Abstract
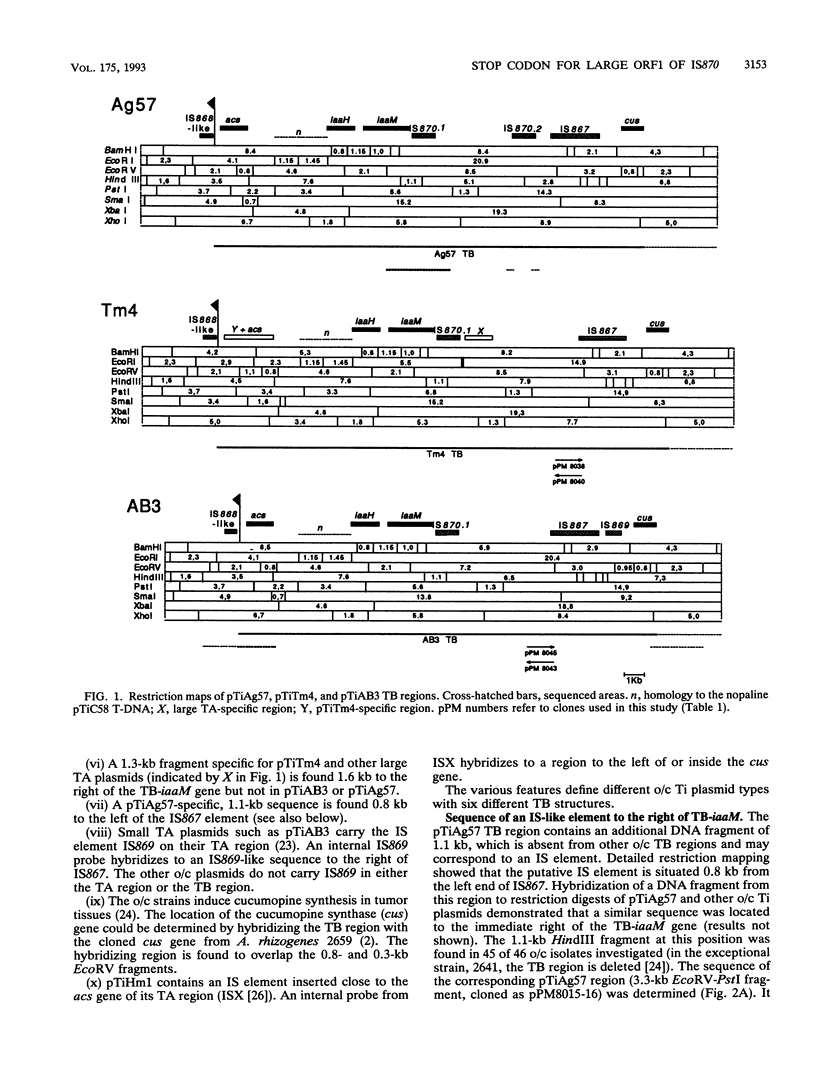
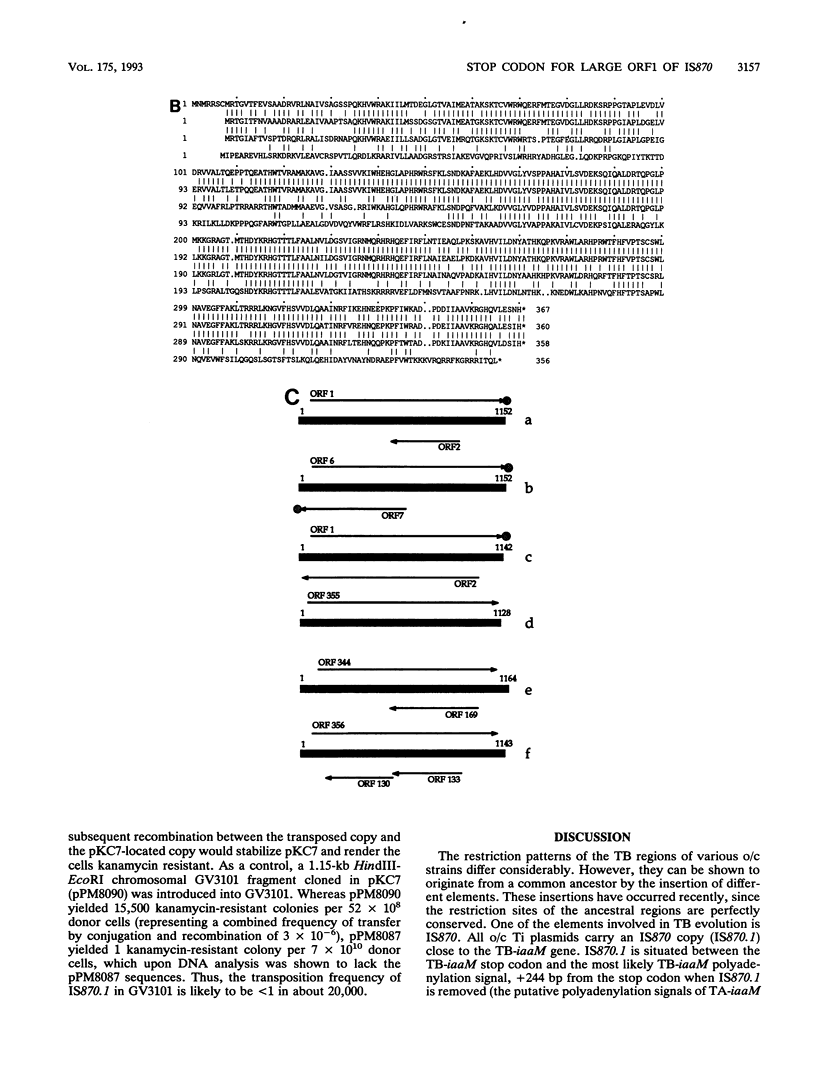
The TB regions of the Agrobacterium vitis octopine/cucumopine Ti plasmids constitute a family of related structures. All contain a bacterial insertion element downstream of the TB-iaaM gene, IS870.1. Whereas 43 isolates with octopine/cucumopine Ti plasmids carry only one IS870 copy, strain Ag57 carries a second copy (IS870.2) 3.9 kb to the right of IS870.1 and part of the same TB region. Two other octopine/cucumopine strains carry an IS870 copy on their chromosome (IS870.3). A study of the unmodified insertion sites of IS870.2 and IS870.3, cloned from closely related strains, enabled us to delimit the IS870 elements. IS870 has a size of 1,152 bp and is terminated by inverted repeats. It contains a large open reading frame without a stop codon. However, a stop codon is generated by insertion into the target sequence 5'-CTAG-3'. IS870 is related to five other insertion sequence elements. For two of these, the stop codon of the largest open reading frame is also created by insertion into a CTAG target site.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bonnard G., Vincent F., Otten L. Sequence of Agrobacterium tumefaciens biotype III auxin genes. Plant Mol Biol. 1991 Apr;16(4):733–738. doi: 10.1007/BF00023438. [DOI] [PubMed] [Google Scholar]
- Buchholz W. G., Thomashow M. F. Comparison of T-DNA oncogene complements of Agrobacterium tumefaciens tumor-inducing plasmids with limited and wide host ranges. J Bacteriol. 1984 Oct;160(1):319–326. doi: 10.1128/jb.160.1.319-326.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos G., De Beuckeleer M., Van Montagu M., Schell J. Restriction endonuclease mapping of the octopine tumor-inducing plasmid pTiAch5 of Agrobacterium tumefaciens. Plasmid. 1981 Sep;6(2):249–253. doi: 10.1016/0147-619x(81)90070-6. [DOI] [PubMed] [Google Scholar]
- Depicker A., De Wilde M., De Vos G., De Vos R., Van Montagu M., Schell J. Molecular cloning of overlapping segments of the nopaline Ti-plasmid pTiC58 as a means to restriction endonuclease mapping. Plasmid. 1980 Mar;3(2):193–211. doi: 10.1016/0147-619x(80)90109-2. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaese P., De Greve H., Decraemer H., Schell J., Van Montagu M. Rapid mapping of transposon insertion and deletion mutations in the large Ti-plasmids of Agrobacterium tumefaciens. Nucleic Acids Res. 1979 Dec 11;7(7):1837–1849. doi: 10.1093/nar/7.7.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen J., De Beuckeleer M., Seurinck J., Deboeck F., De Greve H., Lemmers M., Van Montagu M., Schell J. The complete nucleotide sequence of the TL-DNA of the Agrobacterium tumefaciens plasmid pTiAch5. EMBO J. 1984 Apr;3(4):835–846. doi: 10.1002/j.1460-2075.1984.tb01894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman G. T., Farrand S. K. Agrobacterium plasmids encode structurally and functionally different loci for catabolism of agrocinopine-type opines. Mol Gen Genet. 1990 Sep;223(3):465–473. doi: 10.1007/BF00264455. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Huss B., Tinland B., Paulus F., Walter B., Otten L. Functional analysis of a complex oncogene arrangement in biotype III Agrobacterium tumefaciens strains. Plant Mol Biol. 1990 Feb;14(2):173–186. doi: 10.1007/BF00018558. [DOI] [PubMed] [Google Scholar]
- Joos H., Inzé D., Caplan A., Sormann M., Van Montagu M., Schell J. Genetic analysis of T-DNA transcripts in nopaline crown galls. Cell. 1983 Apr;32(4):1057–1067. doi: 10.1016/0092-8674(83)90290-8. [DOI] [PubMed] [Google Scholar]
- Kaluza K., Hahn M., Hennecke H. Repeated sequences similar to insertion elements clustered around the nif region of the Rhizobium japonicum genome. J Bacteriol. 1985 May;162(2):535–542. doi: 10.1128/jb.162.2.535-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan H. B., Pueppke S. G. Repetitive sequences with homology to Bradyrhizobium japonicum DNA and the T-DNA of Agrobacterium rhizogenes are closely linked to nodABC of Rhizobium fredii USDA257. Mol Plant Microbe Interact. 1991 Sep-Oct;4(5):521–529. doi: 10.1094/mpmi-4-521. [DOI] [PubMed] [Google Scholar]
- Matsutani S., Ohtsubo H., Maeda Y., Ohtsubo E. Isolation and characterization of IS elements repeated in the bacterial chromosome. J Mol Biol. 1987 Aug 5;196(3):445–455. doi: 10.1016/0022-2836(87)90023-4. [DOI] [PubMed] [Google Scholar]
- Otten L., Canaday J., Gérard J. C., Fournier P., Crouzet P., Paulus F. Evolution of agrobacteria and their Ti plasmids--a review. Mol Plant Microbe Interact. 1992 Jul-Aug;5(4):279–287. doi: 10.1094/mpmi-5-279. [DOI] [PubMed] [Google Scholar]
- Paulus F., Canaday J., Otten L. Limited host range Ti plasmids: recent origin from wide host range Ti plasmids and involvement of a novel IS element, IS868. Mol Plant Microbe Interact. 1991 Mar-Apr;4(2):190–197. doi: 10.1094/mpmi-4-190. [DOI] [PubMed] [Google Scholar]
- Paulus F., Canaday J., Vincent F., Bonnard G., Kares C., Otten L. Sequence of the iaa and ipt region of different Agrobacterium tumefaciens biotype III octopine strains: reconstruction of octopine Ti plasmid evolution. Plant Mol Biol. 1991 Apr;16(4):601–614. doi: 10.1007/BF00023425. [DOI] [PubMed] [Google Scholar]
- Paulus F., Huss B., Tinland B., Herrmann A., Canaday J., Otten L. Role of T-region borders in Agrobacterium host range. Mol Plant Microbe Interact. 1991 Mar-Apr;4(2):163–172. doi: 10.1094/mpmi-4-163. [DOI] [PubMed] [Google Scholar]
- Ramseier T. M., Göttfert M. Codon usage and G + C content in Bradyrhizobium japonicum genes are not uniform. Arch Microbiol. 1991;156(4):270–276. doi: 10.1007/BF00262997. [DOI] [PubMed] [Google Scholar]
- Rao R. N., Rogers S. G. Plasmid pKC7: a vector containing ten restriction endonuclease sites suitable for cloning DNA segments. Gene. 1979 Sep;7(1):79–82. doi: 10.1016/0378-1119(79)90044-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slightom J. L., Durand-Tardif M., Jouanin L., Tepfer D. Nucleotide sequence analysis of TL-DNA of Agrobacterium rhizogenes agropine type plasmid. Identification of open reading frames. J Biol Chem. 1986 Jan 5;261(1):108–121. [PubMed] [Google Scholar]
- Tenzen T., Matsutani S., Ohtsubo E. Site-specific transposition of insertion sequence IS630. J Bacteriol. 1990 Jul;172(7):3830–3836. doi: 10.1128/jb.172.7.3830-3836.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzen T., Ohtsubo E. Preferential transposition of an IS630-associated composite transposon to TA in the 5'-CTAG-3' sequence. J Bacteriol. 1991 Oct;173(19):6207–6212. doi: 10.1128/jb.173.19.6207-6212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haute E., Joos H., Maes M., Warren G., Van Montagu M., Schell J. Intergeneric transfer and exchange recombination of restriction fragments cloned in pBR322: a novel strategy for the reversed genetics of the Ti plasmids of Agrobacterium tumefaciens. EMBO J. 1983;2(3):411–417. doi: 10.1002/j.1460-2075.1983.tb01438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmitzer L., Dhaese P., Schreier P. H., Schmalenbach W., Van Montagu M., Schell J. Size, location and polarity of T-DNA-encoded transcripts in nopaline crown gall tumors; common transcripts in octopine and nopaline tumors. Cell. 1983 Apr;32(4):1045–1056. doi: 10.1016/0092-8674(83)90289-1. [DOI] [PubMed] [Google Scholar]
- Yanofsky M., Montoya A., Knauf V., Lowe B., Gordon M., Nester E. Limited-host-range plasmid of Agrobacterium tumefaciens: molecular and genetic analyses of transferred DNA. J Bacteriol. 1985 Jul;163(1):341–348. doi: 10.1128/jb.163.1.341-348.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski P., Tempe J., Schell J. Transfer and function of T-DNA genes from agrobacterium Ti and Ri plasmids in plants. Cell. 1989 Jan 27;56(2):193–201. doi: 10.1016/0092-8674(89)90892-1. [DOI] [PubMed] [Google Scholar]
- van der Meer J. R., Zehnder A. J., de Vos W. M. Identification of a novel composite transposable element, Tn5280, carrying chlorobenzene dioxygenase genes of Pseudomonas sp. strain P51. J Bacteriol. 1991 Nov;173(22):7077–7083. doi: 10.1128/jb.173.22.7077-7083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]


