Abstract
Novel plants with individual maize chromosomes added to a complete oat genome have been recovered via embryo rescue from oat (Avena sativa L., 2n = 6x = 42) × maize (Zea mays L., 2n = 20) crosses. An oat–maize disomic addition line possessing 21 pairs of oat chromosomes and one maize chromosome 9 pair was used to construct a cosmid library. A multiprobe (mixture of labeled fragments used as a probe) of highly repetitive maize-specific sequences was used to selectively isolate cosmid clones containing maize genomic DNA. Hybridization of individual maize cosmid clones or their subcloned fragments to maize and oat genomic DNA revealed that most high, middle, or low copy number DNA sequences are maize-specific. Such DNA markers allow the identification of maize genomic DNA in an oat genomic background. Chimeric cosmid clones were not found; apparently, significant exchanges of genetic material had not occurred between the maize-addition chromosome and the oat genome in these novel plants or in the cloning process. About 95% of clones selected at random from a maize genomic cosmid library could be detected by the multiprobe. The ability to selectively detect maize sequences in an oat background enables us to consider oat as a host for the cloning of specific maize chromosomes or maize chromosome segments. Introgressing maize chromosome segments into the oat genome via irradiation should allow the construction of a library of overlapping fragments for each maize chromosome to be used for developing a physical map of the maize genome.
Keywords: corn, chromosome-specific, cosmid library, repetitive DNA, cloning
Chromosome addition lines of different plant species (1–5) have been generated to introgress valuable genes from wild or cultivated relatives into host plant species. Alien chromosome additions have been used for gene mapping (2, 6, 7) and serve as an enriched source of markers for positional cloning and constructing physical maps of specific chromosomes. The discovery of maize-chromosome retention in oat “haploids” after oat × maize crosses and the recovery of stable maize chromosome-addition oat lines (8, 9) should allow the development of a system of chromosome analysis similar to that available in mammalian hybrid-cell systems (10–12). Such a system may be used for gene assignment, isolation of chromosome-specific probes (13), flow sorting (14) and microdissection of chromosomes (15), development of chromosome-specific “paints” of fluorochrome-labeled DNA fragments (16–18), physical mapping, and selective isolation and mapping of cDNAs of a particular chromosome (19, 20).
In this paper we describe an approach for isolating clones containing large-fragment maize DNA of a single-chromosome origin from the oat–maize chromosome addition lines that will assist in the construction of physical maps for maize chromosomes. The approach is based on cloning genomic DNA of an oat–maize chromosome-addition line in an appropriate vector and subsequent use of maize-specific dispersed repetitive DNA sequences as detection probes to isolate clones carrying maize genomic DNA. Such an approach was successfully applied to the isolation of human-specific DNA fragments in cosmid libraries constructed from DNA of human-rodent hybrid cell lines carrying individual human chromosomes. In those experiments the probes included Alu-repeats (21), a Cot1 DNA fraction (22), or labeled total human DNA (23).
Plant repetitive sequences that are apparently species-specific were first isolated from rye (24, 25) and later found in many other species (26). Comparative studies revealed a strong correlation between the proportion of species-specific repeated families in a genome and phylogenetic relationships (27, 28). About 90–95% of all randomly tested genomic repeated sequences from barley were detected in wheat and rye (27). According to DNA reassociation studies, less-related species such as maize and wheat have less than 10% nucleotide sequences in common (28). The success of isolating maize DNA from oat–maize chromosome-addition lines depends on how many maize-specific (relative to oat) high-copy-number dispersed nucleotide sequences are found in the maize genome. Genome analysis in grasses, including maize and oat, reveals that a major portion of genomic DNA consists of families of repetitive sequences dispersed throughout the genome (26, 29, 30). According to DNA–DNA renaturation data in maize (28), unique sequences of average length 2,100 bp are interspersed with mid-repetitive sequences. Direct analysis of sequences adjacent to several maize genes reveals that these genes are flanked by highly repetitive DNA sequences (31). Thus, maize large DNA inserts cloned in an appropriate vector will likely carry some kind of dispersed maize-specific nucleotide sequence. A cosmid vector was chosen for this project because of the high cloning efficiency and relatively large insertion size. Because the oat genome is about 11,300 Mb, an added maize chromosome with median size of about 250 Mb would constitute about 2–4% of the total nuclear DNA of an oat–maize chromosome addition line.
We found that the major part of the maize genome consists of nucleotide sequences that do not cross-hybridize to oat genomic sequences under standard hybridization conditions. This predominant nonhomology between oat and maize genomic sequences means that many maize genomic sequences can be directly used for Southern blot hybridization on DNA from oat–maize chromosome addition lines to selectively detect sequences of maize origin. A mixture of highly repetitive dispersed DNA sequences of maize was used as a maize-specific multiprobe to screen a cosmid library of the chromosome addition line. A group of maize-specific cosmids with inserts from only maize chromosome 9 was isolated and characterized. Based on the large difference in nucleotide sequence composition between oat and maize and the means to efficiently isolate and identify maize DNA fragments from a chromosome addition line, we propose oat as an effective host for the cloning of maize DNA segments to construct physical maps for maize chromosomes.
MATERIALS AND METHODS
Maize and Oat Strains.
Oat–maize disomic addition lines for maize chromosomes 2, 3, 4, 7, and 9 were derived from plants recovered following sexual crosses of oat by maize (8, 9). In these crosses about one-third of the recovered plants contained a haploid set of 21 oat chromosomes plus one or more maize chromosomes as a result of incomplete maize-chromosome elimination during early embryo development. Partial self-fertility due to the production of unreduced gametes by these plants yielded disomic maize-chromosome-addition oat plants (2n = 42 + 2). The presence of maize chromosomes was verified cytologically for each plant in the current study. The chromosome addition lines, the maize parent lines Seneca 60 and A188, and the oat parent lines Starter-1 and Sun II were used for DNA extraction.
Isolation and Analysis of DNA.
Leaves of 2- to 4-week-old seedlings grown in a growth chamber were used for nuclei isolation in pH 9.5 buffer according to the protocol of Liu and Whitter (32). High molecular weight DNA was purified by phenol extraction from nuclei after lysis of the nuclei suspension in an equal volume of the same buffer supplemented with 2% sarkosyl. After two phenol extractions, DNA was precipitated with two volumes of ethanol in the presence of 0.3 M NaOAc, dissolved in TE buffer (10 mM Tris·Cl/1 mM EDTA, pH 8.0), treated with RNase (50 mg/ml), and extracted with phenol-chloroform.
Sau3A genomic DNA fragments were cloned into the BamHI site of dephosphorylated plasmid vector pBlueScript (pBS) II KS (Stratagene). EcoRI subfragments from cosmid clones were cloned into the EcoRI site of the dephosphorylated pBS KS vector according to standard procedures (33). Insertions were amplified with the help of forward and reverse primers (Stratagene) and purified by agarose gel electrophoresis.
Gel-blot analysis of plant and cosmid DNA was carried out as described by Sambrook et al. (33) with several modifications (34). DNA fragments and total plant DNA were labeled by random primer extension (35).
Seventeen clones carrying maize repetitive nucleotide sequences (30) were kindly provided by J. Bennetzen (Purdue University, West Lafayette, IN). A clone containing the maize 185-bp knob repeat (36) was provided by W. Peacock (Commonwealth Scientific and Industrial Research Organization, Canberra, Australia).
Cosmid Library Construction and Screening.
Cosmid library construction and screening were done using the cosmid vector SuperCos 1 (Stratagene) and packaging extract GigaPack II (Stratagene) with protocols provided by the manufacturer. Total nuclear DNA was partially digested with Sau3A, dephosphorylated, and ligated to the cosmid vector. Ligation products were packaged and the library propagated in Escherichia coli XL1-Blue MR. Cosmid libraries were constructed for genomic DNAs of the parental maize (Seneca 60) and oat (Starter-1) lines, as well as for the oat–maize chromosome 9 addition line.
The size of the insertion in a cosmid clone was determined as the sum of EcoRI subfragments after fractionation in gels with different concentrations of agarose from 0.6% up to 1.5% to achieve satisfactory resolution of long and short DNA subfragments.
RESULTS
Cloning of Maize-Specific Repetitive Sequences.
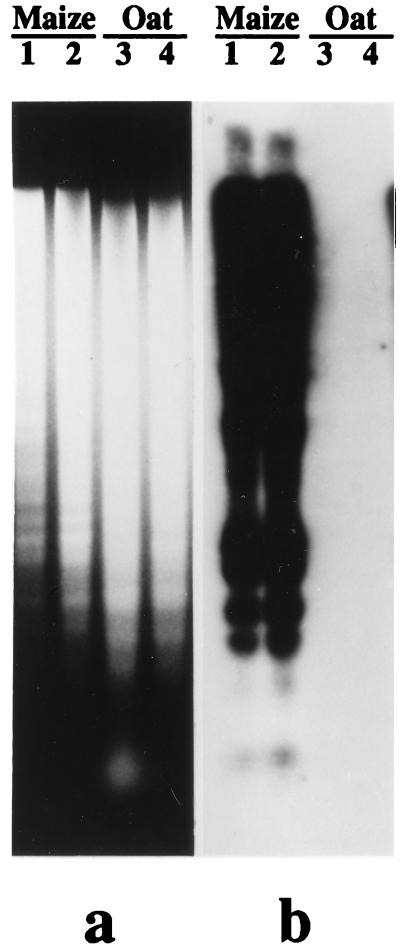
When labeled maize total genomic DNA was used as a probe in blot hybridization, little cross-hybridization to oat genomic DNA occurred under standard conditions (65°C in 6× SSC) in comparison with strong hybridization to maize genomic DNA (data not shown). This hybridization pattern indicated that a significant portion of the repeated nucleotide sequences of maize and oat are not shared. To isolate maize repeated DNA sequences specific to maize relative to oat (hereafter referred to simply as “maize-specific” sequences) a maize plasmid genomic library was constructed with fragments from a complete Sau3A digest and screened with labeled total maize DNA. Clones were isolated that gave a high signal with total labeled maize DNA as the probe. Labeled oat genomic DNA hybridized only to a small extent to the same set of clones on a replica filter. Purified plasmids with Sau3A insertions, together with a group of maize repetitive DNA sequences in plasmids (see Materials and Methods), were cut with appropriate restriction enzymes to release insertions. They were screened by blot hybridization with labeled maize and oat DNA. Clones showing a high signal similar to the signal of an 18S–26S rDNA sequence were considered as highly repetitive. From this screening, 15 plasmids from the Sau3A maize genomic library, 6 Zpr clones (from Bennetzen), and the one 185-bp knob repeat clone were selected as highly repetitive maize-specific nucleotide sequences. Insertions from these 22 selected plasmids were combined to form a composite multiprobe. This multiprobe revealed very strong hybridization to maize genomic DNA on a Southern blot and no detectable hybridization to oat genomic DNA (Fig. 1). The same multiprobe was used to screen cosmid partial libraries of genomic DNA of maize and oat (about 10,000 clones each). Almost all colonies (90–95%) in the maize cosmid library revealed strong or moderate signals. At the same time, only a few colonies, all with relatively weak signals, were detected in the oat cosmid library. Taken together, these results indicate that this composite multiprobe is highly specific for maize DNA and is suitable for detecting maize DNA fragments in a cosmid library made from an oat–maize chromosome-addition line.
Figure 1.
Blot hybridization of a labeled multiprobe, composed of 22 maize-specific repetitive DNA sequences, to genomic DNA digests of 2 maize and 2 oat varieties. Strong signal is seen over lanes with maize DNA but not those with oat DNA. (a) EtdBr-stained 0.8% agarose gel after separation of DNA samples cut with EcoRI. (b) Autoradiogram after hybridization to the P-32 labeled multiprobe.
Isolation of Maize-Specific Cosmids from an Oat–Maize Addition-Line Cosmid Library.
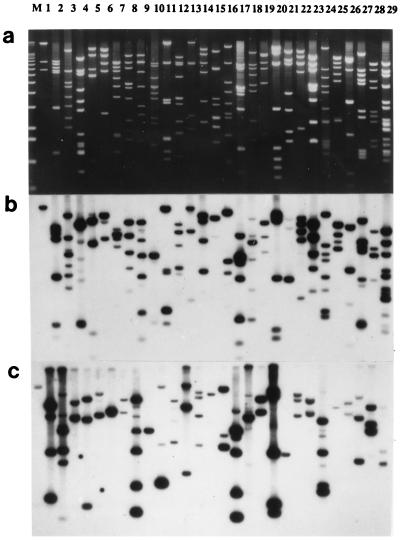
The multiprobe was used to screen a cosmid library made from an oat–maize chromosome addition line carrying maize chromosome 9. The screening and rescreening of about 5,000 clones led to the isolation of 29 hybridization-positive individual colonies. The resulting clones constituted a maize chromosome 9 library. The cloned maize DNA fragments in the selected 29 clones had a median size of about 39 kb. Together they comprised more than 1 Mb of maize genomic DNA. EcoRI restriction enzyme digestion enabled cutting out the cloned DNA fragments from the vector and identification of the number and size of EcoRI subfragments in the original insertions (Fig. 2a). Fractionation of the samples on gels of various agarose concentrations from 0.6 to 1.5% enabled detection of doublets and additional small subfragments below 0.5 kb not detected in the 0.8% agarose gel blots shown.
Figure 2.
Blot panel of 29 maize-specific cosmid clones isolated from a cosmid library of an oat–maize chromosome 9 addition line. (a) Cosmids (1–29) are cut by EcoRI restriction enzyme and size-fractionated in an 0.85% agarose gel stained with EtdBr (M = molecular weight marker, 1-kb ladder). (b) Labeled, total genomic maize DNA as a probe shows strong, medium, or weak hybridization to almost 90% of all EcoRI subfragments present in the cosmid clones. (c) Labeled multiprobe of highly repeated maize DNA sequences shows hybridization to from one to five EcoRI fragments in each lane.
Blot hybridization of labeled oat genomic DNA to purified DNA recovered from each cosmid clone cut by EcoRI did not reveal strong cross-hybridization to any of the EcoRI subfragments (data not shown). Only after substantial overexposure were weak signals revealed and then only for a portion of the DNA fragments. At the same time, most (>90%) of the EcoRI fragments revealed strong, medium, or weak hybridization to labeled maize genomic DNA (Fig. 2b). The difference in hybridization to oat vs. maize genomic DNA indicates that the selected cloned DNA fragments likely originated from the maize chromosome, with no evidence that any were chimeric in origin.
The EcoRI digests of the set of selected cosmid clones were hybridized with the maize-specific repeated-sequence multiprobe that was used to screen the cosmid library (Fig. 2c). The multiprobe highlighted about 100 of the more than 200 EcoRI fragments identified in this set of cosmid clones. On average about three fragments per cosmid were homologous to various repeated DNA sequences present in the multiprobe. At the same time a portion of the EcoRI fragments did not hybridize to the multiprobe nor to total oat DNA but did hybridize to labeled total maize DNA. These fragments may be added to the collection of maize-specific repeated DNA sequences to make the multiprobe an even more efficient screening tool.
Maize-Specific Clones Originate from Maize Chromosome 9.
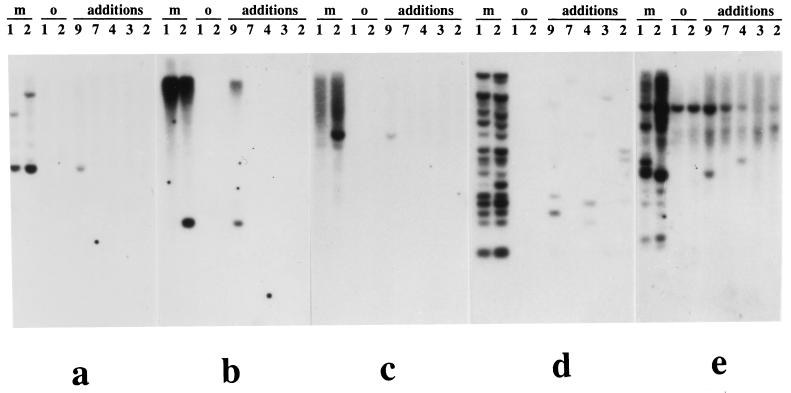
EcoRI subfragments that showed no readily detectable hybridization to maize genomic DNA as the probe were identified in 12 of the 29 cosmid clones. Presumably they are low copy number or unique nucleotide sequences. Altogether they comprise 11% of the EcoRI fragments or about 6.5% of the total amount of maize DNA cloned in this set of cosmid clones. Twenty-one of them were recloned in plasmids and, as illustrated for five fragments in Fig. 3, used as probes on blot panels of DNA samples from addition lines for maize chromosomes 2, 3, 4, 7, and 9. Two parental maize and two parental oat lines were also on this blot panel (Fig. 3). Nine of these 21 EcoRI fragments revealed one or several bands with DNA specific for chromosome 9 of maize and did not hybridize to the DNA of the other four maize chromosomes tested (Fig. 3 a–c). Seven of these nine fragments revealed restriction fragment length polymorphism (RFLP) in the two maize stocks, A188 and Seneca 60 (Fig. 3 a and b). Another 11 EcoRI fragments produced multiple bands with DNA of all maize chromosomes tested, which is characteristic for low-copy-number families of dispersed repeated sequences (10–100 copies) (Fig. 3 d and e). Eight of these sequences produced a chromosome-specific pattern of hybridization and potentially could be used for chromosome identification (Fig. 3 d and e).
Figure 3.
Hybridization of unique and low-copy-number DNA sequences isolated from maize-specific cosmids shown in Fig. 2 to a blot panel of oat–maize chromosome addition lines carrying maize chromosomes 9, 7, 4, 3, and 2, respectively; m1 and m2 are maize stocks A188 and Seneca 60; o1 and o2 are oat stocks Sun II and Starter-1. (a) The 2.5-kb EcoRI fragment from cosmid 15 shows one band on the chromosome 9 addition line. An additional polymorphic band is present in the parental stocks of maize. (b) The 1.8-kb EcoRI fragment from cosmid 28 detects two bands. One band, polymorphic between m1 and m2, is present in the chromosome 9 addition line. An additional nonpolymorphic band is present on chromosome 9 and in both parental stocks. (c) The 2.1-kb EcoRI fragment from cosmid 10 shows one band on chromosome 9. (d) The 1.4-kb EcoRI fragment from cosmid 20 detects about 20 bands in parental maize stocks and several bands among the chromosome addition lines. The band pattern is chromosome-specific. No cross-hybridization occurred to oat DNA in a–d. (e) The 2.9-kb EcoRI fragment from cosmid 6 detects several polymorphic bands in parental maize stocks. One nonpolymorphic band is seen in maize, oat, and chromosomes 9, 7, 4, and 2 addition lines. Additional bands are seen in other chromosome addition lines.
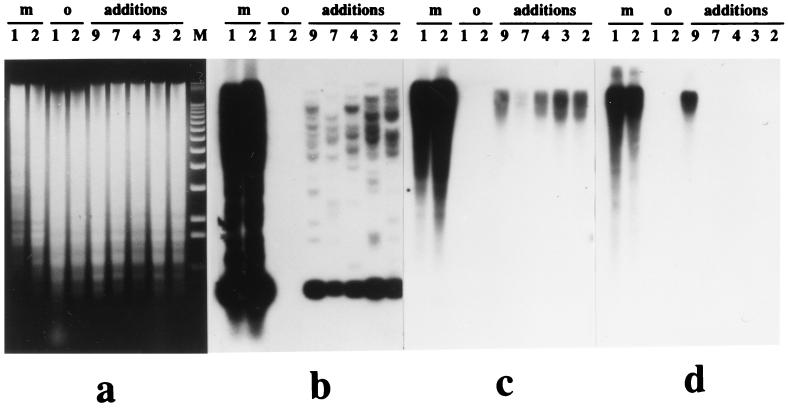
Several EcoRI fragments belonging to medium and highly repetitive classes of nucleotide sequences from different cosmid clones of maize chromosome 9 were used as probes. These revealed a complex pattern of hybridization for all maize chromosomes on the panel of chromosome-addition lines (Fig. 4b). Labeled highly repetitive DNA from cosmid 1 (Fig. 4c), as well as the full set of 29 cosmids (data not shown), gave strong hybridization to all maize chromosomes and did not reveal any chromosome 9-specific repetitive nucleotide sequences. Only the 185-bp knob-repeat revealed apparent chromosome specificity (Fig. 4d). Many copies of 185-bp repeats are located on chromosome 9, and a small fraction is present on chromosome 4 in these materials. Several weak bands are seen with DNA of maize chromosomes 2 and 3. This result is compatible with reports that most 185-bp knob DNA sequences are organized in the form of clusters on different chromosomes and relative knob size can differ greatly among chromosomes in different maize lines (31).
Figure 4.
Hybridization of medium and highly repetitive cloned maize DNA sequences to a blot panel of oat–maize chromosome addition lines carrying maize chromosomes 9, 7, 4, 3, and 2, respectively; m1 and m2 are maize stocks A188 and Seneca 60; o1 and o2 are oat stocks Sun II and Starter-1. (a) EtdBr-stained 0.8% agarose gel; lane M is a 1-kb molecular weight marker ladder. (b) The 0.7-kb EcoRI fragment of cosmid 10 shows multiple bands on different maize chromosomes as well as one common band for all chromosomes. (c) Cosmid 1, with a 40-kb insertion, shows strong hybridization signal over maize (lines m1 and m2) and over chromosome addition lines 9, 7, 4, 3, and 2; no hybridization is seen over oat DNA (lines o1 and o2). (d) With the 185-bp knob repeat, most of the hybridization signal is located on chromosome 9. No hybridization to oat DNA is detected.
Most Cloned Maize Nucleotide Sequences Are Maize-Specific.
Seven out of 11 unique or low-copy-number sequences showed no cross-hybridization to oat genomic DNA (Fig. 3 a–d). This result indicates that a significant portion (>50%) of low-copy-number maize sequences have diverged enough to show no cross-hybridization to oat genomic DNA. Eleven of 14 (80%) middle repetitive sequences (class 10–100 copies) are also maize-specific. Two among the remaining three showed only faint cross-hybridization to oat genomic DNA. Highly repetitive sequences were preselected in this study, and the process of selection showed that most highly repetitive maize sequences are also maize-specific (Fig. 4 b–d).
DISCUSSION
Maize Species-Specific Sequences.
The cross between oat and maize, two species from different subfamilies of the Graminae, represents the widest combination of crop species to date yielding stable, fertile partial hybrids (37). However, little is known about the comparative structure and composition of the two genomes. Highly conserved sequences such as rRNA genes or tubulin cDNAs cross-hybridize to corresponding genes in the maize and oat genomes. Unique maize RFLP probes also have been used to detect sequences in the oat genome (9). A series of cDNA probes has shown 71% conservation of linkage associations between maize and oat according to Van Deynze et al. (38); however, almost 50% of maize PstI unique genomic probes do not hybridize to oat genomic DNA under standard conditions (39).
We report herein that the proportion of maize and oat nucleotide sequences that cross-hybridize to each other is low under standard hybridization conditions (65°C, 6× SCC). Among the set of low-copy-number sequences isolated and tested, more than 50% appeared to be maize specific. The proportion of maize-specific sequences among the middle or highly repetitive maize elements was about 80–95% and among those that do hybridize to oat, most hybridize only weakly; therefore, even total genomic DNA of maize can serve as an efficient probe to identify maize-specific cosmids in an oat genomic background. But total maize DNA is less reliable for this use than the described multiprobe because it gives more false positive clones during primary screening of the cosmid library. The oat–maize chromosome addition lines are especially attractive as a source for isolation of region-specific maize DNA fragments. It is possible to identify even small portions of a maize chromosome in an oat genome by probing with total maize genomic DNA, a multiprobe of maize repeated nucleotide sequences, or many other types of cloned maize sequences. In the case of the highly conserved nucleotide sequences, which are common in both maize and oat genomes, RFLPs may be used to differentially identify maize DNA in the oat genome.
Efficiency of Recovering Maize-Specific Cosmid Clones.
The efficiency of identifying maize-specific cosmid clones depends on copy number and the pattern of distribution of repeated sequences chosen to comprise the multiprobe. Studies of genome structure in maize (28, 40), especially analysis of repeated DNA sequences around several maize genes (30, 31), show that unique sequences with a median size of 2.1 kb are surrounded by a long, complex array of repeated sequences that belong to many different families, many of which appear to be of retrotransposon origin (31).
Our multiprobe highlighted EcoRI subfragments in each of the 29 cosmid clones of chromosome 9 analyzed, detecting about 100 of the total of more than 200 fragments. Each clone contained one or more of the repeated sequences present in the multiprobe. The same group of EcoRI fragments were classified as highly repetitive nucleotide sequences because they showed strong hybridization to labeled total maize genomic DNA. In the same set of clones at least 50 additional EcoRI fragments showed a strong hybridization signal to maize genomic DNA and may be classified as additional highly repetitive nucleotide sequences. These additional sequences could be included in our collection of maize-specific repeated nucleotide sequences to increase the efficiency of the multiprobe used in searching for maize-specific cosmids.
The number and diversity of repeats differ among clones. Some of the cosmids consist entirely of several different repeated sequences; others have only one copy of the tested repeats. Regions of maize chromosomal DNA consisting only of low-copy-number sequences with a composite length exceeding 40 kb, the size of the DNA insertion in most cosmids, would not be detected in a cosmid library by screening with the multiprobe.
Because the 29 analyzed clones of maize chromosome 9 were selected with the help of the multiprobe, they may carry more repeated DNA sequences than a random set of clones. Therefore, a control experiment was conducted to analyze the distribution of repetitive sequences in a set of 38 cosmid clones randomly chosen from a cosmid library constructed from DNA of the parental maize line Seneca 60. Only 2 of 38 maize cosmid clones revealed cross-hybridization to oat genomic DNA, and they were found to be composed of rDNA sequences. Thirty-four of the remaining 36 cosmid clones carried maize-specific highly repetitive sequences detected by the multiprobe (data not shown). This experiment indicates that the probability of recovering maize-specific cosmid clones in a genomic library of maize chromosome 9 with the help of the primary multiprobe is around 95%.
Oat as a Host for Cloning Large Maize Chromosomal DNA Fragments.
Impressive achievements have been made in cloning large (>1 Mb) DNA fragments in yeast artificial chromosomes (41). However, the difficulties encountered make this method of cloning a eukaryotic genome expensive and problematic for wide use. Radiation hybrids (derivative cell lines from irradiated somatic cell hybrids) are an attractive alternative for cloning subfragments of a chromosome. Irradiation to produce chromosome breakage and segregation of genetic material has been applied to a number of mammalian somatic cell hybrids containing individual alien chromosomes (10). A disadvantage of this approach in some cases is that a host genome and an alien chromosome have too many in-common nucleotide sequences, thus making difficult the direct analysis, identification, and isolation of the alien chromosome fragments.
The results of the research reported here allow us to propose the use of oat as a host for cloning maize genomic DNA. The substantial differences in the repetitive DNA composition of oat and maize genomes and the high level of genetic stability of the maize-addition chromosomes make it possible to apply conventional molecular methods to the study of maize genetic material directly in the oat genome. This approach would be modeled after that used in mammalian systems (10). Radiation would be used to induce translocations of maize segments into the oat genome, from which maize DNA could subsequently be isolated. In comparison with other possible maize-cloning systems using phages, bacteria, or yeast, the advantage of this approach is that all maize chromosome fragments will derive from a known chromosome. The high proportion of maize sequences that may serve as maize-specific DNA probes in this system allows the identification of almost any maize chromosome segment in oat or any cloned maize DNA fragment in a library. The current availability of an extensive collection of mapped DNA markers for all maize chromosomes (42) may allow us to identify the boundaries of cloned subfragments and to generate a collection of overlapping subfragments representing the whole chromosome. This approach would allow the cloning in oat of subfragments of maize chromosomes of a wide size range. Chromosomal subfragments 5–30 Mb in size may be considered optimal because they can be aligned relative to the genetic map and, if cloned in bacterial artificial chromosomes or even in cosmid vectors, may be arranged in contigs to generate detailed physical maps for each subfragment. Thus, 10–60 subchromosomal fragments may comprise a collection of overlapping large DNA fragments of one particular maize chromosome. Taking into account the high stability of maize genetic material in the oat background, oat lines with maize DNA represent a continuous, renewable source of large segments of maize chromosomal DNA.
Acknowledgments
This is a joint publication of the University of Minnesota and U.S. Department of Agriculture–Agricultural Research Service and is Paper No. 22,475 in the Scientific Journal Series, Minnesota Agricultural Experiment Station. Mention of a trademark or proprietary product does not constitute a guarantee or warranty by the University of Minnesota or the USDA-ARS and does not imply approval over other products that also may be suitable. This research was funded in part by the Plant Molecular Genetics Institute, University of Minnesota, and The Quaker Oats Company.
ABBREVIATION
- RFLP
restriction fragment length polymorphism
- Mb
megabase(s)
References
- 1.Islam A K M R, Shepherd R, Sparrow D H B. Heredity. 1981;46:161–174. [Google Scholar]
- 2.Riley R, Law C N. Stadler Genet Symp. 1984;16:301–322. [Google Scholar]
- 3.Thomas H, Leggett J M, Jones I T. Euphytica. 1975;24:717–724. [Google Scholar]
- 4.Jena K K, Khush G S. Genome. 1989;32:449–455. [Google Scholar]
- 5.Jiang J, Morris K L, Gill B S. Chromosome Res. 1994;2:3–13. doi: 10.1007/BF01539447. [DOI] [PubMed] [Google Scholar]
- 6.Appels R, Moran L B. Stadler Genet Symp. 1984;16:529–558. [Google Scholar]
- 7.Kanazin V, Ananiev E V, Blake T. Genome. 1993;36:1023–1028. doi: 10.1139/g93-136. [DOI] [PubMed] [Google Scholar]
- 8.Rines H W, Riera-Lizarazu O, Phillips R L. In: Modification of Gene Expression and Non-Mendelian Inheritance. Oono K, Takiwa F, editors. Tsukuba, Japan: National Institute of Agrobiological Resources; 1995. pp. 235–251. [Google Scholar]
- 9.Riera-Lizarazu O, Rines H W, Phillips R L. Theor Appl Genet. 1996;93:123–135. doi: 10.1007/BF00225737. [DOI] [PubMed] [Google Scholar]
- 10.Cox D R, Burmeister M, Price E R, Kim S, Myers R M. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- 11.Reichester M, Parsons B. J Cell Physiol. 1976;88:167–179. doi: 10.1002/jcp.1040880206. [DOI] [PubMed] [Google Scholar]
- 12.Olson M V. Proc Natl Acad Sci USA. 1993;90:4338–4344. doi: 10.1073/pnas.90.10.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke B, Stancombe P, Money T, Foote T, Moore G. Nucleic Acids Res. 1992;20:1289–1292. doi: 10.1093/nar/20.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang M L, Leitch A R, Schwarzacher T, Heslop-Harrison J S, Moore G. Nucleic Acids Res. 1992;20:1897–1901. doi: 10.1093/nar/20.8.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung C, Claussen U, Horsthemke B, Fisher F, Herrmann R G. Plant Mol Biol. 1992;20:503–511. doi: 10.1007/BF00040609. [DOI] [PubMed] [Google Scholar]
- 16.Carter N P. In: Cell Biology: A Laboratory Handbook. Celis J E, editor. San Diego: Academic; 1994. pp. 442–449. [Google Scholar]
- 17.Vega M, Abbo S, Feldman M, Levy A A. Proc Natl Acad Sci USA. 1994;91:12041–12045. doi: 10.1073/pnas.91.25.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs J, Houben A, Brandes A, Shubert I. Chromosoma. 1996;104:315–320. doi: 10.1007/BF00337219. [DOI] [PubMed] [Google Scholar]
- 19.Lovett M, Kere J, Hinton L M. Proc Natl Acad Sci USA. 1991;88:9628–9632. doi: 10.1073/pnas.88.21.9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parimo S, Patanjali S R, Shukla H, Chaplin D D, Weisman S M. Proc Natl Acad Sci USA. 1991;88:9623–9627. doi: 10.1073/pnas.88.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monaco A P, Lam V M S, Zehetner G, Lenon G G, Douglas C, Nizetic D, Goodfellow P N, Lehrach H. Nucleic Acids Res. 1991;19:3315–3318. doi: 10.1093/nar/19.12.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graw S L, Buckler A J, Britt D E, Jackson C L, Taruscio D, Baldini A, Ward D C, Housman D E. Somatic Cell Mol Genet. 1992;18:269–284. doi: 10.1007/BF01233863. [DOI] [PubMed] [Google Scholar]
- 23.Pritchard C A, Casher D, Uglum E, Cox D R, Myers R M. Genomics. 1989;4:408–418. doi: 10.1016/0888-7543(89)90348-0. [DOI] [PubMed] [Google Scholar]
- 24.Bedbrook J R, O’Dell M, Flavell R B. Nature (London) 1980;228:133–320. [Google Scholar]
- 25.Appels R, Reddy P, McIntyre C L, Moran L B, Frankel O H, Clarke B C. Genome. 1989;31:122–133. doi: 10.1139/g89-023. [DOI] [PubMed] [Google Scholar]
- 26.Flavell R B. Philos Trans R Soc Lond. 1986;312:227–242. doi: 10.1098/rstb.1986.0004. [DOI] [PubMed] [Google Scholar]
- 27.Ananiev E V. In: Barley Genetics, Biochemistry, Molecular Biology and Biotechnology. Shewry P R, editor. Wallingford, U.K.: CAB International; 1992. pp. 135–150. [Google Scholar]
- 28.Hake S, Walbot V. Chromosoma. 1980;79:251–270. [Google Scholar]
- 29.Rimpau I, Smith D B, Flavell R B. Heredity. 1980;44:131–149. [Google Scholar]
- 30.Bennetzen J L, Schrick K, Springer P S, Brown W E, SanMiguel P. Genome. 1994;37:565–576. doi: 10.1139/g94-081. [DOI] [PubMed] [Google Scholar]
- 31.San Miguel P, Tikhonov A, Jin Y K, Motchulskaia N, Zakharov D, Melake-Berhan A, Springer P, Edwards K, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Whitter R F. Nucleic Acids Res. 1994;22:2168–2169. doi: 10.1093/nar/22.11.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. pp. 1.60–1.62. [Google Scholar]
- 34.Helentjaris T. UMC Maize RFLP Procedures Manual, UMC Maize RFLP Laboratory. Columbia, MD: University of Missouri-Columbia; 1995. pp. 11–12. [Google Scholar]
- 35.Feinberg A P, Vogelstein B. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 36.Peacock W J, Dennis E S, Rhoades M, Pryor A J. Proc Natl Acad Sci USA. 1981;78:4490–4494. doi: 10.1073/pnas.78.7.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rines H W, Dahleen L S. Crop Sci. 1990;30:1073–1078. [Google Scholar]
- 38.Van Deynze A E, Nelson J C, O’Donoughue L S, Ahn S N, Siripoonwiwat W, Harrington S E, Yglesias E S, Braga D P, McCouch S R, Sorrells M E. Mol Gen Genet. 1995;249:349–356. doi: 10.1007/BF00290536. [DOI] [PubMed] [Google Scholar]
- 39.Yu G X, Bush A L, Wise R P. Genome. 1996;39:155–164. doi: 10.1139/g96-021. [DOI] [PubMed] [Google Scholar]
- 40.Gupta M, Sheperd N S, Bertman I, Saedler H. EMBO J. 1984;3:133–139. doi: 10.1002/j.1460-2075.1984.tb01773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haldi M, Perrot V, Saumier M, Desai T, Cohen D, Cherif D, Ward D, Lander E S. Genomics. 1995;24:478–484. doi: 10.1006/geno.1994.1656. [DOI] [PubMed] [Google Scholar]
- 42.Gardiner J M, Coe E H, Melia-Hancock S, Hoisington D A, Chao S. Genetics. 1993;134:917–930. doi: 10.1093/genetics/134.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]