Abstract
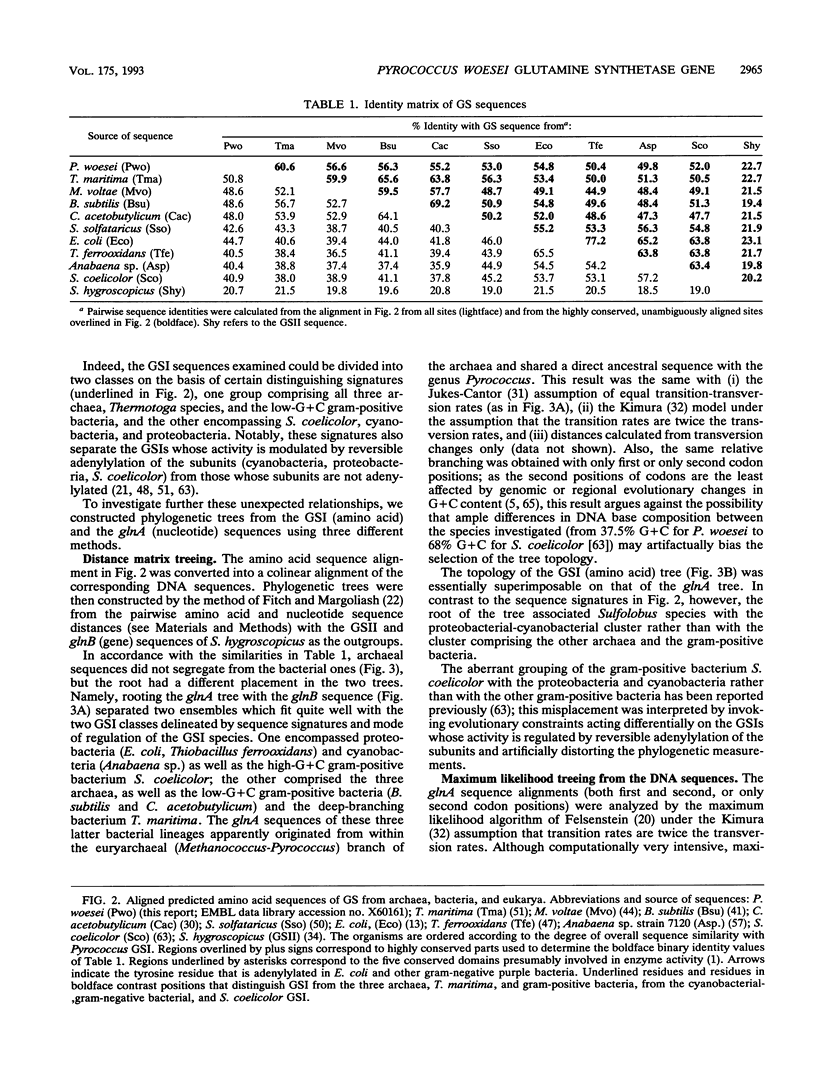
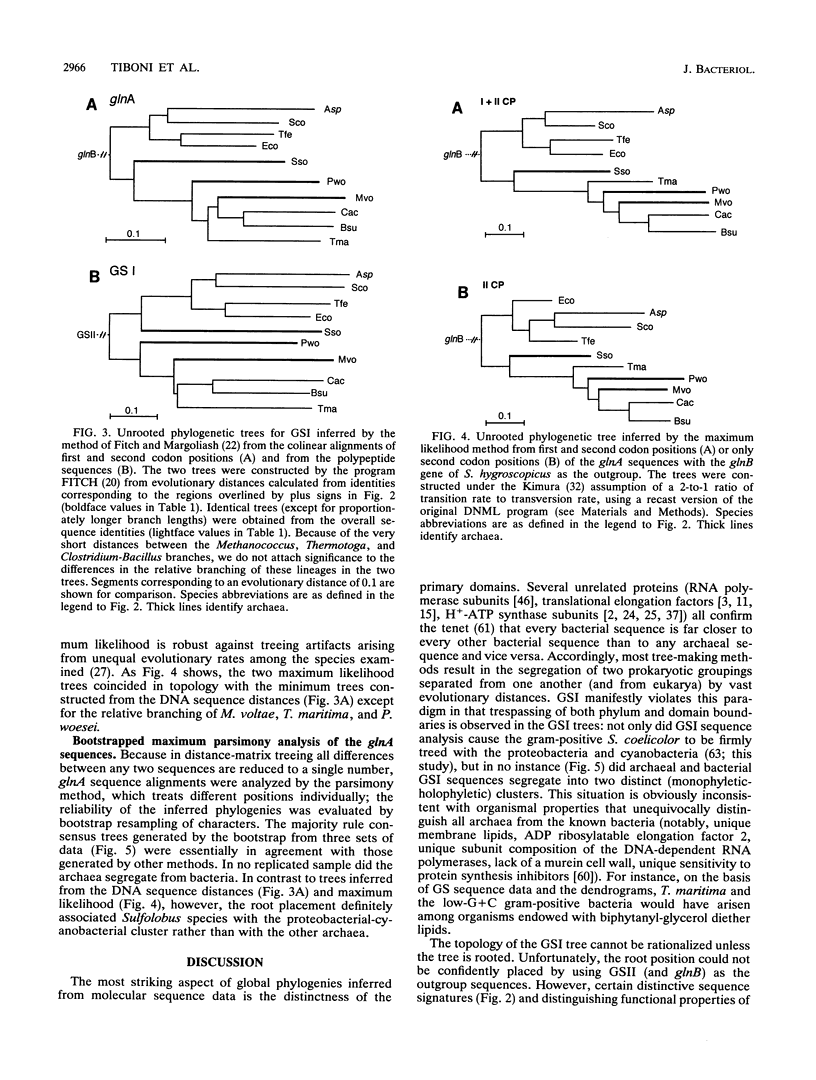
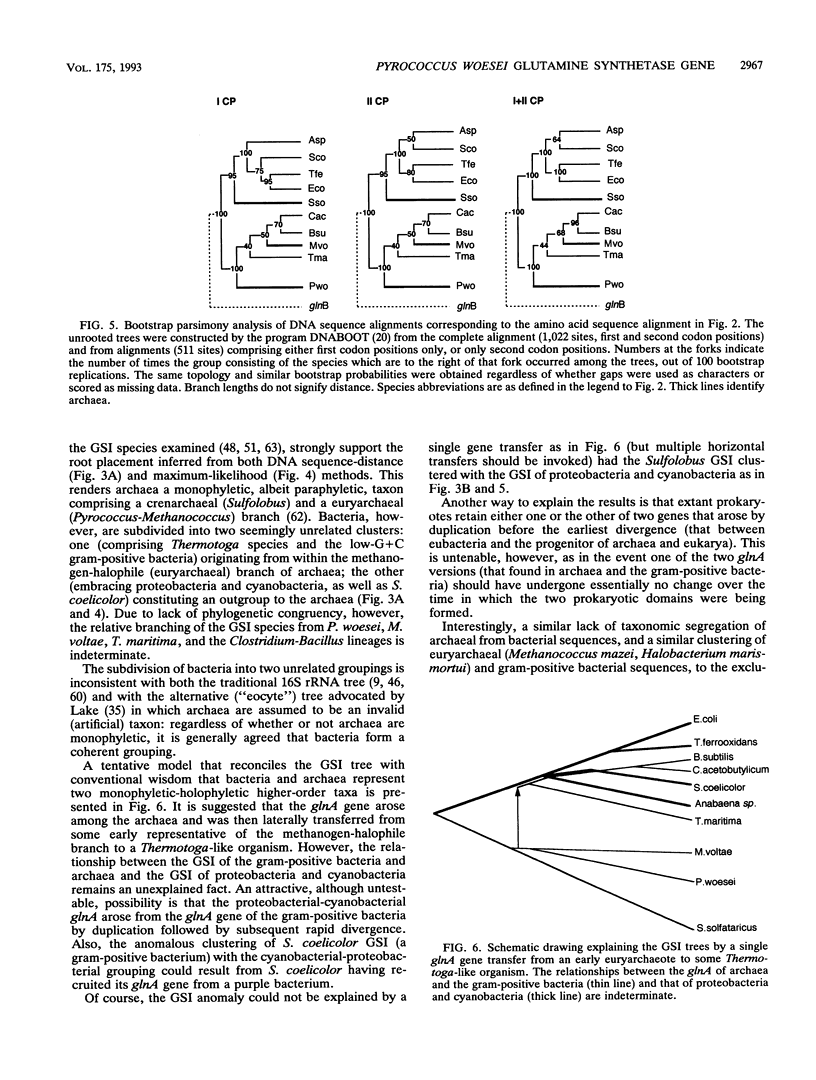
The gene glnA encoding glutamine synthetase I (GSI) from the archaeum Pyrococcus woesei was cloned and sequenced with the Sulfolobus solfataricus glnA gene as the probe. An operon reading frame of 448 amino acids was identified within a DNA segment of 1,528 bp. The encoded protein was 49% identical with the GSI of Methanococcus voltae and exhibited conserved regions characteristic of the GSI family. The P. woesei GSI was aligned with available homologs from other archaea (S. solfataricus, M. voltae) and with representative sequences from cyanobacteria, proteobacteria, and gram-positive bacteria. Phylogenetic trees were constructed from both the amino acid and the nucleotide sequence alignments. In accordance with the sequence similarities, archaeal and bacterial sequences did not segregate on a phylogeny. On the basis of sequence signatures, the GSI trees could be subdivided into two ensembles. One encompassed the GSI of cyanobacteria and proteobacteria, but also that of the high-G + C gram-positive bacterium Streptomyces coelicolor (all of which are regulated by the reversible adenylylation of the enzyme subunits); the other embraced the GSI of the three archaea as well as that of the low-G + C gram-positive bacteria (Clostridium acetobutilycum, Bacillus subtilis) and Thermotoga maritima (none of which are regulated by subunit adenylylation). The GSIs of the Thermotoga and the Bacillus-Clostridium lineages shared a direct common ancestor with that of P. woesei and the methanogens and were unrelated to their homologs from cyanobacteria, proteobacteria, and S. coelicolor. The possibility is presented that the GSI gene arose among the archaea and was then laterally transferred from some early methanogen to a Thermotoga-like organism. However, the relationship of the cyanobacterial-proteobacterial GSIs to the Thermotoga GSI and the GSI of low-G+C gram-positive bacteria remains unexplained.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almassy R. J., Janson C. A., Hamlin R., Xuong N. H., Eisenberg D. Novel subunit-subunit interactions in the structure of glutamine synthetase. 1986 Sep 25-Oct 1Nature. 323(6086):304–309. doi: 10.1038/323304a0. [DOI] [PubMed] [Google Scholar]
- Amann R., Ludwig W., Schleifer K. H. Beta-subunit of ATP-synthase: a useful marker for studying the phylogenetic relationship of eubacteria. J Gen Microbiol. 1988 Oct;134(10):2815–2821. doi: 10.1099/00221287-134-10-2815. [DOI] [PubMed] [Google Scholar]
- Behrmann I., Hillemann D., Pühler A., Strauch E., Wohlleben W. Overexpression of a Streptomyces viridochromogenes gene (glnII) encoding a glutamine synthetase similar to those of eucaryotes confers resistance against the antibiotic phosphinothricyl-alanyl-alanine. J Bacteriol. 1990 Sep;172(9):5326–5334. doi: 10.1128/jb.172.9.5326-5334.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G., Bernardi G. Compositional constraints and genome evolution. J Mol Evol. 1986;24(1-2):1–11. doi: 10.1007/BF02099946. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brown J. W., Daniels C. J., Reeve J. N. Gene structure, organization, and expression in archaebacteria. Crit Rev Microbiol. 1989;16(4):287–338. doi: 10.3109/10408418909105479. [DOI] [PubMed] [Google Scholar]
- Burggraf S., Olsen G. J., Stetter K. O., Woese C. R. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992 Aug;15(3):352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- Cammarano P., Palm P., Creti R., Ceccarelli E., Sanangelantoni A. M., Tiboni O. Early evolutionary relationships among known life forms inferred from elongation factor EF-2/EF-G sequences: phylogenetic coherence and structure of the archaeal domain. J Mol Evol. 1992 May;34(5):396–405. doi: 10.1007/BF00162996. [DOI] [PubMed] [Google Scholar]
- Colombo G., Villafranca J. J. Amino acid sequence of Escherichia coli glutamine synthetase deduced from the DNA nucleotide sequence. J Biol Chem. 1986 Aug 15;261(23):10587–10591. [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988 Nov 25;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creti R., Citarella F., Tiboni O., Sanangelantoni A., Palm P., Cammarano P. Nucleotide sequence of a DNA region comprising the gene for elongation factor 1 alpha (EF-1 alpha) from the ultrathermophilic archaeote Pyrococcus woesei: phylogenetic implications. J Mol Evol. 1991 Oct;33(4):332–342. doi: 10.1007/BF02102864. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Anderson K. L., Alberro M. R. A naturally occurring horizontal gene transfer from a eukaryote to a prokaryote. J Mol Evol. 1990 Nov;31(5):383–388. doi: 10.1007/BF02106053. [DOI] [PubMed] [Google Scholar]
- Edmands J., Noridge N. A., Benson D. R. The actinorhizal root-nodule symbiont Frankia sp. strain CpI1 has two glutamine synthetases. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6126–6130. doi: 10.1073/pnas.84.17.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17(6):368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Fisher R., Tuli R., Haselkorn R. A cloned cyanobacterial gene for glutamine synthetase functions in Escherichia coli, but the enzyme is not adenylylated. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3393–3397. doi: 10.1073/pnas.78.6.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Fuchs R. L., Keister D. L. Comparative properties of glutamine synthetases I and II in Rhizobium and Agrobacterium spp. J Bacteriol. 1980 Nov;144(2):641–648. doi: 10.1128/jb.144.2.641-648.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten J. P., Kibak H., Dittrich P., Taiz L., Bowman E. J., Bowman B. J., Manolson M. F., Poole R. J., Date T., Oshima T. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten J. P., Rausch T., Bernasconi P., Kibak H., Taiz L. Molecular evolution of H+-ATPases. I. Methanococcus and Sulfolobus are monophyletic with respect to eukaryotes and Eubacteria. Z Naturforsch C. 1989 Jul-Aug;44(7-8):641–650. doi: 10.1515/znc-1989-7-816. [DOI] [PubMed] [Google Scholar]
- Gupta R. S., Singh B. Cloning of the HSP70 gene from Halobacterium marismortui: relatedness of archaebacterial HSP70 to its eubacterial homologs and a model for the evolution of the HSP70 gene. J Bacteriol. 1992 Jul;174(14):4594–4605. doi: 10.1128/jb.174.14.4594-4605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Saitou N. On the maximum likelihood method in molecular phylogenetics. J Mol Evol. 1991 May;32(5):443–445. doi: 10.1007/BF02101285. [DOI] [PubMed] [Google Scholar]
- Higuchi R., Stang H. D., Browne J. K., Martin M. O., Huot M., Lipeles J., Salser W. Human ribosomal RNA gene spacer sequences are found interspersed elsewhere in the genome. Gene. 1981 Nov;15(2-3):177–186. doi: 10.1016/0378-1119(81)90127-x. [DOI] [PubMed] [Google Scholar]
- Hill R. T., Parker J. R., Goodman H. J., Jones D. T., Woods D. R. Molecular analysis of a novel glutamine synthetase of the anaerobe Bacteroides fragilis. J Gen Microbiol. 1989 Dec;135(12):3271–3279. doi: 10.1099/00221287-135-12-3271. [DOI] [PubMed] [Google Scholar]
- Janssen P. J., Jones W. A., Jones D. T., Woods D. R. Molecular analysis and regulation of the glnA gene of the gram-positive anaerobe Clostridium acetobutylicum. J Bacteriol. 1988 Jan;170(1):400–408. doi: 10.1128/jb.170.1.400-408.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kumada Y., Takano E., Nagaoka K., Thompson C. J. Streptomyces hygroscopicus has two glutamine synthetase genes. J Bacteriol. 1990 Sep;172(9):5343–5351. doi: 10.1128/jb.172.9.5343-5351.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature. 1988 Jan 14;331(6152):184–186. doi: 10.1038/331184a0. [DOI] [PubMed] [Google Scholar]
- Linkkila T. P., Gogarten J. P. Tracing origins with molecular sequences: rooting the universal tree of life. Trends Biochem Sci. 1991 Aug;16(8):287–288. doi: 10.1016/0968-0004(91)90117-e. [DOI] [PubMed] [Google Scholar]
- Marck C. 'DNA Strider': a 'C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988 Mar 11;16(5):1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Cerff R. Prokaryotic features of a nucleus-encoded enzyme. cDNA sequences for chloroplast and cytosolic glyceraldehyde-3-phosphate dehydrogenases from mustard (Sinapis alba). Eur J Biochem. 1986 Sep 1;159(2):323–331. doi: 10.1111/j.1432-1033.1986.tb09871.x. [DOI] [PubMed] [Google Scholar]
- Nakano Y., Tanaka E., Kato C., Kimura K., Horikoshi K. The complete nucleotide sequence of the glutamine synthetase gene (glnA) of Bacillus subtilis. FEMS Microbiol Lett. 1989 Jan 1;48(1):81–86. doi: 10.1016/0378-1097(89)90151-1. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Pace N. R., Nuell M., Kaine B. P., Gupta R., Woese C. R. Sequence of the 16S rRNA gene from the thermoacidophilic archaebacterium Sulfolobus solfataricus and its evolutionary implications. J Mol Evol. 1985;22(4):301–307. doi: 10.1007/BF02115685. [DOI] [PubMed] [Google Scholar]
- Pesole G., Bozzetti M. P., Lanave C., Preparata G., Saccone C. Glutamine synthetase gene evolution: a good molecular clock. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):522–526. doi: 10.1073/pnas.88.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possot O., Sibold L., Aubert J. P. Nucleotide sequence and expression of the glutamine synthetase structural gene, glnA, of the archaebacterium Methanococcus voltae. Res Microbiol. 1989 Jul-Aug;140(6):355–371. doi: 10.1016/0923-2508(89)90012-0. [DOI] [PubMed] [Google Scholar]
- Potts M., Angeloni S. V., Ebel R. E., Bassam D. Myoglobin in a cyanobacterium. Science. 1992 Jun 19;256(5064):1690–1691. doi: 10.1126/science.256.5064.1690. [DOI] [PubMed] [Google Scholar]
- Pühler G., Leffers H., Gropp F., Palm P., Klenk H. P., Lottspeich F., Garrett R. A., Zillig W. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings D. E., Jones W. A., O'Neill E. G., Woods D. R. Nucleotide sequence of the glutamine synthetase gene and its controlling region from the acidophilic autotroph Thiobacillus ferrooxidans. Gene. 1987;53(2-3):211–217. doi: 10.1016/0378-1119(87)90009-6. [DOI] [PubMed] [Google Scholar]
- Rochefort D. A., Benson D. R. Molecular cloning, sequencing, and expression of the glutamine synthetase II (glnII) gene from the actinomycete root nodule symbiont Frankia sp. strain CpI1. J Bacteriol. 1990 Sep;172(9):5335–5342. doi: 10.1128/jb.172.9.5335-5342.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanangelantoni A. M., Barbarini D., Di Pasquale G., Cammarano P., Tiboni O. Cloning and nucleotide sequence of an archaebacterial glutamine synthetase gene: phylogenetic implications. Mol Gen Genet. 1990 Apr;221(2):187–194. doi: 10.1007/BF00261719. [DOI] [PubMed] [Google Scholar]
- Sanangelantoni A. M., Forlani G., Ambroselli F., Cammarano P., Tiboni O. The glnA gene of the extremely thermophilic eubacterium Thermotoga maritima: cloning, primary structure, and expression in Escherichia coli. J Gen Microbiol. 1992 Feb;138(2):383–393. doi: 10.1099/00221287-138-2-383. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatters R. G., Kahn M. L. Glutamine synthetase II in Rhizobium: reexamination of the proposed horizontal transfer of DNA from eukaryotes to prokaryotes. J Mol Evol. 1989 Nov;29(5):422–428. doi: 10.1007/BF02602912. [DOI] [PubMed] [Google Scholar]
- Smith M. W., Doolittle R. F. Anomalous phylogeny involving the enzyme glucose-6-phosphate isomerase. J Mol Evol. 1992 Jun;34(6):544–545. doi: 10.1007/BF00160467. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wakabayashi S., Matsubara H., Webster D. A. Primary sequence of a dimeric bacterial haemoglobin from Vitreoscilla. 1986 Jul 31-Aug 6Nature. 322(6078):481–483. doi: 10.1038/322481a0. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray L. V., Jr, Fisher S. H. Cloning and nucleotide sequence of the Streptomyces coelicolor gene encoding glutamine synthetase. Gene. 1988 Nov 30;71(2):247–256. doi: 10.1016/0378-1119(88)90041-8. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E. On the molecular evolutionary clock. J Mol Evol. 1987;26(1-2):34–46. doi: 10.1007/BF02111280. [DOI] [PubMed] [Google Scholar]


