Abstract
Site-directed mutagenesis of selected residues of mammalian protein phosphatase 1 (PP-1) has been carried out to further define the mechanism of catalysis, activation by divalent cations, and inhibition by toxins and inhibitory proteins. Mutation of active site residues predicted to bind metals (N124D and H248N) resulted in a large loss of enzyme activity and decreased affinity for metal ions; mutation of residues predicted to bind phosphosubstrate (R96A or R221S) led to a large loss of enzyme activity; and mutation of active site residues (D95A and D208A) resulted in a large loss of enzyme activity. Mutants N124D, H248N, R96A, and R221S exhibited large decreases in sensitivity to the toxins calyculin A, okadaic acid, and microcystin and to thiophospho-DARPP-32. Mutation of Y272 (Y272F) had little effect on activity but resulted in a large decrease in sensitivity to okadaic acid and calyculin A. Mutant D208A exhibited a decrease in sensitivity to okadaic acid and calyculin A, but, paradoxically, the sensitivity to inhibition by thiophospho-DARPP-32 was increased. Mutation of acidic groove residues (E256R, E275R, E252A:D253A, and E252A:D253A:E256R) exhibited little change in enzyme activity and no change in sensitivity to toxins, but increased sensitivity to thiophospho-DARPP-32. These results suggest that toxins and phospho-DARPP-32 interact at the active site of PP-1 in a similar fashion despite their differences in structure. In addition, acidic groove residues appear to influence the interaction of the phosphoinhibitor with the active site of PP-1.
Based on their biochemical properties, in particular substrate specificity and sensitivity to divalent cations and protein inhibitors, serine/threonine protein phosphatases (PPases) have been classified into four major types (PP-1, -2A, -2B, and -2C). The catalytic subunits of PP-1, -2A, and -2B (referred to here as the PPase family) show a high degree of sequence identity (40–50%) within a region of ≈30 kDa, suggesting that these enzymes share a conserved structure and mechanism of catalysis (1–3). In contrast, the amino acid sequence of PP-2C is unrelated to any of the PPase family members and is likely to have a distinct structure and enzyme mechanism. The catalytic subunits of the PPases are subject to regulation by a variety of interacting subunits, targeting proteins and inhibitors (1, 3–5). For example, PP-1 is regulated by the heat-stable proteins, inhibitor-1, the related homolog DARPP-32 (dopamine and cAMP-regulated phosphoprotein of Mr 32,000), and inhibitor-2. Phosphorylation of inhibitor-1 (at Thr-35) or DARPP-32 (at Thr-34) by cAMP-dependent protein kinase (PKA) converts them into potent inhibitors of PP-1. PP-1 and PP-2A, but not PP-2B, are also highly sensitive to inhibition by a number of tumor-promoting natural toxins, for example, okadaic acid, calyculin A, and microcystin (3). In addition, PP-1 is inhibited by phosphorylation of a threonine residue (Thr-320) in the COOH-terminal domain of the protein by cyclin-dependent protein kinase (6, 7). PP-2B exists as a heterodimer of the catalytic A subunit and a calmodulin-like B subunit and is regulated by binding of Ca2+ to the B-subunit as well as by binding of Ca2+/calmodulin to a regulatory domain that follows the catalytic domain in the A-subunit (1, 3).
A detailed understanding of the mechanism of catalysis and regulation of the PPase family has resulted from the recent elucidation of the x-ray crystal structure of the catalytic subunit of mammalian PP-1 complexed with microcystin (8), PP-1 complexed with a phosphate analog, tungstate (9), and the A-subunit of PP-2B complexed with the B-subunit, with and without FK506/FKBP (10, 11). The overall structural features of the catalytic domains of the two enzymes appear to be very similar. An NH2-terminal subdomain contains two metal atoms embedded at the core of a conserved phosphoesterase motif, DXH(X)nGDXXD(X)nGNHD/E (n = 25), found in the PPases and other phosphoesterases (12). The COOH-terminal subdomain sits on the surface of the NH2-terminal subdomain, forming three surface grooves, the hydrophobic groove, the acidic groove, and the COOH-terminal groove, with the active site situated at the bifurcation point of the three grooves (8).
To evaluate the roles(s) played by specific amino acid residues in catalysis and in the interaction with toxins and protein inhibitors, we have, based on the crystal structure of PP-1, prepared a series of mutants in which residues in the active site and in the acidic groove have been changed. The results indicate that active site residues not only play a critical role in catalysis, but also interact with all toxins analyzed, as well as with the protein inhibitor, DARPP-32. Surface residues that are not part of the active site also appear to influence the binding of protein inhibitors. Together with the information obtained from the crystal structures, these results help to further define the structure and regulation of this important class of enzymes.
MATERIALS AND METHODS
Site-Directed Mutagenesis of PP-1α.
Rabbit skeletal muscle PP-1α cDNA was a generous gift from N. Bernt (13). The PP-1α cDNA was used as a template for site-directed mutagenesis using PCR. PCR products were gel-purified, digested with appropriate restriction enzymes (XmaI and AflII), and used to replace the wild-type fragment of PP-1α.
Purification of Recombinant PP-1.
Escherichia coli (DH5α), harboring the wild-type or mutated cDNA, was grown in the presence of 1 mM MnCl2 as described (14). Cells were resuspended in lysis buffer containing 20 mM Tris·HCl (pH 7.0), 1 mM MnCl2, 20 μg/ml pepstatin A, 20 μg/ml leupeptin, 4 mM benzamidine, 0.2 mM phenylmethylsulfonyl fluoride, and 0.1% 2-mercaptoethanol and lysed using a French press. Insoluble material was removed by centrifugation (20,000 × g, 20 min), and PP-1 in the supernatant was purified by chromatography using heparin-Sepharose, phenyl-Sepharose, Sephacryl S-200, and Mono-Q (Pharmacia Biotech; H.-B.H., unpublished work). MnCl2 (1 mM) was included in all purification buffers and in the final enzyme storage buffer.
PP-1 Assays.
PP-1 was assayed using [32P]phosphorylase a as substrate essentially as described (15). Unless indicated, assays contained 1 mM Mn2+. When added, toxins and protein inhibitors were diluted in Tris·HCl (pH 7.0) and preincubated with PP-1 at 30°C for 10 min. For kinetic analyses, 5–30 μM substrate (final concentration) was used. Km and Vmax values were determined from Lineweaver–Burk plots. The pH optima of wild-type PP-1 and mutant D95A were measured using Mes·NaOH (pH 5–7), Tris·HCl (pH 7–9), and glycine-NaOH (pH 9.5–10). All reactions were performed in duplicate. With the exception of the D95A mutant, which was prepared once, all mutants were prepared at least twice and similar results were obtained from the different preparations. Okadaic acid, microcystin-LR, and calyculin A were obtained from LC Services (Woburn, MA). Native PP-1 was purified from rabbit skeletal muscle essentially as described (15). CF-2 was prepared and phosphorylated with PKA as described (16).
Preparation of Phosphorylated and Thiophosphorylated DARPP-32.
Phosphorylated recombinant DARPP-32 was prepared essentially as described (17). DARPP-32 (2 mg) was dissolved in 1 ml of 50 mM Hepes (pH 7.4), 1 mM EGTA, 10 mM magnesium acetate, and 1 mM ATP. The phosphorylation reaction was started by addition of 2 μg of PKA catalytic subunit (18) and the incubation was carried out for 60 min. For preparation of [32P]DARPP-32, [γ-32P]ATP replaced ATP. For preparation of thiophosphorylated DARPP-32, 1 mM γ-thiophospho-ATP (Boehringer Mannheim) replaced ATP, and the reaction was carried out at 30°C for 5 days, with fresh PKA (2 μg) and γ-thiophospho-ATP (1 mM final concentration) added every 24 h. Phosphorylated DARPP-32 and thiophospho-DARPP-32 were purified by HPLC using a C-18 column as described (17).
RESULTS
Expression of PP-1 Mutants.
For amino acids that are conserved in the PPases and that, based on the crystal structure of PP-1, were predicted to be in or close to the active site, substitutions were chosen in an attempt not to disrupt the overall structure of the enzyme. Substitution of acidic groove residues was made either to the equivalent residues found in either PP-2A or PP-2B, or to alanine. The yield of the R96A and E252A:D253A mutants was similar to that of the wild type (0.6–1.5 mg/liter); N124D, H248N, and E256R were ≈80% that of wild type; E256R, E252A:D253A:E256R, E275R, Y272F, D220V, and R221S were ≈60% that of wild type; D208A was ≈40% that of wild type; and D95A was ≈5% that of wild type. All PP-1 mutants were assayed immediately following purification, because replacement of some active site residues resulted in instability of enzyme activity over a period of 7 days at 4°C.
Kinetic Analysis of the Dephosphorylation of [32P]Phosphorylase a by PP-1 Mutants.
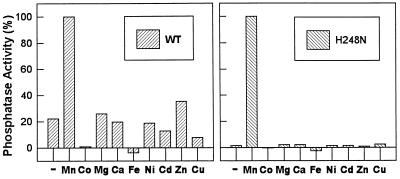
With the exception of the mutants C127S and Y272F, all mutations at the active site, D95A, R96A, N124D, D208A, R221S, and H248N, resulted in a significant decrease in specific activity (Table 1). Mutation of residues predicted to be metal ligands (N124D and H248N) resulted in large decreases in catalytic efficiency (≈20- to 30-fold), primarily as a result of reductions in kcat. Following removal of Mn2+ from the enzyme storage solution by ion-exchange chromatography on a Mono-Q FPLC column, the metal dependence of wild-type PP-1 and the N124D and H248N mutants was examined. Wild-type PP-1 retained ≈20% of the original activity, suggesting that the enzyme retained some bound metal. Addition of 1 mM Mn2+, but not of several other metals tested, fully restored enzyme activity (Fig. 1), with an apparent Km for Mn2+ of 2.9 μM (data not shown). After ion-exchange chromatography, the N124D (≈10%) and H248N (≈2%) mutants retained relatively less basal activity than the wild-type enzyme, when normalized to activity measured in the presence of Mn2+, suggesting that metals were easier to remove from the mutants (Fig. 1 and data not shown). The apparent Km for Mn2+ was increased >10-fold for either mutant (40 μM for N124D and 37 μM for H248N). As was observed for the wild-type enzyme, of the metals tested, only Mn2+ activated the mutants. In contrast to a recent study (19), combinations of metals also failed to activate any of the PP-1 preparations (data not shown).
Table 1.
Kinetic analysis of PP-1 mutants
| PP-1 | Specific activity, units/mg | Km, μM | kcat, s−1 | kcat/Km, s−1·M−1 × 10−6 | Vmax, μmol·mg−1·min−1 |
|---|---|---|---|---|---|
| Wild-type | 34 | 10.6 | 39 | 3.63 | 63 |
| N124D | 2.0 | 19.8 | 3.1 | 0.16 | 5.0 |
| H248N | 0.7 | 5.1 | 0.5 | 0.10 | 0.8 |
| D95A | 0.03 | 5.8 | 0.02 | 0.003 | 0.03 |
| R96A | 0.08 | 6.9 | 0.09 | 0.013 | 0.2 |
| R221S | 0.05 | 105.0 | 0.2 | 0.002 | 0.6 |
| D208A | 8.0 | 23.3 | 13.3 | 0.57 | 22 |
| C127S | 49 | 10.4 | 58 | 5.56 | 94 |
| Y272F | 26 | 5.0 | 14 | 2.82 | 23 |
| D220V | 30 | 8.1 | 30 | 3.70 | 49.3 |
| E256R | 36 | ND | ND | ND | ND |
| E275R | 69 | 13.2 | 86 | 6.50 | 140 |
| E252A:D253A | 37 | ND | ND | ND | ND |
| E252A:D253A:E256R | 12 | ND | ND | ND | ND |
ND, not determined.
Figure 1.
Metal ion dependence of wild-type PP-1 and the H248N mutant. Metals were removed by Mono-Q chromatography before assay. Each enzyme was diluted in 50 mM Tris·HCl buffer (pH 7.0), containing 10% glycerol, 0.1% 2-mercaptoethanol, and 2 mM EDTA. The enzyme was eluted with a gradient of 0.1–0.5 M NaCl in the equilibration buffer, and the fractions containing the most Mn2+-dependent enzyme activity were pooled. Enzyme activity was measured in the absence (−) or presence (+) of 1 mM metal chloride as indicated. (Left) Wild type; (Right) H248N. Enzyme activities were normalized (see Table 1 for values).
The crystal structures of PP-1 suggest that catalysis involves nucleophilic attack by a metal-activated water molecule. Furthermore, H125 is likely to act as a general acid protonating the leaving group serine (or threonine). D95 is buried at the active site with a carboxyl oxygen forming a hydrogen bond with the Nδ atom of H125; the location of D95 suggests a catalytic role in confining the position of H125 and stabilizing its protonated state. Consistent with this, the D95A mutant exhibited a large decrease in kcat (>2000-fold) with no significant change in Km.
The structures of PP-1 and PP-2B suggest that three residues, R96, N124 (one of the metal ligands), and R221 bind to the substrate oxygens and may contribute to substrate binding as well to stabilization of a pentacoordinate transition state. D208 is also likely to help maintain the conformation of R221 via a salt bridge. The R96A mutant exhibited a large decrease in kcat (>400-fold) but no significant change in Km for substrate. The R221S mutant exhibited a large reduction in kcat (≈200-fold), but in contrast to the R96A mutant, showed a significant reduction in affinity for substrate (≈10-fold increase in Km). The D208A mutant exhibited a smaller decrease in both kcat (≈3-fold) and affinity for substrate (≈2-fold increase in Km), consistent with its role in stabilizing R221.
Inhibition of PP-1 Mutants by Toxins.
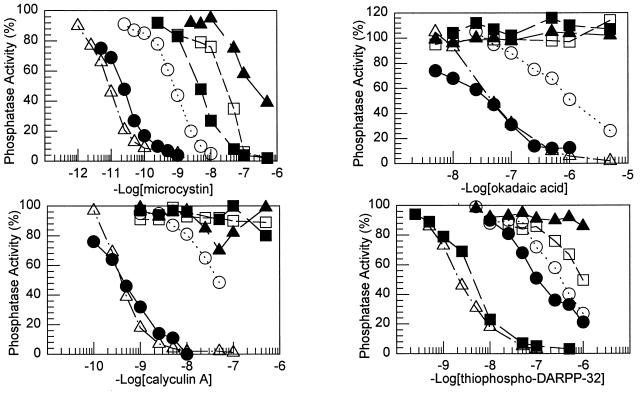
Dose–response curves for the inhibition of the PP-1 mutants by microcystin, okadaic acid, and calyculin A were obtained (Fig. 2), and the IC50 values were calculated (Table 2). None of the acidic groove mutants exhibited changes in sensitivity to any of the toxins. In contrast, mutation of all the active site residues analyzed greatly decreased the sensitivity of each mutant to microcystin, okadaic acid, and calyculin A. The changes in sensitivity of the PP-1 active site mutants to the cyclic heptapeptide, microcystin-LR, were completely consistent with that predicted by the PP-1/microcystin structure (8). For example, R96 hydrogen bonds with the carboxylate group of the β-linked d-erythro-β-methyl aspartic acid (Masp) side chain of microcystin. Mutation of R96 (R96A) increased the IC50 for microcystin ≈2000-fold. Mutation of Y272 (Y272F) increased the IC50 35-fold, consistent with a contribution of the phenolic hydroxyl group of Y272 to the interaction with microcystin. Notably, the Y272F mutant also exhibited decreased sensitivity to both okadaic acid (≈26-fold) and calyculin A (107-fold). No inhibition was observed for mutants N124D, D208A, R221S, or H248N by concentrations of okadaic acid up to 5000 nM or of calyculin A up to 500 nM. The R96A mutant exhibited a decrease in sensitivity to both okadaic acid (≈6-fold) and calyculin A (≈130-fold). However, this decrease was not as great as that of the R221S and D208A mutants.
Figure 2.
Inhibition of wild-type PP-1 and PP-1 mutants by microcystin, okadaic acid, calyculin A, and thiophospho-DARPP-32. Phosphatase activity is expressed as a percentage of activity for each enzyme in the absence of any toxin. The points shown are averages of closely agreeing duplicate measurements. •, Wild type; ▴, N124D; ▪, D208A; □, R221S; ○, Y272F; and ▵, E275R. Results for some mutants were omitted for clarity (see Table 2).
Table 2.
Inhibition of PP-1 mutants by toxins and DARPP-32
| PP-1 | IC50, nM
|
|||
|---|---|---|---|---|
| Microcystin-LR | Okadaic acid | Calyculin A | S-DARPP-32 | |
| Wild-type | 0.02 | 45 | 0.45 | 115 |
| N124D | 200 | >5000 | >500 | >1000 |
| H248N | 35 | >5000 | >500 | >1000 |
| D95A | ND | ND | ND | ND |
| R96A | 35 | 260 | 58 | 640 |
| R221S | 29 | >5000 | >500 | 960 |
| D208A | 4.9 | >5000 | >500 | 4.2 |
| C127S | 0.56 | 78 | 2.2 | 250 |
| Y272F | 0.80 | 1150 | 48 | 310 |
| D220V | 0.02 | 28 | 0.14 | 210 |
| E256R | ND | ND | ND | 11 |
| E275R | 0.01 | 48 | 0.4 | 2.2 |
| E252A;D253A | ND | ND | ND | 8.6 |
| E252A:D253A:E256R | ND | ND | ND | 7.0 |
ND, not determined.
Inhibition of Wild-Type PP-1 and PP-1 Mutants by Thiophosphorylated DARPP-32.
Recombinant PP-1 expressed in E. coli differs from the native enzyme with respect to a number of important criteria, including the fact that it is dependent on added Mn2+ for activity and is able to dephosphorylate phosphotyrosine-containing substrates and the artificial substrate, para-nitrophenyl phosphate (9, 14). Recombinant E. coli PP-1 also exhibits a lower sensitivity to phospho-DARPP-32 than the native enzyme (IC50, ≈450 nM for E. coli PP-1; IC50, 1.7 nM for native PP-1). Similar results have been reported for phospho-inhibitor-1 (20). Native PP-1 does not dephosphorylate phospho-DARPP-32 at any significant rate. However, E. coli PP-1 was found to efficiently dephosphorylate phospho-DARPP-32 (Fig. 3) and phospho-inhibitor-1 (H.-B.H. and Y.-S. Kwon, unpublished work), which is likely to explain the lower sensitivity of E. coli PP-1 to these two inhibitor proteins. Thiophosphorylation renders DARPP-32 fully resistant to dephosphorylation by E. coli PP-1 (H.-B.H., unpublished work). Nevertheless, E. coli PP-1 still exhibits a lower sensitivity to thiophospho-DARPP-32 than the native enzyme (IC50, 1.5 nM for native PP-1; IC50, 115 nM for E. coli PP-1; Table 2).
Figure 3.
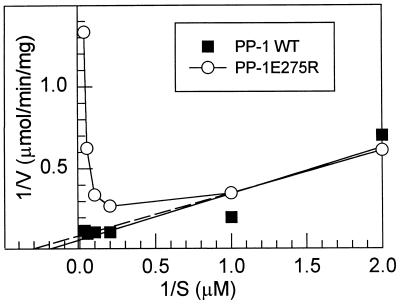
Kinetic analysis of the dephosphorylation of [32P]DARPP-32 by PP-1. Dephosphorylation of [32P]DARPP-32 was measured at various concentrations of substrate. The data obtained were plotted using the Lineweaver–Burk method. ▪, Wild-type recombinant PP-1 (Km, 5.0 μM; Vmax, 12.8 μmol·min−1·mg−1); and ○, the values for Km and Vmax for the E275R mutant were determined by extrapolation (dashed line) of the data obtained at 0.5 and 1.0 μM substrate (Km, 2.9 μM; Vmax, 11.1 μmol·min−1·mg−1). The C127S mutant also efficiently dephosphorylated [32P]DARPP-32 (Km, 4.2 μM; Vmax, 30.8 μmol·min−1·mg−1), exhibiting linear kinetics (data not shown).
Dose–response curves for the inhibition of the PP-1 mutants by thiophospho-DARPP-32 were obtained (Fig. 2), and the IC50 values were calculated (Table 2). Mutation of active site residues N124 and H248 caused a large increase in the IC50; the R96A and R221S mutants (≈6- to 8-fold) and the C127S and Y272F mutants (≈2- to 3-fold) were also less sensitive to thiophospho-DARPP-32. Of the acidic groove residues, D220, E256, and E275 are found in PP-1 but not PP-2A or PP-2B, whereas D210, E252, and D253 (or equivalent acidic residues) are conserved in PP-1 and one of the other two PPases. D208, an active site residue conserved in all the PPases, is situated close to the acidic groove. With the exception of the D220V mutant, all of the acidic groove mutants exhibited a paradoxical increase in sensitivity to thiophospho-DARPP-32. These reductions in IC50 were such that the mutant enzymes resembled native PP-1 in their sensitivity to thiophospho-DARPP-32. The D208A and E275R mutants were further analyzed for their ability to be inhibited by phospho-DARPP-32 and to dephosphorylate [32P]DARPP-32 (Fig. 3 and data not shown). The D208A mutant most clearly resembled native PP-1, exhibiting increased sensitivity to phospho-DARPP-32 (IC50, 17 nM) and thiophospho-DARPP-32 (IC50, 4.2 nM), which was paralleled by an inability to dephosphorylate [32P]DARPP-32. The E275R mutant showed intermediate properties, exhibiting a somewhat increased sensitivity to phospho-DARPP-32 (IC50, 150 nM) and increased sensitivity to thiophospho-DARPP-32 (IC50, 2.2 nM). This enzyme also displayed nonlinear kinetics for dephosphorylation of [32P]DARPP-32 that were characteristic of substrate inhibition (Fig. 3).
Substrate Specificity of PP-1 Mutants.
The catalytic subunits of PP-1, PP-2A, and PP-2B display distinct substrate specificities, although the structural basis for this has not been elucidated. Possibly, acidic groove residues that are specific for PP-1 might influence substrate specificity. We therefore analyzed the dephosphorylation of CF-2, an intracellular domain of the chloride channel, CFTR, which is phosphorylated at multiple sites by PKA and which is dephosphorylated efficiently by PP-2A but not at all by native PP-1 (A.C.N. and D. Gadsby, unpublished results). None of the PP-1 mutants, were able to dephosphorylate CF-2 (data not shown). Finally, of the PP-1 mutants prepared, only the R221A mutant exhibited a change in affinity for [32P]phosphorylase (Table 1).
DISCUSSION
The crystal structures of PP-1 (8, 9) and PP-2B (10, 11) resolved many but not all questions concerning the function of active site residues. In the present study, we have used site-directed mutagenesis to further evaluate structure–function relationships of selected amino acids in PP-1. The results obtained highlight the roles of active site residues, not only in metal binding and catalysis, but in binding to both toxins and the phosphorylated inhibitor protein, phospho-DARPP-32. The results also suggest that acidic groove residues influence in some way the interaction of phospho-DARPP-32 with the active site of PP-1.
Analysis of bacteriophage λ PPase resulted in the first mechanistic assessment of metal-binding residues, and the definition of the active site metallo-phosphoesterase motif (12). This phosphoesterase motif is also found in purple acid phosphatase (PAP), which contains almost identical metal-binding ligands and active site geometry despite having no overall amino acid sequence conservation (21). The results obtained are generally consistent with those obtained from mutagenesis studies of bacteriophage λ as well as from detailed mechanistic studies of PAP (22). The results obtained are also in general agreement with site-directed mutagenesis studies reported for PP-1 by Lee and coworkers (23). H125 and D95 are part of an active site aspartate–histidine pair, with H125 predicted to act as a general acid that protonates the leaving dephosphorylated peptide. Mutation of D95 resulted in a large decrease in kcat with no change in Km for the substrate, consistent with a critical functional and structural role for this residue. Mutation of any one of the metal-binding ligands analyzed in the various studies (D64, H66, D92, N124, and H248) results in decreased affinity for metal ion that most likely explains the large decrease in kcat values observed. The PP-1 structures indicated that the amino group of N124 binds to one of the phosphorous oxygens. The retention of the carbonyl function in the N124D mutant probably contributes to its relatively higher activity compared with the other metal-binding mutants. However, we did observe a similar reduction in metal affinity for both the N124D and H248N mutants. The results obtained do not provide any insight into the identity of the metal(s) in the active site of PP-1. The crystal structures suggest that in recombinant PP-1, Fe2+ and Mn2+ are likely to be metals 1 and 2, respectively (8, 9). PP-2B is believed to contain Fe2+ and Zn2+ and recent studies of PP-1 have suggested that these two metals may also be present in native PP-1 (19). However, in the present study, addition of Fe2+ or Zn2+ alone, or in combination with each other, did not activate any of the PP-1 preparations.
R96 and R221 are predicted to contribute to substrate binding as well as to be involved in stabilization of a pentacoordinate transition state. There was a large reduction in kcat for both the R96A and R221S mutants, but only the R221S mutant exhibited a significant reduction in Km for peptide substrate. This is suggestive of a more important role for R96 in stabilization of the transition state than in the initial binding of substrate, although changes in polypeptide Km need not necessarily be expected, since the amino acid residues adjacent to the phospho-serine or -threonine residue presumably interact with the PP-1 at multiple sites. In this respect, it is is notable that mutation of R53 of bacteriophage λ (equivalent to R96 of PP-1) decreased significantly the affinity for the nonpeptide substrate, para-nitrophenyl-phosphate (12). Notably, in PAP, two histidine residues replace R96 and R221, and it has been suggested that these substitutions might explain the lower pH optimum of PAP (22). However, we did not examine the pH optima of the R96A and R221S mutants. The present study, as well as that of Lee and coworkers (24), indicate that mutation of Y272 has no effect on enzyme activity. This was somewhat surprising, considering that this active site tyrosine is conserved in all the PPases. It has been suggested that Y272 in the β12/β13 loop is in a different position in native PP-1 than that found in the structure of the bacterial preparation (9). However, the structure of the active site of native PP-2B, including an equivalent tyrosine, appears to be essentially the same as in PP-1 (10, 11).
Mutation of all active site residues analyzed, but not any of the acidic groove residues, resulted in reduced sensitivity to microcystin, okadaic acid, and calyculin A. The results obtained for microcystin were consistent with the structure of PP-1/microcystin (8). In general, the fold reductions in sensitivity to okadaic acid and calyculin A were greater than that for microcystin. Previous studies have highlighted an important role of the β12/β13 loop (residues 272–276 in PP-1) in the inhibition of PP-1 and PP-2A by toxins (25, 26). In particular, recent mutagenesis analysis of PP-1 has shown that Y272 is involved in the binding to the toxins analyzed in the present study, and also to nodularin and tautomycin (24). Together the results strongly suggest that all of these toxins interact with a number of active site residues, including residues in the β12/β13 loop. Thus, despite their dissimilar structures, the toxins are all likely to present a conserved inhibitory surface that makes multiple contacts with the active site of PP-1. This conclusion is consistent with competition binding studies (27). In addition, modeling studies of PP-1, PP-2A, and a number of different toxins support this idea (R. Chamberlin, personal communication). The relative specificity of the various toxins for PP-1 and PP-2A appears to be explained in part by specific interactions with variable amino acid side chains in the β12/β13 loop (24, 25). However, it is less clear why PP-2B is relatively resistant to inhibition, since all the residues analyzed in this study are conserved in PP-2B as well as other PPases.
The results of the present study provide insight into the interaction of PP-1 with phospho-DARPP-32 (and by analogy phospho-inhibitor-1). Our previous studies have suggested that two domains of phospho-DARPP-32 are important for its high potency as an inhibitor (17, 28). One domain includes the phospho-threonine residue (phospho-serine was not effective), which is likely to interact with or close to the active site of the enzyme. The second domain consists of a short stretch of amino acids NH2-terminal to the phosphorylation site that is likely to interact with a region of PP-1 away from the active site. An important question that remains to be answered relates to why phospho-DARPP-32 is not dephosphorylated by native PP-1 yet is efficiently dephosphorylated by either recombinant E. coli PP-1 or by native PP-2A or native PP-2B. In the present study, mutation of several active site residues (N124D, H248N, R96A, and R221S) resulted in large decreases in the sensitivity to thiophospho-DARPP-32, consistent with the idea that the phosphate moiety of the inhibitor interacts at the active site of PP-1. Elucidation of the structure of PP-1 indicated that several acidic amino acids were present in a surface grove close to the active site of PP-1, but these amino acids were not conserved in PP-2A and PP-2B, suggesting that they might be important in binding to four arginine residues preceding the phospho threonine of DARPP-32 (8). Based on these initial modeling studies (8), we might have expected that alteration of these residues of PP-1 would have decreased the sensitivity of the mutant enzymes to thiophospho-DARPP-32. However, the mutant enzymes (D208A, E256R, E275R, E252A:D253A, and E252A:D253A:E256R) paradoxically exhibited increased sensitivity to thiophospho-DARPP-32, while exhibiting no significant alteration of enzyme activity.
As discussed above, bacterially expressed PP-1 differs from the native enzyme in several ways, including the fact that it is dependent on added Mn2+ for activity and can efficiently dephosphorylate phospho- but not thiophospho-DARPP-32. Notably, mutation of the acidic groove residues either to uncharged amino acids (alanine) or to basic amino acids (arginine) generated enzymes that more closely resembled native PP-1 in their inability to dephosphorylate phospho-DARPP-32 and in their sensitivity to the phosphoprotein as an inhibitor. In the absence of detailed structural information, it is not possible to fully evaluate these results. Arginine residues preceding the phospho threonine of DARPP-32 may ion pair with the acidic groove aspartate and glutamate residues present only in PP-1. For some reason, one or more of these interactions may be perturbed in E. coli PP-1, resulting in the ability of the recombinant enzyme to dephosphorylate the phospho-inhibitor. Consequently mutation of the aspartate or glutamate residues causes a reorientation of phospho-DARPP-32 within the acidic groove. Alternatively, mutations in the acidic groove indirectly affect the structure of the active site, leading to changes in its interaction with phospho-DARPP-32. In either case, the results indicate that apparently subtle changes in PP-1 structure can result in the orientation of the phosphate moiety of phospho-DARPP-32 within the active site such that it can serve either as a substrate or as an inhibitor. Modeling studies have indicated that the phospho-threonine of DARPP-32 can be accommodated in the active site in several different conformations (J.G., unpublished work). In one conformation the “leaving group” threonine is positioned such that it would be less efficiently protonated by H125. Alternatively, the phosphate group could displace the catalytic water, in a manner analogous to that proposed for inhibition of PAP by phosphate (22). If this is the case, several features of the binding of phospho-DARPP-32 to PP-1, including potential interactions with the acidic groove, as well as with a binding domain distant from the active site, are all likely to contribute to the orientation of the phosphate in the active site, and therefore to its high potency and specificity as an inhibitor of PP-1.
The molecular basis for the interaction of the PPases with substrate has not yet been established. Interaction with elements of the peptide in addition to phospho-serine or -threonine is clearly critical, since Km values for protein substrates are much lower than that of artificial substrates like para-nitrophenyl phosphate (29). Unlike the protein kinases, where short peptides encompassing the phosphorylation site contain the structural elements necessary for efficient enzyme binding, similar studies with the PPases have been much less informative (30). Notably, the catalytic subunit of PP-1 does not dephosphorylate short peptides, while PP-2A does so with high efficiency, and PP-2B is intermediate. With the exception of the R221S mutant, the affinity for phosphorylase was not affected by any of the mutations studied. Furthermore, the general substrate specificity of the acidic groove mutants was unchanged. These results raise the possibility that basic amino acids in phosphorylation sites of substrates such as phosphorylase may not bind to the acidic groove of PP-1. In this respect, mutation of D71, located in the COOH-terminal groove, has been found to reduce the affinity for phosphorylase (23). Furthermore, the autoinhibitory region of PP-2B, which is believed to be a competitive inhibitor, is located close to the hydrophobic groove, the active site, and the β12/β13 loop, although there are not extensive contacts outside of the active site (11). Potentially, different substrates may therefore interact in a variety of conformations with PP-1 or the other PPases.
In conclusion, these studies provide added insight into the mechanism of catalysis of the PPases and the interaction of toxins and protein inhibitors with PP-1. The results also suggest potential strategies for the design of partially inactive enzymes that could be used as dominant negative molecules. Alternatively, mutants like D208A or Y272F that retain substantial catalytic activity could be used in the presence of concentrations of toxins that would inhibit wild-type PP-1 or PP-2A.
Acknowledgments
This work was supported by U.S. Public Health Service Grant MH 40899 (to A.C.N. and P.G.) and National Science Council (Taiwan) Grant NSC 86-2314-B-320-002 (to H.-B.H.).
ABBREVIATIONS
- PPase
protein phosphatase
- PP-1
-2A, -2B, and -2C, PPase 1, 2A, 2B, and 2C, respectively
- PKA
cAMP-dependent protein kinase
- PAP
purple acid phosphatase
References
- 1.Shenolikar S, Nairn A C. Adv Second Messenger Phosphoprotein Res. 1991;23:1–121. [PubMed] [Google Scholar]
- 2.Cohen P. Annu Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 3.Shenolikar S. Annu Rev Cell Biol. 1994;10:55–86. doi: 10.1146/annurev.cb.10.110194.000415. [DOI] [PubMed] [Google Scholar]
- 4.Hubbard M J, Cohen P. Trends Biochem Sci. 1993;18:172–177. doi: 10.1016/0968-0004(93)90109-z. [DOI] [PubMed] [Google Scholar]
- 5.Faux M C, Scott J D. Trends Biochem Sci. 1996;21:312–315. [PubMed] [Google Scholar]
- 6.Dohadwala M, Da Cruz e Silva E F, Hall F L, Williams R T, Carbonaro-Hall D A, Nairn A C, Greengard P, Berndt N. Proc Natl Acad Sci USA. 1994;91:6408–6412. doi: 10.1073/pnas.91.14.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamano H, Ishii K, Yanagida M. EMBO J. 1994;13:5310–5318. doi: 10.1002/j.1460-2075.1994.tb06865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg J, Huang H-B, Kwon Y-G, Greengard P, Nairn A C, Kuriyan J. Nature (London) 1995;376:745–753. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- 9.Egloff M P, Cohen P T W, Reinemer P, Barford D. J Mol Biol. 1995;254:942–959. doi: 10.1006/jmbi.1995.0667. [DOI] [PubMed] [Google Scholar]
- 10.Griffith J P, Kim J L, Kim E E, Sintchak M D, Thomson J A, Fitzgibbon M J, Fleming M A, Caron P R, Hsiao K, Navia M A. Cell. 1995;82:507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 11.Kissinger C R, Parge H E, Knighton D R, Lewis C T, Pelletier L A, Tempczyk A, Kalish V J, Tucker K D, Showalter R E, Moomaw E W, Gastinel L N, Habuka N, Chen X H, Maldonado F, Barker J E, Bacquet R, Villafranca J E. Nature (London) 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 12.Zhuo S, Clemens J C, Stone R L, Dixon J E. J Biol Chem. 1994;269:26234–26238. [PubMed] [Google Scholar]
- 13.Berndt N, Campbell D G, Caudwell F B, Cohen P, da Cruz e Silva E F, da Cruz e Silva O B, Cohen P T. FEBS Lett. 1987;223:340–346. doi: 10.1016/0014-5793(87)80316-2. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Bai G, Deans-Zirattu S, Browner M F, Lee E Y C. J Biol Chem. 1992;267:1484–1490. [PubMed] [Google Scholar]
- 15.Cohen P, Alemany S, Hemmings B A, Resink T J, Stralfors P, Tung H Y. Methods Enzymol. 1988;159:390–408. doi: 10.1016/0076-6879(88)59039-0. [DOI] [PubMed] [Google Scholar]
- 16.Picciotto M R, Cohn J A, Bertuzzi G, Greengard P, Nairn A C. J Biol Chem. 1992;267:12742–12752. [PubMed] [Google Scholar]
- 17.Desdouits F, Cheetham J J, Huang H-B, Kwon Y-G, da Cruz e Silva E F, Denefle P, Ehrlich M E, Nairn A C, Greengard P, Girault J-A. Biochem Biophys Res Commun. 1995;206:652–658. doi: 10.1006/bbrc.1995.1092. [DOI] [PubMed] [Google Scholar]
- 18.Kaczmarek L K, Jennings K R, Strumwasser F, Nairn A C, Walter U, Wilson F D, Greengard P. Proc Natl Acad Sci USA. 1980;77:7487–7491. doi: 10.1073/pnas.77.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu Y F, Lee E Y C, Schlender K K. J Biol Chem. 1996;271:2574–2577. doi: 10.1074/jbc.271.5.2574. [DOI] [PubMed] [Google Scholar]
- 20.Alessi D R, Street A J, Cohen P, Cohen P T W. Eur J Biochem. 1993;213:1055–1066. doi: 10.1111/j.1432-1033.1993.tb17853.x. [DOI] [PubMed] [Google Scholar]
- 21.Sträter N, Klabunde T, Tucker P, Witzel H, Krebs B. Science. 1995;268:1489–1492. doi: 10.1126/science.7770774. [DOI] [PubMed] [Google Scholar]
- 22.Klabunde, T., Sträter, N., Frohlich, R., Witzel, H. & Krebs, B. (1996) J. Mol. Biol. 737–748. [DOI] [PubMed]
- 23.Zhang J, Zhang Z J, Brew K, Lee E Y C. Biochemistry. 1996;35:6276–6282. doi: 10.1021/bi952954l. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L F, Zhang Z J, Long F X, Lee E Y C. Biochemistry. 1996;35:1606–1611. doi: 10.1021/bi9521396. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Zhao S, Long F, Zhang L, Bai G, Shima H, Nagao M, Lee E Y C. J Biol Chem. 1994;269:16997–17000. [PubMed] [Google Scholar]
- 26.Shima H, Tohda H, Aonuma S, Nakayasu M, DePaoli-Roach A A, Sugimura T, Nagao M. Proc Natl Acad Sci USA. 1994;91:9267–9271. doi: 10.1073/pnas.91.20.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takai A, Sasaki K, Nagai H, Mieskes G, Isobe M, Isono K, Yasumoto T. Biochem J. 1995;306:657–665. doi: 10.1042/bj3060657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hemmings H C, Jr, Nairn A C, Elliott J I, Greengard P. J Biol Chem. 1990;265:20369–20376. [PubMed] [Google Scholar]
- 29.Li H C. Eur J Biochem. 1979;102:363–374. doi: 10.1111/j.1432-1033.1979.tb04251.x. [DOI] [PubMed] [Google Scholar]
- 30.Pinna L A, Donella-Deana A. Biochim Biophys Acta. 1994;1222:415–431. doi: 10.1016/0167-4889(94)90050-7. [DOI] [PubMed] [Google Scholar]