Abstract
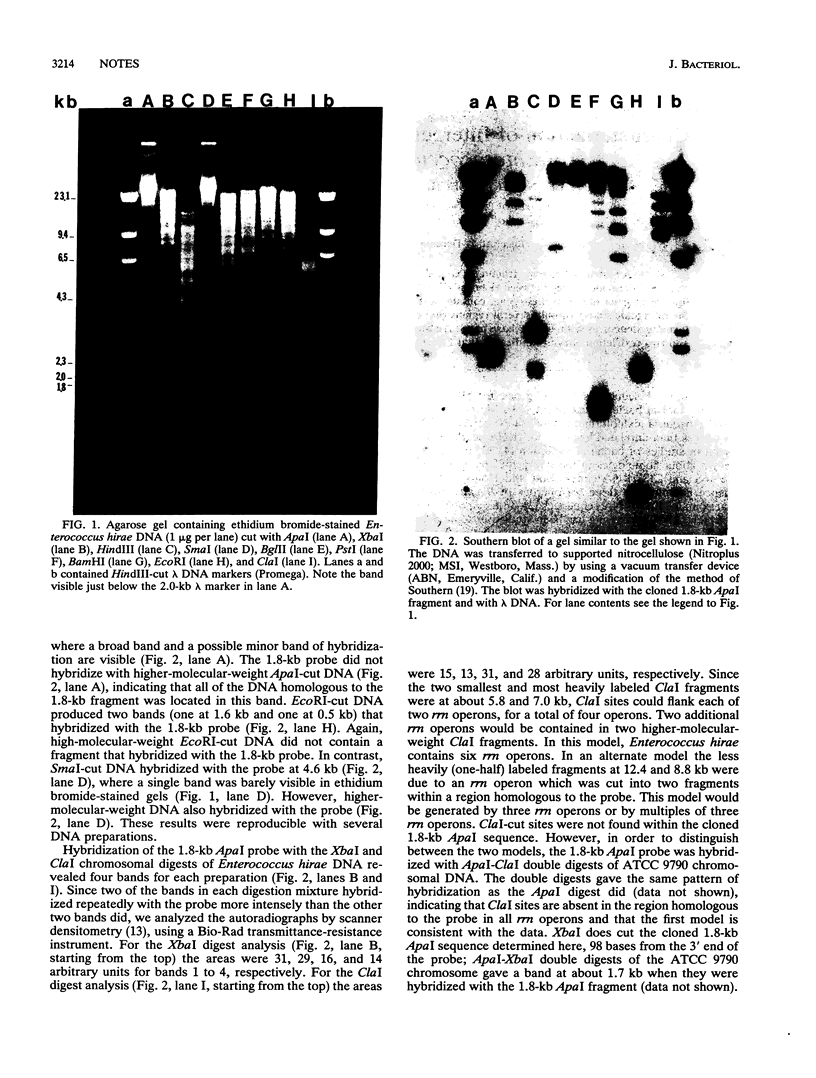
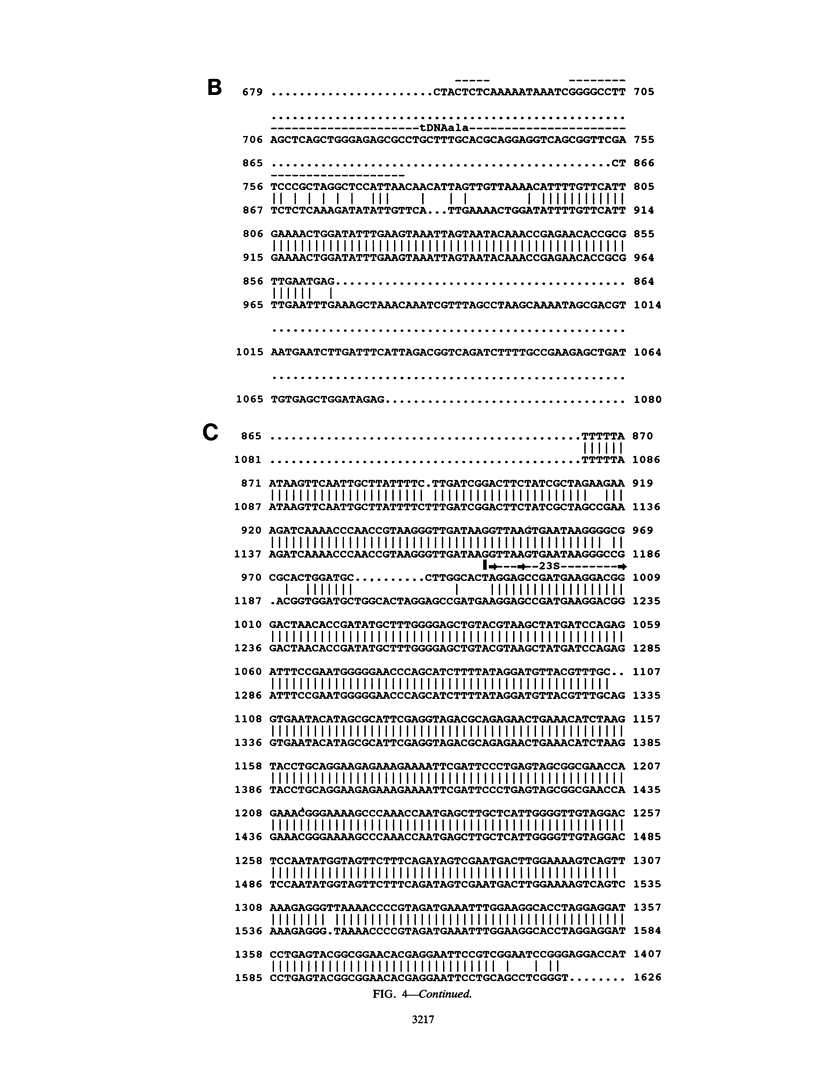
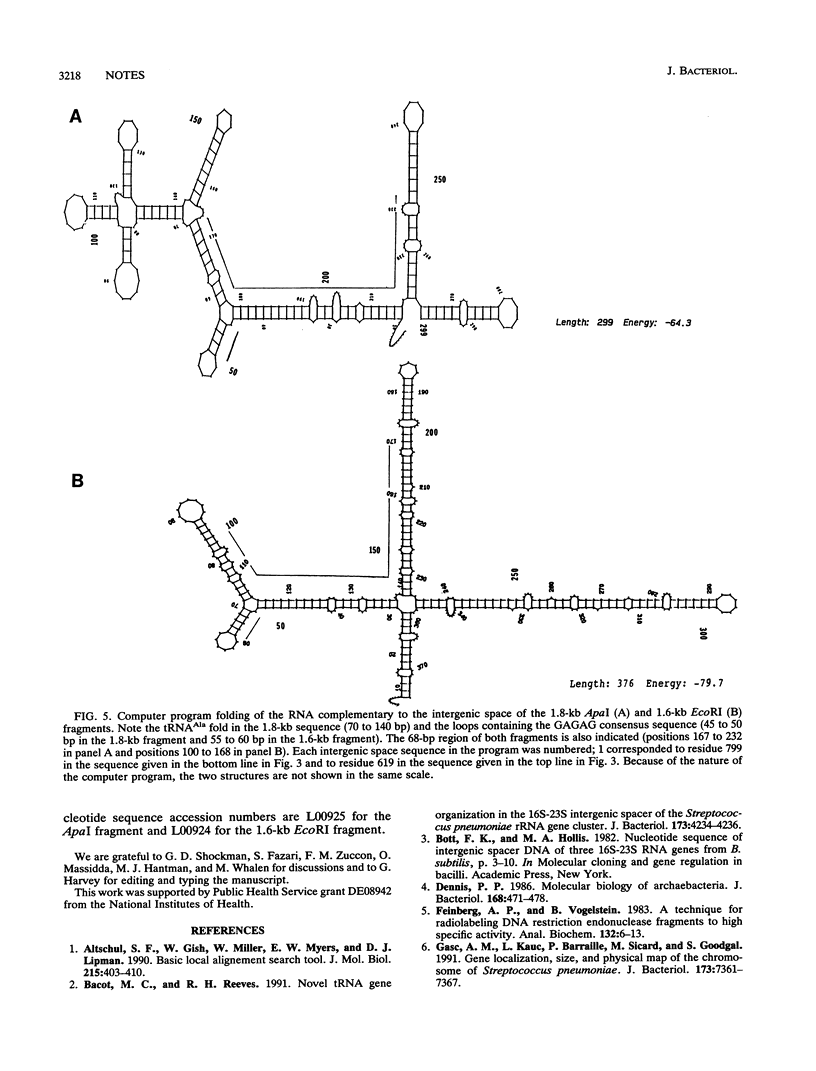
Two DNA restriction enzyme fragments coding for the 3' termini of 16S rRNA, the 5' termini of 23S rRNA, and the intergenic spaces between them in Enterococcus hirae ATCC 9790 were cloned and sequenced. The intergenic space of one of these genes contains a tRNA(Ala) sequence, whereas the other does not. Nevertheless, the intergenic spaces contain several regions that exhibit high levels of sequence homology and are capable of forming structures with similar base pairs. An analysis of Southern blots of chromosomal DNA cut with one and two restriction enzymes indicated that E. hirae has a total of six rrn operons.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bacot C. M., Reeves R. H. Novel tRNA gene organization in the 16S-23S intergenic spacer of the Streptococcus pneumoniae rRNA gene cluster. J Bacteriol. 1991 Jul;173(13):4234–4236. doi: 10.1128/jb.173.13.4234-4236.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P. P. Molecular biology of archaebacteria. J Bacteriol. 1986 Nov;168(2):471–478. doi: 10.1128/jb.168.2.471-478.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gasc A. M., Kauc L., Barraillé P., Sicard M., Goodgal S. Gene localization, size, and physical map of the chromosome of Streptococcus pneumoniae. J Bacteriol. 1991 Nov;173(22):7361–7367. doi: 10.1128/jb.173.22.7361-7367.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. J., Stewart G. C., Hollis M. A., Vold B. S., Bott K. F. Nucleotide sequence of the Bacillus subtilis ribosomal RNA operon, rrnB. Gene. 1985;37(1-3):261–266. doi: 10.1016/0378-1119(85)90281-1. [DOI] [PubMed] [Google Scholar]
- Hui I., Dennis P. P. Characterization of the ribosomal RNA gene clusters in Halobacterium cutirubrum. J Biol Chem. 1985 Jan 25;260(2):899–906. [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- Krawiec S., Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990 Dec;54(4):502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bouguenec C., Horodniceanu T. Conjugative R plasmids in Streptococcus faecium (group D). Antimicrob Agents Chemother. 1982 May;21(5):698–705. doi: 10.1128/aac.21.5.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. J., Smith H. O., Redfield R. J. Organization of the Haemophilus influenzae Rd genome. J Bacteriol. 1989 Jun;171(6):3016–3024. doi: 10.1128/jb.171.6.3016-3024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Dahlberg J. E., Lindahl L., Jaskunas S. R., Dennis P. P., Nomura M. Transfer RNA genes between 16S and 23S rRNA genes in rRNA transcription units of E. coli. Cell. 1976 Feb;7(2):165–177. doi: 10.1016/0092-8674(76)90016-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Srivastava A. K., Schlessinger D. Escherichia coli 16S rRNA 3'-end formation requires a distal transfer RNA sequence at a proper distance. EMBO J. 1989 Oct;8(10):3159–3166. doi: 10.1002/j.1460-2075.1989.tb08470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. K., Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- Srivastava A. K., Schlessinger D. Structure and organization of ribosomal DNA. Biochimie. 1991 Jun;73(6):631–638. doi: 10.1016/0300-9084(91)90042-y. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]