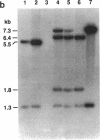
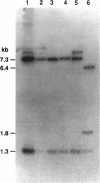
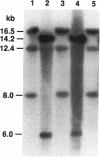
Abstract
SLP1 is a 17.2-kbp genetic element indigenous to the Streptomyces coelicolor chromosome. During conjugation, SLP1 can undergo excision and subsequent site-specific integration into the chromosomes of recipient cells. We report here the localization, nucleotide sequences, and initial characterization of the genes mediating these recombination events. A region of SLP1 adjacent to the previously identified site of integration, attP, was found to be sufficient to promote site-specific integration of an unrelated Streptomyces plasmid. Nucleotide sequence analysis of a 2.2-kb segment of this region reveals two open reading frames that are adjacent to and transcribed toward the attP site. One of these, the 1,365-bp int gene of SLP1, encodes a predicted 50.6-kDa basic protein having substantial amino acid sequence similarity to a family of site-specific recombinases that includes the Escherichia coli bacteriophage lambda integrase. A linker insertion in the 5' end of the cloned int gene prevents integration, indicating that Int is essential for promoting integration. An open reading frame (orf61) lying immediately 5' to int encodes a predicted 7.1-kDa basic peptide showing limited sequence similarity to the excisionase (xis) genes of other site-specific recombination systems.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J., Limberger R., Schneider S. J., Campbell A. Recombination and modular exchange in the genesis of new lambdoid phages. New Biol. 1991 Mar;3(3):297–308. [PubMed] [Google Scholar]
- Benigni R., Petrov P. A., Carere A. Estimate of the genome size by renaturation studies in Streptomyces. Appl Microbiol. 1975 Aug;30(2):324–326. doi: 10.1128/am.30.2.324-326.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Ward J. M., Kieser T., Cohen S. N., Hopwood D. A. Excision of chromosomal DNA sequences from Streptomyces coelicolor forms a novel family of plasmids detectable in Streptomyces lividans. Mol Gen Genet. 1981;184(2):230–240. doi: 10.1007/BF00272910. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccard F., Smokvina T., Pernodet J. L., Friedmann A., Guérineau M. The integrated conjugative plasmid pSAM2 of Streptomyces ambofaciens is related to temperate bacteriophages. EMBO J. 1989 Mar;8(3):973–980. doi: 10.1002/j.1460-2075.1989.tb03460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch M. A., Cohen S. N. Excisive recombination of the SLP1 element in Streptomyces lividans is mediated by Int and enhanced by Xis. J Bacteriol. 1993 May;175(10):3075–3082. doi: 10.1128/jb.175.10.3075-3082.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. P., Idler K. B., Katz L. Characterization of the genetic elements required for site-specific integration of plasmid pSE211 in Saccharopolyspora erythraea. J Bacteriol. 1990 Apr;172(4):1877–1888. doi: 10.1128/jb.172.4.1877-1888.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colloms S. D., Sykora P., Szatmari G., Sherratt D. J. Recombination at ColE1 cer requires the Escherichia coli xerC gene product, a member of the lambda integrase family of site-specific recombinases. J Bacteriol. 1990 Dec;172(12):6973–6980. doi: 10.1128/jb.172.12.6973-6980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. W. DNA sequence of the int-xis-Pi region of the bacteriophage lambda; overlap of the int and xis genes. Nucleic Acids Res. 1980 Apr 25;8(8):1765–1782. doi: 10.1093/nar/8.8.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist L. W., Kikuchi A., Weisberg R. A. The role of lambda integrase in integration and excision. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1115–1120. doi: 10.1101/sqb.1979.043.01.124. [DOI] [PubMed] [Google Scholar]
- Fawcett T. W., Bartlett G. An effective method for eliminating "artifact banding" when sequencing double-stranded DNA templates. Biotechniques. 1990 Jul;9(1):46–48. [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S. R., Lee S. C., Kendall K., Cohen S. N. Identification and characterization of a locus inhibiting extrachromosomal maintenance of the Streptomyces plasmid SLP1. Mol Gen Genet. 1989 Jun;217(2-3):324–331. doi: 10.1007/BF02464900. [DOI] [PubMed] [Google Scholar]
- Gronostajski R. M., Sadowski P. D. The FLP recombinase of the Saccharomyces cerevisiae 2 microns plasmid attaches covalently to DNA via a phosphotyrosyl linkage. Mol Cell Biol. 1985 Nov;5(11):3274–3279. doi: 10.1128/mcb.5.11.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R. H., Abremski K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol. 1985 Feb 5;181(3):351–362. doi: 10.1016/0022-2836(85)90224-4. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Foeller C., Bidwell K., Landy A. Site-specific recombination functions of bacteriophage lambda: DNA sequence of regulatory regions and overlapping structural genes for Int and Xis. Proc Natl Acad Sci U S A. 1980 May;77(5):2482–2486. doi: 10.1073/pnas.77.5.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. A., Kieser T., Wright H. M., Bibb M. J. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J Gen Microbiol. 1983 Jul;129(7):2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- Kendall K. J., Cohen S. N. Complete nucleotide sequence of the Streptomyces lividans plasmid pIJ101 and correlation of the sequence with genetic properties. J Bacteriol. 1988 Oct;170(10):4634–4651. doi: 10.1128/jb.170.10.4634-4651.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson J. L., Hershberger C. L. The minimal replicon of a streptomycete plasmid produces an ultrahigh level of plasmid DNA. Plasmid. 1986 May;15(3):199–209. doi: 10.1016/0147-619x(86)90038-7. [DOI] [PubMed] [Google Scholar]
- Lee S. C., Omer C. A., Brasch M. A., Cohen S. N. Analysis of recombination occurring at SLP1 att sites. J Bacteriol. 1988 Dec;170(12):5806–5813. doi: 10.1128/jb.170.12.5806-5813.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S. E., Oser A. B., Lesser C. F., Youderian P., Susskind M. M., Landy A. Structural and regulatory divergence among site-specific recombination genes of lambdoid phage. J Mol Biol. 1986 Jun 20;189(4):603–616. doi: 10.1016/0022-2836(86)90491-2. [DOI] [PubMed] [Google Scholar]
- Lydiate D. J., Malpartida F., Hopwood D. A. The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene. 1985;35(3):223–235. doi: 10.1016/0378-1119(85)90001-0. [DOI] [PubMed] [Google Scholar]
- Marko M. A., Chipperfield R., Birnboim H. C. A procedure for the large-scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal Biochem. 1982 Apr;121(2):382–387. doi: 10.1016/0003-2697(82)90497-3. [DOI] [PubMed] [Google Scholar]
- Omer C. A., Cohen S. N. Plasmid formation in Streptomyces: excision and integration of the SLP1 replicon at a specific chromosomal site. Mol Gen Genet. 1984;196(3):429–438. doi: 10.1007/BF00436190. [DOI] [PubMed] [Google Scholar]
- Omer C. A., Cohen S. N. Structural analysis of plasmid and chromosomal loci involved in site-specific excision and integration of the SLP1 element of Streptomyces coelicolor. J Bacteriol. 1986 Jun;166(3):999–1006. doi: 10.1128/jb.166.3.999-1006.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer C. A., Stein D., Cohen S. N. Site-specific insertion of biologically functional adventitious genes into the Streptomyces lividans chromosome. J Bacteriol. 1988 May;170(5):2174–2184. doi: 10.1128/jb.170.5.2174-2184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pargellis C. A., Nunes-Düby S. E., de Vargas L. M., Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988 Jun 5;263(16):7678–7685. [PubMed] [Google Scholar]
- Poyart-Salmeron C., Trieu-Cuot P., Carlier C., Courvalin P. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 1989 Aug;8(8):2425–2433. doi: 10.1002/j.1460-2075.1989.tb08373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad P. V., Young L. J., Jayaram M. Mutations in the 2-microns circle site-specific recombinase that abolish recombination without affecting substrate recognition. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2189–2193. doi: 10.1073/pnas.84.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrempf H., Bujard H., Hopwood D. A., Goebel W. Isolation of covalently closed circular deoxyribonucleic acid from Streptomyces coelicolor A3(2). J Bacteriol. 1975 Feb;121(2):416–421. doi: 10.1128/jb.121.2.416-421.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair R. B., Bibb M. J. Transcriptional analysis of the repressor gene of the temperate Streptomyces phage phi C31. Gene. 1989 Dec 28;85(2):275–282. doi: 10.1016/0378-1119(89)90419-8. [DOI] [PubMed] [Google Scholar]
- Stein D. S., Kendall K. J., Cohen S. N. Identification and analysis of transcriptional regulatory signals for the kil and kor loci of Streptomyces plasmid pIJ101. J Bacteriol. 1989 Nov;171(11):5768–5775. doi: 10.1128/jb.171.11.5768-5775.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohl W. R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992 Mar 11;20(5):961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. J., Ward J. M., Hopwood D. A. Cloning of antibiotic resistance and nutritional genes in streptomycetes. J Bacteriol. 1982 Aug;151(2):668–677. doi: 10.1128/jb.151.2.668-677.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtli M., Cohen S. N. The chromosomal integration site for the Streptomyces plasmid SLP1 is a functional tRNA(Tyr) gene essential for cell viability. Mol Microbiol. 1992 Oct;6(20):3041–3050. doi: 10.1111/j.1365-2958.1992.tb01762.x. [DOI] [PubMed] [Google Scholar]
- Yagil E., Dolev S., Oberto J., Kislev N., Ramaiah N., Weisberg R. A. Determinants of site-specific recombination in the lambdoid coliphage HK022. An evolutionary change in specificity. J Mol Biol. 1989 Jun 20;207(4):695–717. doi: 10.1016/0022-2836(89)90238-6. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Ye Z. H., Buranen S. L., Lee C. Y. Sequence analysis and comparison of int and xis genes from staphylococcal bacteriophages L54a and phi 11. J Bacteriol. 1990 May;172(5):2568–2575. doi: 10.1128/jb.172.5.2568-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]