Abstract
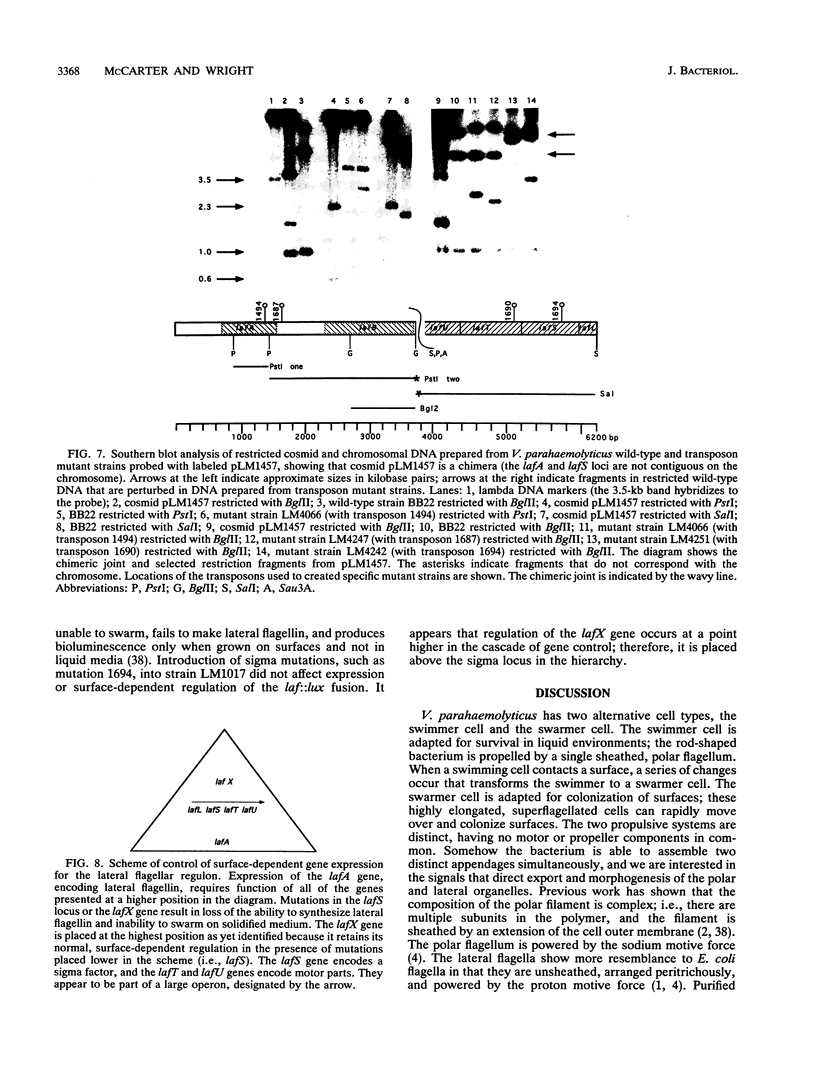
Vibrio parahaemolyticus possesses two distinct motility systems, the polar system used for swimming in liquid environments and the lateral system used for swarming over surfaces. Growth on surfaces induces swarmer cell differentiation and expression of the lateral motility system. Mutants, created by transposon mutagenesis of a clone expressing lateral flagellin and gene disruption in V. parahaemolyticus, were unable to swarm and failed to make lateral flagellin; therefore, unlike the case for the polar system, there is one gene (lafA) encoding lateral flagellin. In addition to lafA, other genes required for swarming but not for swimming were identified by gene replacement mutagenesis. The nucleotide sequence of the clone determined open reading frames (ORFs) and deduced amino acid sequences showed similarities to flagellar components of other bacteria: flagellin, hook-associated protein (HAP2), motor components, and flagellar sigma factor (sigma 28). Many sigma 28 factors have been shown to recognize cognate promoters; however, expression of lafA in Escherichia coli required LafS, and E. coli sigma 28 did not substitute. Also, there were no sequences preceding genes encoding flagellin or HAP2 resembling the sigma 28 consensus promoter. The product of the sigma-like gene seems to be a unique member of the sigma 28 cluster. It appears the result of requiring expression for immunodetection of flagellin clones was that the sigma locus was fortuitously cloned, since the sigma and lafA loci were not contiguous in the chromosome. This work initiates identification and placement of genes in a scheme of control for swarmer cell differentiation; three levels have been identified in the transcriptional hierarchy.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Caramori T., Crabb W. D., Scoffone F., Galizzi A. The flaA locus of Bacillus subtilis is part of a large operon coding for flagellar structures, motility functions, and an ATPase-like polypeptide. J Bacteriol. 1991 Jun;173(11):3573–3579. doi: 10.1128/jb.173.11.3573-3579.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R. D., Baumann P. Structure and arrangement of flagella in species of the genus Beneckea and Photobacterium fischeri. J Bacteriol. 1971 Jul;107(1):295–302. doi: 10.1128/jb.107.1.295-302.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Atsumi T., McCarter L., Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature. 1992 Jan 9;355(6356):182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- Belas R., Simon M., Silverman M. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J Bacteriol. 1986 Jul;167(1):210–218. doi: 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair D. F., Berg H. C. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990 Feb 9;60(3):439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- Broach J., Neumann C., Kustu S. Mutant strains (nit) of Salmonella typhimurium with a pleiotropic defect in nitrogen metabolism. J Bacteriol. 1976 Oct;128(1):86–98. doi: 10.1128/jb.128.1.86-98.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J., Plaskitt K. A., Buttner M. J., Méndez C., Helmann J. D. The developmental fate of S. coelicolor hyphae depends upon a gene product homologous with the motility sigma factor of B. subtilis. Cell. 1989 Oct 6;59(1):133–143. doi: 10.1016/0092-8674(89)90876-3. [DOI] [PubMed] [Google Scholar]
- Chun S. Y., Parkinson J. S. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988 Jan 15;239(4837):276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- Dean G. E., Macnab R. M., Stader J., Matsumura P., Burks C. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984 Sep;159(3):991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984 Oct 15;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Gutierrez C., Barondess J., Manoil C., Beckwith J. The use of transposon TnphoA to detect genes for cell envelope proteins subject to a common regulatory stimulus. Analysis of osmotically regulated genes in Escherichia coli. J Mol Biol. 1987 May 20;195(2):289–297. doi: 10.1016/0022-2836(87)90650-4. [DOI] [PubMed] [Google Scholar]
- Helmann J. D. Alternative sigma factors and the regulation of flagellar gene expression. Mol Microbiol. 1991 Dec;5(12):2875–2882. doi: 10.1111/j.1365-2958.1991.tb01847.x. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Márquez L. M., Chamberlin M. J. Cloning, sequencing, and disruption of the Bacillus subtilis sigma 28 gene. J Bacteriol. 1988 Apr;170(4):1568–1574. doi: 10.1128/jb.170.4.1568-1574.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen J. Bacterial surface translocation: a survey and a classification. Bacteriol Rev. 1972 Dec;36(4):478–503. doi: 10.1128/br.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., DeRosier D. J., Macnab R. M. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J Mol Biol. 1990 Jun 20;213(4):819–832. doi: 10.1016/S0022-2836(05)80266-9. [DOI] [PubMed] [Google Scholar]
- Homma M., Fujita H., Yamaguchi S., Iino T. Regions of Salmonella typhimurium flagellin essential for its polymerization and excretion. J Bacteriol. 1987 Jan;169(1):291–296. doi: 10.1128/jb.169.1.291-296.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Kutsukake K., Hasebe M., Iino T., Macnab R. M. FlgB, FlgC, FlgF and FlgG. A family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1990 Jan 20;211(2):465–477. doi: 10.1016/0022-2836(90)90365-S. [DOI] [PubMed] [Google Scholar]
- Joys T. M. The flagellar filament protein. Can J Microbiol. 1988 Apr;34(4):452–458. doi: 10.1139/m88-078. [DOI] [PubMed] [Google Scholar]
- Keen N. T., Tamaki S., Kobayashi D., Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988 Oct 15;70(1):191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- Kihara M., Homma M., Kutsukake K., Macnab R. M. Flagellar switch of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989 Jun;171(6):3247–3257. doi: 10.1128/jb.171.6.3247-3257.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y., Iino T. Regulation of expression of the flagellin gene (hag) in Escherichia coli K-12: analysis of hag-lac gene fusions. J Bacteriol. 1979 Sep;139(3):721–729. doi: 10.1128/jb.139.3.721-729.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y. Transcriptional control of flagellar genes in Escherichia coli K-12. J Bacteriol. 1986 Dec;168(3):1315–1318. doi: 10.1128/jb.168.3.1315-1318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S. C., Koshland D. E., Jr Sequence of the flaA (cheC) locus of Escherichia coli and discovery of a new gene. J Bacteriol. 1986 Jun;166(3):1007–1012. doi: 10.1128/jb.166.3.1007-1012.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K., Ohya Y., Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol. 1990 Feb;172(2):741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima G., Kawagishi I., Homma M., Asaka J., Kondo E., Macnab R. M. Export of an N-terminal fragment of Escherichia coli flagellin by a flagellum-specific pathway. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4953–4957. doi: 10.1073/pnas.86.13.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lonetto M., Gribskov M., Gross C. A. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992 Jun;174(12):3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R. M., Parkinson J. S. Genetic analysis of the bacterial flagellum. Trends Genet. 1991 Jun;7(6):196–200. doi: 10.1016/0168-9525(91)90436-t. [DOI] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Showalter R., Silverman M. Identification of a locus controlling expression of luminescence genes in Vibrio harveyi. J Bacteriol. 1989 May;171(5):2406–2414. doi: 10.1128/jb.171.5.2406-2414.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L. L., Silverman M. Phosphate regulation of gene expression in Vibrio parahaemolyticus. J Bacteriol. 1987 Aug;169(8):3441–3449. doi: 10.1128/jb.169.8.3441-3449.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L., Hilmen M., Silverman M. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell. 1988 Jul 29;54(3):345–351. doi: 10.1016/0092-8674(88)90197-3. [DOI] [PubMed] [Google Scholar]
- McCarter L., Silverman M. Iron regulation of swarmer cell differentiation of Vibrio parahaemolyticus. J Bacteriol. 1989 Feb;171(2):731–736. doi: 10.1128/jb.171.2.731-736.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L., Silverman M. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol Microbiol. 1990 Jul;4(7):1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Newton A., Ohta N. Regulation of the cell division cycle and differentiation in bacteria. Annu Rev Microbiol. 1990;44:689–719. doi: 10.1146/annurev.mi.44.100190.003353. [DOI] [PubMed] [Google Scholar]
- Nishimura A., Hirota Y. A cell division regulatory mechanism controls the flagellar regulon in Escherichia coli. Mol Gen Genet. 1989 Apr;216(2-3):340–346. doi: 10.1007/BF00334374. [DOI] [PubMed] [Google Scholar]
- Ohnishi K., Kutsukake K., Suzuki H., Iino T. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet. 1990 Apr;221(2):139–147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sar N., McCarter L., Simon M., Silverman M. Chemotactic control of the two flagellar systems of Vibrio parahaemolyticus. J Bacteriol. 1990 Jan;172(1):334–341. doi: 10.1128/jb.172.1.334-341.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda S., Okamoto K. Formation and function of Vibrio parahaemolyticus lateral flagella. J Bacteriol. 1977 Mar;129(3):1266–1271. doi: 10.1128/jb.129.3.1266-1271.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Showalter R., McCarter L. Genetic analysis in vibrio. Methods Enzymol. 1991;204:515–536. doi: 10.1016/0076-6879(91)04026-k. [DOI] [PubMed] [Google Scholar]
- Stader J., Matsumura P., Vacante D., Dean G. E., Macnab R. M. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J Bacteriol. 1986 Apr;166(1):244–252. doi: 10.1128/jb.166.1.244-252.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starnbach M. N., Lory S. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol Microbiol. 1992 Feb;6(4):459–469. doi: 10.1111/j.1365-2958.1992.tb01490.x. [DOI] [PubMed] [Google Scholar]
- Ulitzur S. Induction of swarming in Vibrio parahaemolyticus. Arch Microbiol. 1974;101(4):357–363. doi: 10.1007/BF00455952. [DOI] [PubMed] [Google Scholar]
- Yu J., Shapiro L. Early Caulobacter crescentus genes fliL and fliM are required for flagellar gene expression and normal cell division. J Bacteriol. 1992 May;174(10):3327–3338. doi: 10.1128/jb.174.10.3327-3338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]