Abstract
We have tested the hypothesis that the promotion of flowering by prolonged exposure to low temperatures (vernalization) is mediated by DNA demethylation [Burn, J. E., Bagnall, D. J., Metzger, J. M., Dennis, E. S. & Peacock, W. J. (1993) Proc. Natl. Acad. Sci. USA 90, 287–291]. Arabidopsis plants that have reduced levels of DNA methylation because of the presence of a methyltransferase (METI) antisense gene flowered earlier than untransformed control plants, without the need for a cold treatment. Decreased DNA methylation mutants (ddm1) also flowered earlier than the wild-type progenitor under conditions where they respond to vernalization. We conclude that demethylation of DNA is sufficient to cause early flowering, and we have found that the promotion of flowering is directly proportional to the decrease in methylation in METI antisense lines. The early-flowering phenotype was inherited in sexual progeny, even when the antisense transgene had been lost by segregation. Methyltransferase antisense plants with low DNA methylation levels responded to a low-temperature treatment by flowering even earlier than their untreated siblings indicating that the promotion of flowering by cold and by demethylation was additive when neither treatment saturated the early-flowering response. As in untransformed control plants, the cold-induced early-flowering signal was reset in progeny of METI antisense plants. These observations suggest that the demethylation brought about by a METI antisense can account for some properties of vernalization, but not for the need for revernalization in each generation.
Keywords: Arabidopsis, methyltransferase antisense, ddm1, FRI, FLC
Many plants growing at high latitudes require exposure, as germinating seeds or vegetatively growing plants, to prolonged periods at low temperatures (vernalization) before they will initiate flowering. This ensures that flowering will occur in the warm days of spring and summer, which are favorable for pollination and seed development. Whereas some plants have an absolute requirement for vernalization, others, such as the late-flowering ecotypes of Arabidopsis, show a facultative requirement for vernalization, and will eventually flower even in the absence of a cold treatment (for review see ref. 1). The phenomenon of vernalization was first described in the middle of the 19th century (Kleppart, cited in ref. 2), but only recently has a hypothesis suggesting a molecular mechanism for the low-temperature promotion of flowering been proposed (3).
The cold treatment is perceived by cells, mitotically active at the time of treatment; early flowering results when these cells, or their descendants, form the floral/inflorescence meristem, indicating that the vernalization signal is inherited mitotically rather than being transmitted from “vernalized cells” to the apex (4, 5). The vernalization signal is not transmitted to sexual progeny (1). The perceived parallels between the inheritance of the vernalization signal and of DNA methylation patterns led to the hypothesis that the vernalization response, in Arabidopsis and other plants, is mediated by changes in DNA methylation (3). Specifically it was proposed that the cold treatment results in demethylation of the promoter region(s) and subsequent activation of a gene or genes critical for initiating reproductive development. Mitotic inheritance of the vernalization signal is consistent with clonal inheritance of DNA methylation patterns (6), but it is now known that, in plants, methylation patterns may not be reset between generations (7–9), suggesting that factors other than DNA methylation may be involved in resetting the vernalization signal.
Treatment of plants with the demethylating agent, 5-azacytidine (5-azaC) resulted in early flowering (3, 10). This early-flowering response was restricted to plants that normally respond to vernalization, including certain late-flowering ecotypes and mutants of Arabidopsis and winter wheat. Spring wheats and other late-flowering Arabidopsis mutants that are insensitive to vernalization did not develop an early-flowering phenotype after 5-azaC treatment. These observations suggest that this response was specific to the vernalization-dependent pathway to flowering (3, 10, 11) rather than being a nonspecific effect of the treatment.
In addition to causing demethylation, 5-azaC is a general inhibitor of transcription (12), so it is possible that the promotion of flowering by 5-azaC resulted from effects other than demethylation of DNA. To discriminate between these possibilities we have used Arabidopsis plants in which methylation levels have been reduced by an antisense methyltransferase transgene (9, 13) or by mutation at the DDM1 (decreased DNA methylation) locus (8). Plants with low levels of DNA methylation flowered early without vernalization, indicating that demethylation was sufficient to cause early flowering. The promotion of flowering was correlated with the extent of demethylation in independent lines of antisense plants. The promotive effects on flowering time of demethylation and of the cold treatment were additive. Both antisense and untransformed control plants reset the vernalization signal in the following generation, but the demethylation-induced early-flowering phenotype was inherited by the progeny.
MATERIALS AND METHODS
Plant Lines Used in These Studies.
The construction and characterization of the methyltransferase antisense families has been described previously (9). In brief, the METI antisense transgene consisted of a 2.8-kb fragment encoding the entire methyltransferase domain and about 300 aa of the amino-terminal domain (13) fused, in the antisense direction, to a constitutive promoter. Family no. 10 has three copies of the antisense transgene inserted at a single locus. Plant T2 10.5 was homozygous for the transgenes at this locus whereas plant T2 10.1 was hemizygous; this locus was segregating in the T3 progeny of 10.1. Family no. 22-6 has four copies of the transgene, three of which cosegregate while the remaining copy is unlinked. Plant T2 22-6.11 was homozygous for all four copies of the transgene; plant T2 22-6.9 was homozygous for the three linked copies and hemizygous for the transgene that is unlinked to this locus. The third family, no. 39, contains five copies of the transgene, but because of very low fertility of this family, no segregation data are available.
PCR Assay for the Transgene.
The presence of the transgene was determined by using a PCR assay to detect the selectable NptII marker gene (9), using template DNA prepared from a single leaf (14).
Determination of Flowering Time.
Seeds were surface-sterilized and placed in individual, sterile, glass tubes containing 7 ml of growth medium consisting of 1× MS iron, macro- and microelements, 0.2× MS vitamins (15), 1.5% sucrose, and 0.75% Noble agar, pH 7.0. Forty tubes for each line were placed at 4°C for 23 days in the dark. Two days before the end of the cold treatment another 40 tubes were prepared for each line and placed at 4°C for 2 days to ensure even germination. At the end of the cold treatments all tubes were transferred to a growth cabinet at 22°C with 8 hr (short day, SD) or 16 hr (long day, LD) of fluorescent light. The 2-day cold treatment did not promote flowering relative to untreated plants. The light intensity varied between experiments, but ranged from 220 μM⋅m−2 ⋅ s−1 in the center to 160 μM ⋅ m−2 ⋅ s−1 at the sides of the cabinet. The tubes were rotated daily so that all plants received equal illumination. The plants were observed daily; flowering time was measured as the time (days) from germination, defined here as emergence of cotyledons, to first elongation of the primary inflorescence. The number of rosette leaves at the time of bolting was also recorded; these data, which parallel the flowering time data, are not presented.
Three types of fluorescent tubes were used: Philips TLMF 140W/33RD (Philips Electronic Instruments, Mahwah, NJ) and, when these were no longer available, Osram 58W/84 (Osram, Berlin) or Sylvania GTE F58W/133 (Sylvania Electric Products, Fall River, MA). The spectral qualities of the light from these tubes differed, and flowering was delayed under Sylvania or Osram tubes. Metal arc lamps were used in one experiment. The promotion of flowering in response to a vernalization treatment varies with the growth conditions subsequent to the 23-day cold treatment resulting in the variations seen between Tables 2 and 3.
Table 2.
A comparison of flowering time of untransformed C24 plants and line T3 10.5, a transgenic line homozygous for the METI antisense, grown in different conditions
| Plant line | Irradiance
|
Flowering time
|
||
|---|---|---|---|---|
| Photoperiod | Lamp (intensity) | Unvern | Vern | |
| C24 | 8 hr L/16 hr D | Philips (180 μE) | 45.1 ± 1.5 | 23.4 ± 0.3 |
| T3 10.5 | 8 hr L/16 hr D | Philips (180 μE) | 34.1 ± 0.7 | 25.3 ± 0.4 |
| C24 | 8 hr L/16 hr D | Osram (150 μE) | 76.3 ± 2.3 | 34.5 ± 0.5 |
| T3 10.5 | 8 hr L/16 hr D | Osram (150 μE) | 43.1 ± 1.1 | 30.4 ± 0.7 |
| C24 | 16 hr L/8 hr D | Philips (100 μE) | 30.4 ± 1.3 | 14 ± 0.2 |
| T3 10.5 | 16 hr L/8 hr D | Philips (100 μE) | 21.4 ± 1.2 | 14.5 ± 0.3 |
| C24 | 16 hr L/8 hr D | Philips (180 μE) | 27.1 ± 0.7 | 10.0 ± 0.2 |
| T3 10.5 | 16 hr L/8 hr D | Philips (180 μE) | 21.3 ± 0.4 | 13.1 ± 0.3 |
| C24 | 16 hr L/8 hr D | M. Arc (200 μE) | 27.2 ± 0.5 | ND |
| T3 10.5 | 16 hr L/8 hr D | M. Arc (200 μE) | 21.5 ± 0.6 | ND |
Flowering time in days ± SE. μE, μM⋅m−2⋅s−1. ND, not done. L, light. D, dark.
Table 3.
The promotion of flowering time in plants with reduced levels of DNA methylation is proportional to the extent of demethylation
| Plant line | Flowering time (SD)
|
% C24 vern response in unvern a/s | Methylation level, % C24 | |
|---|---|---|---|---|
| Unvern | Vern | |||
| C24 | 86.6 ± 1.5 | 33.8 ± 0.9 | 100 | |
| T3 22-6.9 | 79.0 ± 1.9 | 37.7 ± 0.9 | 14.4 | 56.9 ± 0.8 |
| T3 22-6.11 | 75.0 ± 2.5 | 34.8 ± 1.0 | 22.1 | 45.6 ± 3.2 |
| T3 10.1 | 58.2 ± 2.5 | 28.0 ± 1.1 | 53.8 | 19.4 ± 4.2 |
| T4 10.1.8 | 54.6 ± 1.9 | 27.8 ± 1.0 | 60.7 | 34.6* |
| T4 10.1.4 | 47.3 ± 2.4 | 26.3 ± 1.1 | 74.4 | 20.1 ± 1.8 |
| T3 39.35 | 30.5 ± 1.0 | 19.7 ± 0.7 | 106.8 | 32.2 ± 1.7 |
Flowering time is in days ± SE. a/s, METI antisense.
Average of two measurements. SD, short days.
Estimation of DNA Methylation.
Methylation of cytosine residues in TaqI sites was measured by a thin-layer chromatography assay (9). Radioactivity in individual dNMPs was quantitated by using a Molecular Dynamics PhosphorImager and imagequant software. Uncut DNA, for each sample, was treated in parallel to determine background radioactivity incorporated into dmCMP and dCMP because of sheared DNA; the background was subtracted before calculating 5-methylcytosine levels, by the formula dmCMP/dmCMP + dCMP, which were then normalized to wild type.
RESULTS
Vernalization Decreases DNA Methylation.
One prediction from the hypothesis that vernalization is mediated through demethylation is that a cold temperature treatment will decrease the level of DNA methylation. Imbibed seeds of untransformed C24 were placed at 8°C for 4 or 8 weeks; after 4 weeks at 8°C the radicle had emerged, and by 8 weeks the cotyledons had emerged but were not fully expanded. DNA was extracted from these seedlings and from untreated control seedlings judged visually to be at the same stage of development. Vernalization for 4 or 8 weeks reduced DNA methylation to 86.3 ± 0.9% of the level in control seedlings. Loss of methylation was transient; after 7 days’ growth at 21°C vernalized seedlings had levels of DNA methylation comparable to those of untreated plantlets (Table 1).
Table 1.
Recovery of DNA methylation after a 4-week vernalization treatment
| Days at 21°C post-cold treatment | Methylation level | |
|---|---|---|
| Unvernalized | Vernalized | |
| 0 | 83.3 ± 1.6 | 74.0 ± 2.5 |
| 4 | 80.3 ± 2.4 | 77.7 ± 3.0 |
| 7 | 86.7 ± 3.3 | 83.7 ± 3.2 |
| 14 | 102.1 ± 3.6 | 95.2 ± 3.5 |
| 21 | 95.8 ± 1.8 | 98.0 ± 3.8 |
Methylation level is expressed as % ± SE, relative to a standard, isolated from mature leaves. Methylation estimates were done at least three times for each sample. The difference in methylation between cold-treated and untreated seedlings was replicated in six independent experiments; the recovery of methylation was observed in two independent experiments.
Decreased DNA Methylation Causes Early Flowering.
If demethylation substitutes for vernalization by activating the vernalization-dependent pathway, then METI antisense plants with reduced levels of DNA methylation should have an early-flowering phenotype without exposure to low temperatures. Because C24 plants grown in short-day photoperiods respond to vernalization, we used these conditions to compare the flowering time of transgenic plants containing a methyltransferase (METI) antisense construct and untransformed C24 plants. Plants from line 10.5, which is homozygous for the antisense and which has only 15% of normal methylation (9), flowered significantly earlier than control C24. In the absence of a vernalization treatment, METI antisense plants showed a promotion in flowering equivalent to 50% of that caused by vernalization of the control (Table 2, rows 1 and 2).
Because the time of flowering in Arabidopsis also is affected by length of photoperiod and by both spectral quality and intensity of light (16, 17), we tested the effect of these variables on the flowering time of line 10.5 METI antisense plants. Plants were grown under an 8-hr or a 16-hr photoperiod, under lights of different intensity and/or spectral properties in separate experiments. Under all conditions tested, the unvernalized antisense plants flowered earlier than the controls (Table 2). The promotion of flowering by demethylation ranged from 33 to 80% of the vernalization response of C24. In general, when C24 was most responsive to vernalization, demethylation with a METI antisense gave the greatest promotion of flowering; this was observed across all antisense families examined.
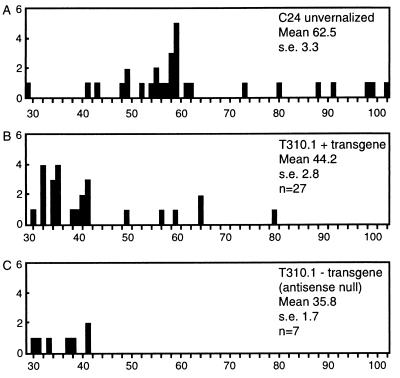
We measured the flowering time of T3 progeny from a T2 plant hemizygous for the transgene (plant 10.1) to determine whether the early-flowering phenotype segregated with the presence of the antisense transgene. The mean flowering time for the nonvernalized progeny of this line was 42.2 ± 2.2 days compared with 62.5 ± 3.3 days for the untransformed C24. The presence of the transgene was scored and the flowering time of plants, with or without the transgene, was compared (Fig. 1). Flowering time of the progeny ranged from 30 to 79 days; plants that did not inherit the transgene (antisense-null) flowered as early as sibling plants that carried the transgene. These antisense-null plants have methylation levels that are substantially reduced compared with untransformed plants, indicating that many sequences were not remethylated after loss of the antisense (9). These observations suggest that low methylation and not the presence of the transgene causes early flowering.
Figure 1.
The early-flowering phenotype segregates with low levels of DNA methylation but not with the METI antisense. The T3 progeny of T2 plant 10.1, which was hemizygous for the METI antisense transgene, flowered early even when they had lost the transgene by segregation. (A) Flowering times of unvernalized C24. (B) Flowering times of progeny that inherited the transgene; flowering times of progeny that did not inherit the transgene are shown in C. Methylation levels of a subset of plants from B were estimated, and in general, the level of methylation (35–45% of normal) correlated with the flowering time. The plant that flowered after 78 days (B) had a higher level of methylation (55% of normal), which may account for the later flowering time.
The Promotion of Flowering Is Correlated with the Extent of Demethylation.
The flowering times of plants from three independent methyltransferase antisense families, which differed in the magnitude of DNA demethylation (9), were compared. With no cold treatment, plants from all three families flowered significantly earlier than controls. The flowering time of plants from these antisense families differed, both between families and between lines within a family, but, in general, the promotion of flowering correlated with the reduction in DNA methylation (Table 3). For example, plants from line 22–6.11, homozygous for four copies of the antisense gene, flowered earlier than those from sibling line 22–6.9, which had a higher level of DNA methylation (Table 3). The latter is homozygous for three copies of the transgene and hemizygous for the remaining copy of the transgene. One antisense-null line, 10.1.8, flowered earlier than predicted by the level of DNA methylation.
The 23-day cold treatment used to vernalize plants in these experiments was chosen because germination did not occur during the treatment. In short-day photoperiods, the treatment resulted in a 50–60% reduction of flowering time for C24 but did not saturate the vernalization response. Demethylation in family 39 is more effective at promoting flowering than a 23-day cold treatment (Table 3), which is consistent with the conclusion that this treatment did not result in maximal promotion of flowering in control plants.
A Decreased DNA Methylation (ddm1) Mutant Is Early Flowering Under Short Days.
Mutants of DDM1 (decreased DNA methylation) have lower levels of DNA methylation than Columbia controls (8). DDM1 does not encode a DNA methyltransferase, but it is required in vivo for DNA methylation (18). It has been reported that ddm1 mutant plants flowered later than the wild-type Columbia progenitor when grown in long-day photoperiods, and that flowering became progressively later in successive generations of progeny from self-pollinated plants (18, 19). We have compared the flowering time of ddm1 homozygotes, from both an early generation and after six generations of self-pollination, with that of controls and have confirmed these observations (Table 4). Under these conditions neither wild-type Columbia nor ddm1 mutants responded to vernalization, suggesting that under long days this ecotype does not rely on the vernalization-dependent pathway for flowering.
Table 4.
Demethylation by mutation at the DDM1 locus promotes flowering of plants grown in short days, but delays flowering when plants are grown in long days
| Plant line | Flowering time (SD)
|
||||
|---|---|---|---|---|---|
| Unvern | Vern | % Col vern response in ddm1 mutant | Unvern | Vern | |
| Col (expt 1) | 40.8 ± 1.5* | 35.7 ± 1.8 | 14.5 ± 0.3† | 13.0 ± 0.2 | |
| ddm1 (expt 1)§ | 37.2 ± 0.8* | 34.3 ± 0.9 | 70.4 | 16.4 ± 0.3† | 15.0 ± 0.4 |
| Col (expt 2) | 70.0 ± 3.4 | 40.7 ± 3.0 | ND | ND | |
| ddm1 (expt 2)§ | 48.9 ± 2.3 | 37.6 ± 2.8 | 72 | ND | ND |
| Col (expt 3) | 72‡ | 50‡ | 19.1 ± 0.4 | 18.2 ± 0.3 | |
| ddm1 (expt 3)§ | 44‡ | 32‡ | >100% | 32.1 ± 1.0 | 33.9 ± 1.1 |
Flowering time is in days ± SE. SD, short days. LD, long days. ND, not done.
Mann–Whitney U test of ranks gave a standard normal deviate Z = 1.69 (probability of Z ≥ |1.69| is 0.091, not statistically significant).
Mann–Whitney U test of ranks gave a standard normal deviate Z = 4.65 (probability of Z ≥ |4.65| is <0.001).
Days for 50% of plants to flower (some plants had not flowered before day 85 when the experiment was terminated because the growth medium had dehydrated).
Plants used in expt 1 and expt 2 were from independent Col and ddm1 lines; the latter had been selfed fewer than four times. In expt 3 ddm1 plants were seventh generation self-progeny, from a line identified as late flowering.
In short-day photoperiods, early-generation mutant plants flowered 3 days earlier than the Columbia control (Table 4, expt 1). When flowering of the wild type was delayed further by changing the spectral quality of the light for the short-day photoperiod (see Materials and Methods), the promotion of flowering in the ddm1 mutant line was even more pronounced (Table 4, expt 2). In short days, with light of either spectral quality, the promotion of flowering in unvernalized ddm1 (19.1 ± 0.9% normal methylation) represented about 70% of the promotion caused by vernalization of the Columbia parent. This is comparable to the promotion of flowering seen in those methyltransferase antisense lines that had a similar level of DNA methylation, such as line T4 10.1.4, which gave 74% of the wild-type vernalization response (Table 3).
Seventh generation ddm1 mutants, which flowered 13 days later than the wild type when grown in long days, flowered at least 25 days earlier than the wild type when grown in short photoperiods. Flowering of these plants occurred at about the same time as the vernalized wild type, indicating that in these plants demethylation caused a greater promotion of flowering than in the early generation ddm1 mutants.
Plants with Reduced DNA Methylation Respond to Vernalization.
We compared the flowering time of cold-treated and untreated sibling methyltransferase antisense plants to determine whether plants that have reduced levels of DNA methylation respond to a vernalization treatment. Cold treatment of antisense plants promoted flowering compared with untreated siblings in all growth conditions tested, indicating that demethylation did not prevent a vernalization response (Table 2). Under some conditions, the flowering time of vernalized antisense and C24 plants differed by a few days, with effects on flowering time of a cold treatment and demethylation being either synergistic or antagonistic. When flowering of C24 was delayed (short days with Osram fluorescent tubes), vernalized antisense plants flowered earlier than the vernalized C24 control (Tables 2 and 3), but in long days or favorable lighting vernalized antisense plants flowered a few days later than cold-treated C24 (Table 2).
Plants from the earliest-flowering antisense family, 39, still showed a vernalization response, suggesting that the demethylation of DNA in these plants had not saturated the early-flowering response. However, the magnitude of the vernalization response (35% reduction in flowering time) in these plants was smaller than that seen for other lines in the same experiment (Table 3). The additive effect of vernalization and demethylation is consistent with either a common mechanism for the promotion of flowering by demethylation of DNA and cold temperatures, or two separate mechanisms, both of which contribute to early flowering.
When grown in long days, neither wild-type Columbia nor ddm1 mutant plants showed a vernalization response. In contrast, both Columbia and the ddm1 mutant showed a vernalization response when grown in short photoperiods (Table 4).
The Vernalization Signal Is Reset in Plants with Reduced DNA Methylation.
If vernalization is mediated by demethylation of specific DNA sequences, then resetting the signal for early flowering may require remethylation of these sequences during meiosis or zygote development. To determine whether this process is disrupted in plants containing a methyltransferase antisense construct, we tested whether methyltransferase antisense plants transmitted the vernalization signal to their progeny.
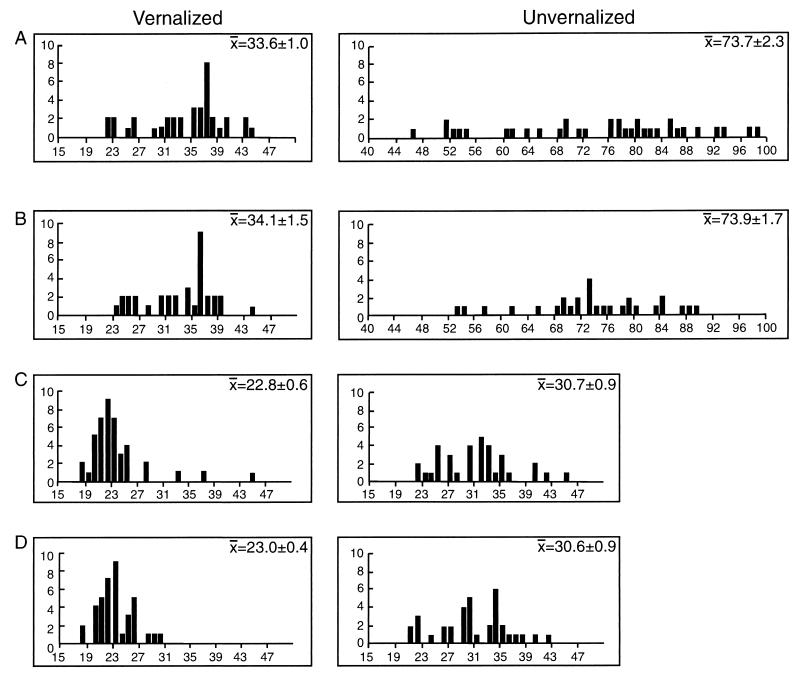
As reported in a previous study (3), progeny from vernalized and unvernalized C24 plants flowered at the same time, indicating that the vernalization signal was not transmitted to the next generation (Fig. 2). We compared the flowering time for progeny of cold-treated and untreated METI antisense plants. There was no significant difference in the flowering time of these progeny, whether or not the parental plants had been vernalized (Fig. 2). Furthermore, flowering of progeny of both vernalized and unvernalized plants was promoted by a cold treatment, confirming that the vernalization signal had been reset in this generation.
Figure 2.
Flowering time of progeny from vernalized and unvernalized C24 or antisense plants. The progeny of vernalized and unvernalized plants flowered at the same time, indicating the cold-induced signal for early flowering was not inherited in progeny of the vernalized antisense plants. All plants in the progeny generation responded to a cold treatment by flowering earlier than untreated sibs, confirming that the vernalization signal had been reset. (A) Progeny of vernalized C24 plants. (B) Progeny of unvernalized C24 plants. (C) Progeny of vernalized T3 10.5 antisense plants. (D) Progeny of unvernalized T3 10.5 antisense plants. For A–D, the left graph shows flowering time of plants vernalized in this generation, and the right graph shows flowering time of plants not vernalized in this generation.
DISCUSSION
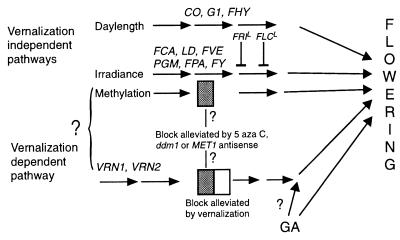
Flowering in Arabidopsis can be initiated by more than one pathway; these pathways are regulated by external or endogenous signals such as day length, temperature, photosynthate, or the hormone gibberellin (Fig. 3) (for reviews see refs. 20 and 21). Although flowering probably can be initiated by the action of a single pathway, it occurs earlier if more than one pathway is active (23). The relative contribution to flowering by each of these pathways depends on environmental conditions and the genotype of the plant (21). The vernalization-dependent pathway is not essential for the initiation of flowering unless one or more of the other pathways is blocked by mutation or environmental conditions (11); when the signals promoting flowering contributed by other pathways are limiting, a cold treatment promotes flowering by activation of the vernalization-dependent pathway.
Figure 3.
Proposed pathways to flowering in Arabidopsis. Some of the genes in the pathways (11, 21, 22) are indicated, but no order of function is intended from the order in which the genes are listed. There may be some functional interaction and/or overlap of these pathways that is not indicated in this simplified model. The step in the vernalization pathway that is blocked, in the absence of a cold treatment, is indicated by the box. The block represented by the shaded part of the box can be alleviated by demethylation, but the unshaded portion is only alleviated by cold. The methylation block may lie within the vernalization pathway and/or in a parallel pathway, as indicated.
Although several genes that are concerned with flowering have now been cloned (24–26), none of these functions within the vernalization-dependent pathway. Mutations that reduce the vernalization response recently have been identified (27), but the corresponding genes, which may encode steps of the vernalization-dependent pathway, have not been isolated. The vernalization pathway has several unique properties; unlike day length, which is perceived by leaves (28), a cold treatment is perceived by the mitotically active cells that ultimately form the apex from which the inflorescence develops (for reviews see refs. 1 and 11). The early-flowering response to a cold treatment can be separated temporally from the treatment by many cell generations, unlike the immediate response to long days (29). These properties strongly suggest that cold-induction of early flowering operates epigenetically, and this led to the hypothesis that vernalization is mediated by changes in DNA methylation (3).
Demethylation by 5-azaC treatment (3), by introduction of a METI antisense transgene or by mutation at the DDM1 locus, promotes flowering in vernalization-responsive Arabidopsis. An early-flowering response was observed only in conditions where wild-type plants showed a vernalization response, suggesting that demethylation and vernalization may activate the same pathway. Decreased methylation promoted flowering in both long and short photoperiods for C24, but only in short photoperiods for plants from the ecotype Columbia (Col). In long days, wild-type Col plants did not respond to vernalization (Table 4) and both Col antisense and ddm1 mutants flowered later than the wild type (8, 29) (Table 4). Genetic regulation of the late-flowering phenotype of ddm1 plants in long days has been mapped close to the FWA locus (30); fwa mutants are hypomethylated over at least 5 Mb spanning FWA (21). However, although fwa mutants do not respond to vernalization in long or short photoperiods (31), ddm1 mutants do respond to vernalization under short photoperiods, suggesting that the delay in flowering in ddm1 and fwa mutants may differ.
The difference in response to vernalization of plants from ecotypes C24 and Col is probably because of allelic differences at two loci, FRI and FLC (23, 32), that regulate flowering time in naturally occurring ecotypes of Arabidopsis (for review see ref. 21). C24 has an early-flowering allele at FLC (32) whereas Col has a late allele at this locus (23). However, because plants with a late-flowering allele of FRI (FRIL), which delays flowering by the postulated irradiance pathway (Fig. 3), show a large response to vernalization (11, 23), it seems more likely that allelic differences at the FRI locus, rather than the FLC locus, are important. Early flowering of antisense C24 plants in long days, when the irradiance pathway is partially blocked by FRIL, supports the idea that demethylation activates another flowering pathway, perhaps the same pathway that is activated by cold. Wild-type Col plants, which have an early-flowering allele of FRI (23), do not flower early in response to vernalization or to reduced DNA methylation when grown in long days. Although there are other differences between the ecotypes C24 and Col, the difference in vernalization responsiveness may account for the difference in the flowering response to demethylation in long days.
Our data suggest that demethylation of DNA can substitute, at least in part, for a cold treatment. Growth at vernalizing temperatures was associated with some reduction of DNA methylation, but the demethylation was transient and normal methylation levels were restored when the seedlings were transferred to warmer temperatures (Table 1). If the demethylation caused by low temperatures is essential for cold-induced early flowering, then sequences critical for this process may be susceptible to demethylation at vernalizing temperatures and be protected from subsequent remethylation, either by binding of transcription factors or by chromatin structure.
Vernalization may preferentially affect the methylation status of sequences that are important for early flowering, in contrast to the more general demethylation assumed to occur in antisense plants. A 70% reduction of methylation occurred in antisense plants in which the promotion in flowering was comparable to that of a vernalizing treatment that resulted in only a 15% decrease in DNA methylation.
The promotion of flowering correlated directly with the extent of demethylation in different C24 METI antisense lines. This parallels the observation that, in Arabidopsis and other plant species, the promotion of flowering is proportional to the length of the cold treatment, until the vernalization response is saturated (33, 34). Cell division is required for vernalization (4, 5); presumably more cells would undergo cell division and adopt the “vernalized state” as the length of cold treatment increases (5). Cold-induced demethylation probably occurs during cell division by the uncoupling of DNA replication and maintenance methylation (35). Failure of maintenance methylation leads to demethylation of cytosines on the newly synthesized DNA strand (6), which, after a second round of replication, results in double-stranded demethylation of DNA in one daughter cell.
Mitotic inheritance of the vernalization signal could be achieved by maintenance of an altered pattern of DNA methylation, characteristic of the “vernalized state.” The vernalization signal could be reset in sexual progeny by de novo methylation during gametogenesis or embryo development; this process may be inhibited in METI antisense plants. Antisense plants responded to a low-temperature treatment by flowering earlier than untreated siblings. However, the antisense transgene did not inhibit the resetting process because the cold-induced signal for early flowering was not inherited by sexual progeny (Fig. 2).
The early-flowering response because of antisense-induced demethylation was inherited by progeny of an antisense-null line (Table 3). Early flowering persisted through at least two generations after loss of the antisense (not shown). These plants flowered earlier than predicted on the basis of the level of DNA methylation (Table 3), suggesting that remethylation of the sites involved in the early-flowering phenotype occurred more slowly than the restoration of methylation at many other sites. In contrast, the early-flowering response to 5-azaC treatment was not inherited in progeny (3), suggesting that methylation patterns were restored in these plants.
These observations on inheritance of demethylation- and cold-induced early-flowering signals indicate that demethylation caused by a METI antisense does not replicate all aspects of vernalization. Our data suggest that if demethylation of DNA is integral to vernalization, then the promotion of flowering is not solely a result of METI-associated demethylation of sequences critical for the transition to flowering, but that there are two processes, one of which is subject to resetting each generation.
It is possible that additional methyltransferases, which methylate different sites, are involved. There is good evidence for the activity of other methyltransferases in METI antisense plants (36); low temperatures could inhibit the activity of all methyltransferases, only one of which is affected in METI antisense plants. Other epigenetic mechanisms, such as the formation of mitotically stable chromatin structures in response to low temperatures, could also be involved (for review see ref. 37). The isolation of mutations that show an altered vernalization response, but that have no other effect on flowering, and the subsequent cloning and characterization of the genes involved will elucidate further the processes associated with the vernalization response.
Acknowledgments
The authors thank E. J. Richards and T. J. Kakutani for providing seed from early and late generations of the ddm1 mutant lines, and Lyndall Thorpe for excellent technical assistance. R.K.G. and K.K. were supported by Australian Postgraduate Awards.
ABBREVIATION
- 5-azaC
5-azacytidine
References
- 1.Lang A. In: Encyclopedia of Plant Physiology. Ruhland W, editor. Berlin: Springer; 1965. pp. 1489–1536. [Google Scholar]
- 2.Whyte R O. In: Vernalization and Photoperiodism. Murneek A E, Whyte R O, editors. Waltham, MA: Chronica Botanica; 1948. pp. 1–38. [Google Scholar]
- 3.Burn J E, Bagnall D J, Metzger J M, Dennis E S, Peacock W J. Proc Natl Acad Sci USA. 1993;90:287–291. doi: 10.1073/pnas.90.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wellensiek S J. Nature (London) 1962;195:307–308. [Google Scholar]
- 5.Wellensiek S J. Plant Physiol. 1964;39:832–835. doi: 10.1104/pp.39.5.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird A P. J Mol Biol. 1978;118:49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- 7.Chandler V L, Walbot V. Proc Natl Acad Sci USA. 1986;83:1767–1771. doi: 10.1073/pnas.83.6.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vongs A, Kakutani T, Martienssen R A, Richards E J. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 9.Finnegan E J, Peacock W J, Dennis E S. Proc Natl Acad Sci USA. 1996;93:8449–8454. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brock R D, Davidson J L. Environ Exp Bot. 1994;34:195–199. [Google Scholar]
- 11.Dennis E S, Finnegan E J, Bilodeau P, Chaudhury A, Genger R, Helliwell C A, Sheldon C C, Bagnall D J, Peacock W J. Semin Cell Dev Biol. 1996;7:441–448. [Google Scholar]
- 12.Cedar H. Cell. 1988;52:3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- 13.Finnegan E J, Dennis E S. Nucleic Acids Res. 1993;21:2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilmyuk V I, Carroll B J, Thomas C M, Jones J D G. Plant J. 1993;3:493–494. doi: 10.1111/j.1365-313x.1993.tb00169.x. [DOI] [PubMed] [Google Scholar]
- 15.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 16.Martinez-Zapater J M, Somerville C R. Plant Physiol. 1990;92:770–776. doi: 10.1104/pp.92.3.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bagnall D J. Ann Bot. 1993;71:75–83. [Google Scholar]
- 18.Kakutani T, Jeddeloh J, Richards E J. Nucleic Acids Res. 1995;23:130–137. doi: 10.1093/nar/23.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kakutani T. Plant J. 1998;12:1447–1451. doi: 10.1046/j.1365-313x.1997.12061447.x. [DOI] [PubMed] [Google Scholar]
- 20.Bernier G, Havelange A, Houssa C, Petijean A, Lejeune P. Plant Cell. 1993;5:1147–1155. doi: 10.1105/tpc.5.10.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koornneef M, Alonso-Blanco C, Peeters A J M, Soppe W. In: Annual Review of Plant Physiology and Plant Molecular Biology. Jones R L, Somerville C R, Walbot V, editors. Palo Alto, CA: Annual Reviews; 1998. , in press. [DOI] [PubMed] [Google Scholar]
- 22.Simon R, Coupland G. Sem Cell Dev Biol. 1996;7:419–425. [Google Scholar]
- 23.Lee I, Amasino R A. Plant Physiol. 1995;108:157–162. doi: 10.1104/pp.108.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee I, Aukerman M J, Gore S L, Lohman K N, Michaels S D, Weaver L M, John M C, Feldmann K, Amasino R. Plant Cell. 1994;6:75–83. doi: 10.1105/tpc.6.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Putterill J, Robson F, Lee K, Simon R, Coupland G. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- 26.MacKnight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C. Cell. 1997;89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- 27.Chandler J, Wilson A, Dean C. Plant J. 1996;10:637–644. doi: 10.1046/j.1365-313x.1996.10040637.x. [DOI] [PubMed] [Google Scholar]
- 28.Zeevaart J A D. In: Light and the Flowering Process. Vince-Prue D, Thomas B, Cockshull B, Orlando K E, editors. London: Academic; 1984. pp. 137–142. [Google Scholar]
- 29.Ronemus M J, Galbiati M, Ticknor C, Chen J, Dellaporta S L. Science. 1996;273:654–657. doi: 10.1126/science.273.5275.654. [DOI] [PubMed] [Google Scholar]
- 30.Kakutani T. Plant J. 1997;12:1447–1451. doi: 10.1046/j.1365-313x.1997.12061447.x. [DOI] [PubMed] [Google Scholar]
- 31.Koornneef M, Hanhart C J, van der Veen J H. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- 32.Sanda S L, Amasino R M. Weeds World. 1995;2:2–8. [Google Scholar]
- 33.Purvis O N, Gregory F G. Ann Bot. 1937;1:569–591. [Google Scholar]
- 34.Napp-Zinn K. Planta. 1957;50:177–210. [Google Scholar]
- 35.Leonhardt H, Page A W, Weier H-U, Bestor T H. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 36.Jacobsen S E, Meyerowitz E M. Science. 1997;277:1100–1103. doi: 10.1126/science.277.5329.1100. [DOI] [PubMed] [Google Scholar]
- 37.Paro R, Harte P J. In: Epigenetics Mechanisms of Gene Regulation. Russo V E A, Martienssen R A, Riggs A D, editors. New York: Cold Spring Harbor Lab. Press; 1996. pp. 507–528. [Google Scholar]