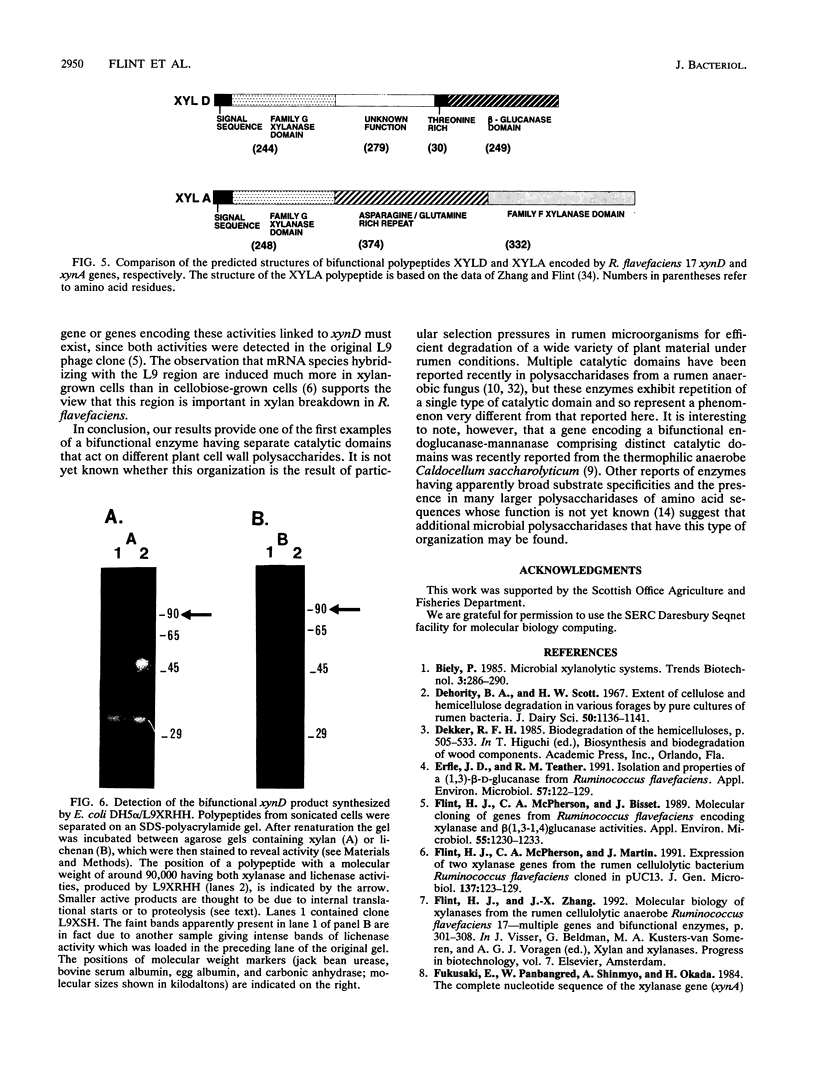
Adjacent regions of a Ruminococcus flavefaciens 17 DNA fragment were found to encode xylanase and beta(1,3-1,4)-glucanase activities. Sequencing of this fragment showed that both activities are encoded by a single 2,406-bp open reading frame corresponding to the xynD gene. The predicted product has a characteristic signal sequence that is followed by an amino-terminal domain related to family G xylanases, while the carboxyterminal domain is related to beta(1,3-1,4)-glucanases from several other bacterial species. These two domains are connected by a region of unknown function that consists of 309 amino acids and includes a 30-amino-acid threonine-rich sequence. A polypeptide having a molecular weight of approximately 90,000 and exhibiting xylanase and beta(1,3-1,4)-glucanase activities was detected in Escherichia coli cells carrying the cloned xynD gene. This is one of the first cases in which a microbial polysaccharidase has been shown to carry separate catalytic domains active against different plant cell wall polysaccharides within the same polypeptide. xynD is one of a family of related genes in R. flavefaciens that encode enzymes having multiple catalytic domains, and the amino terminus of XYLD exhibits a high degree of similarity with the corresponding regions of another xylanase, XYLA, which carries two different xylanase catalytic domains.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Erfle J. D., Teather R. M. Isolation and properties of a (1,3)-beta-D-glucanase from Ruminococcus flavefaciens. Appl Environ Microbiol. 1991 Jan;57(1):122–129. doi: 10.1128/aem.57.1.122-129.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint H. J., McPherson C. A., Bisset J. Molecular cloning of genes from Ruminococcus flavefaciens encoding xylanase and beta(1-3,1-4)glucanase activities. Appl Environ Microbiol. 1989 May;55(5):1230–1233. doi: 10.1128/aem.55.5.1230-1233.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint H. J., McPherson C. A., Martin J. Expression of two xylanase genes from the rumen cellulolytic bacterium Ruminococcus flavefaciens 17 cloned in pUC13. J Gen Microbiol. 1991 Jan;137(1):123–129. doi: 10.1099/00221287-137-1-123. [DOI] [PubMed] [Google Scholar]
- Gibbs M. D., Saul D. J., Lüthi E., Bergquist P. L. The beta-mannanase from "Caldocellum saccharolyticum" is part of a multidomain enzyme. Appl Environ Microbiol. 1992 Dec;58(12):3864–3867. doi: 10.1128/aem.58.12.3864-3867.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert H. J., Hazlewood G. P., Laurie J. I., Orpin C. G., Xue G. P. Homologous catalytic domains in a rumen fungal xylanase: evidence for gene duplication and prokaryotic origin. Mol Microbiol. 1992 Aug;6(15):2065–2072. doi: 10.1111/j.1365-2958.1992.tb01379.x. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalbes M. J., Pérez-González J. A., González R., Navarro A. Two beta-glycanase genes are clustered in Bacillus polymyxa: molecular cloning, expression, and sequence analysis of genes encoding a xylanase and an endo-beta-(1,3)-(1,4)-glucanase. J Bacteriol. 1991 Dec;173(23):7705–7710. doi: 10.1128/jb.173.23.7705-7710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991 Dec 1;280(Pt 2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. F., Forsberg C. W., Rattray J. B. Purification and Characterization of Two Endoxylanases from Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1987 Apr;53(4):644–650. doi: 10.1128/aem.53.4.644-650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloberas J., Perez-Pons J. A., Querol E. Molecular cloning, expression and nucleotide sequence of the endo-beta-1,3-1,4-D-glucanase gene from Bacillus licheniformis. Predictive structural analyses of the encoded polypeptide. Eur J Biochem. 1991 Apr 23;197(2):337–343. doi: 10.1111/j.1432-1033.1991.tb15916.x. [DOI] [PubMed] [Google Scholar]
- Murphy N., McConnell D. J., Cantwell B. A. The DNA sequence of the gene and genetic control sites for the excreted B. subtilis enzyme beta-glucanase. Nucleic Acids Res. 1984 Jul 11;12(13):5355–5367. doi: 10.1093/nar/12.13.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul D. J., Williams L. C., Grayling R. A., Chamley L. W., Love D. R., Bergquist P. L. celB, a gene coding for a bifunctional cellulase from the extreme thermophile "Caldocellum saccharolyticum". Appl Environ Microbiol. 1990 Oct;56(10):3117–3124. doi: 10.1128/aem.56.10.3117-3124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimming S., Schwarz W. H., Staudenbauer W. L. Structure of the Clostridium thermocellum gene licB and the encoded beta-1,3-1,4-glucanase. A catalytic region homologous to Bacillus lichenases joined to the reiterated domain of clostridial cellulases. Eur J Biochem. 1992 Feb 15;204(1):13–19. doi: 10.1111/j.1432-1033.1992.tb16600.x. [DOI] [PubMed] [Google Scholar]
- Teather R. M., Erfle J. D. DNA sequence of a Fibrobacter succinogenes mixed-linkage beta-glucanase (1,3-1,4-beta-D-glucan 4-glucanohydrolase) gene. J Bacteriol. 1990 Jul;172(7):3837–3841. doi: 10.1128/jb.172.7.3837-3841.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teather R. M., Wood P. J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982 Apr;43(4):777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Wong K. K., Tan L. U., Saddler J. N. Multiplicity of beta-1,4-xylanase in microorganisms: functions and applications. Microbiol Rev. 1988 Sep;52(3):305–317. doi: 10.1128/mr.52.3.305-317.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G. P., Gobius K. S., Orpin C. G. A novel polysaccharide hydrolase cDNA (celD) from Neocallimastix patriciarum encoding three multi-functional catalytic domains with high endoglucanase, cellobiohydrolase and xylanase activities. J Gen Microbiol. 1992 Nov;138(11):2397–2403. doi: 10.1099/00221287-138-11-2397. [DOI] [PubMed] [Google Scholar]
- Yahata N., Watanabe T., Nakamura Y., Yamamoto Y., Kamimiya S., Tanaka H. Structure of the gene encoding beta-1,3-glucanase A1 of Bacillus circulans WL-12. Gene. 1990 Jan 31;86(1):113–117. doi: 10.1016/0378-1119(90)90122-8. [DOI] [PubMed] [Google Scholar]
- Zhang J. X., Flint H. J. A bifunctional xylanase encoded by the xynA gene of the rumen cellulolytic bacterium Ruminococcus flavefaciens 17 comprises two dissimilar domains linked by an asparagine/glutamine-rich sequence. Mol Microbiol. 1992 Apr;6(8):1013–1023. doi: 10.1111/j.1365-2958.1992.tb02167.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Transcending the impenetrable: how proteins come to terms with membranes. Biochim Biophys Acta. 1988 Jun 9;947(2):307–333. doi: 10.1016/0304-4157(88)90013-5. [DOI] [PubMed] [Google Scholar]