Abstract
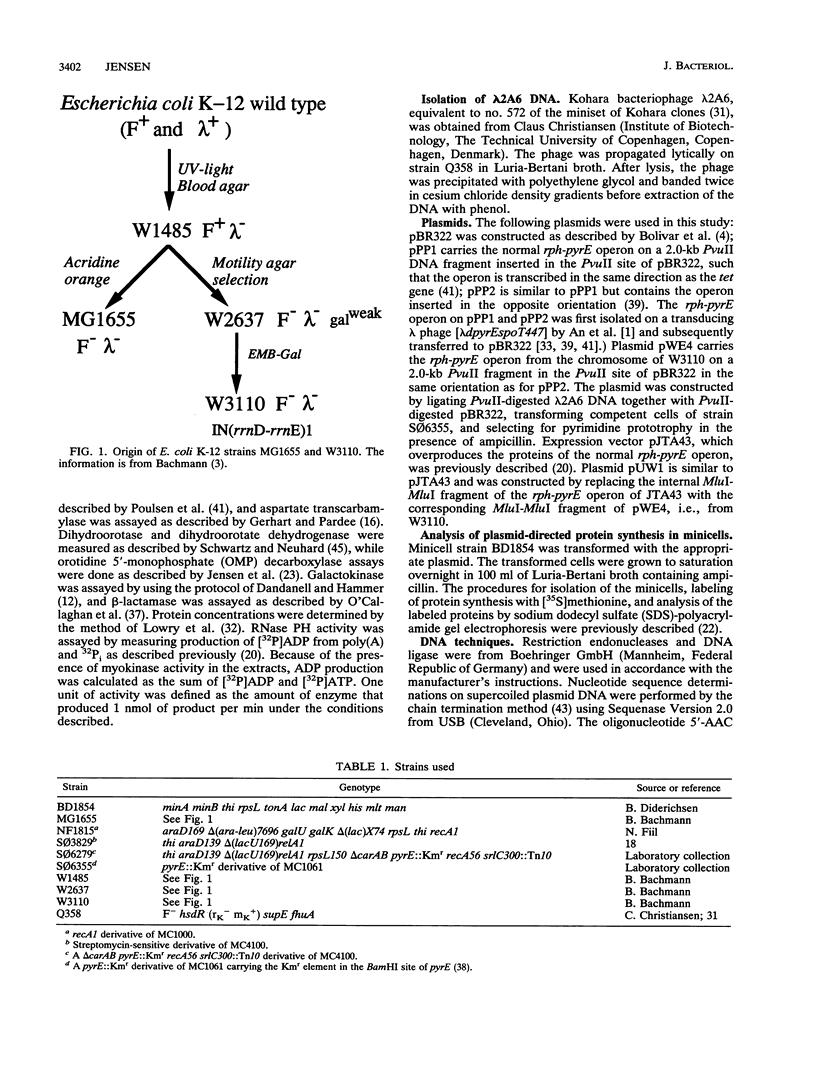
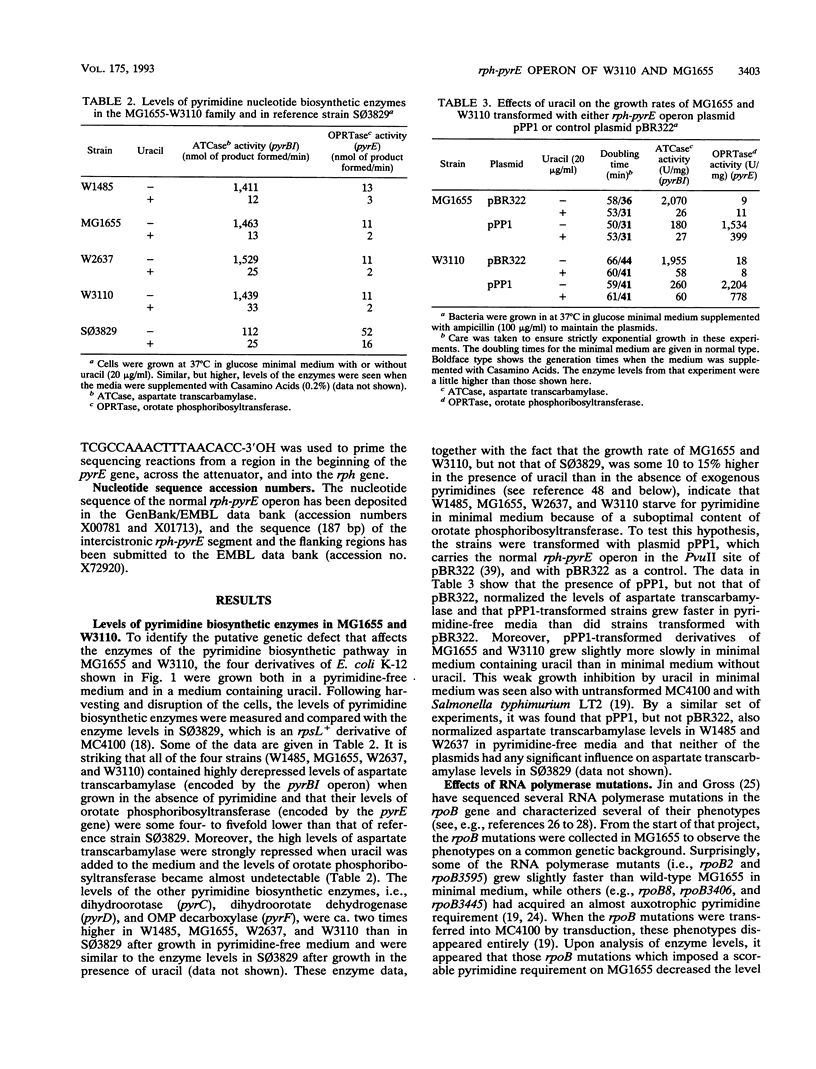
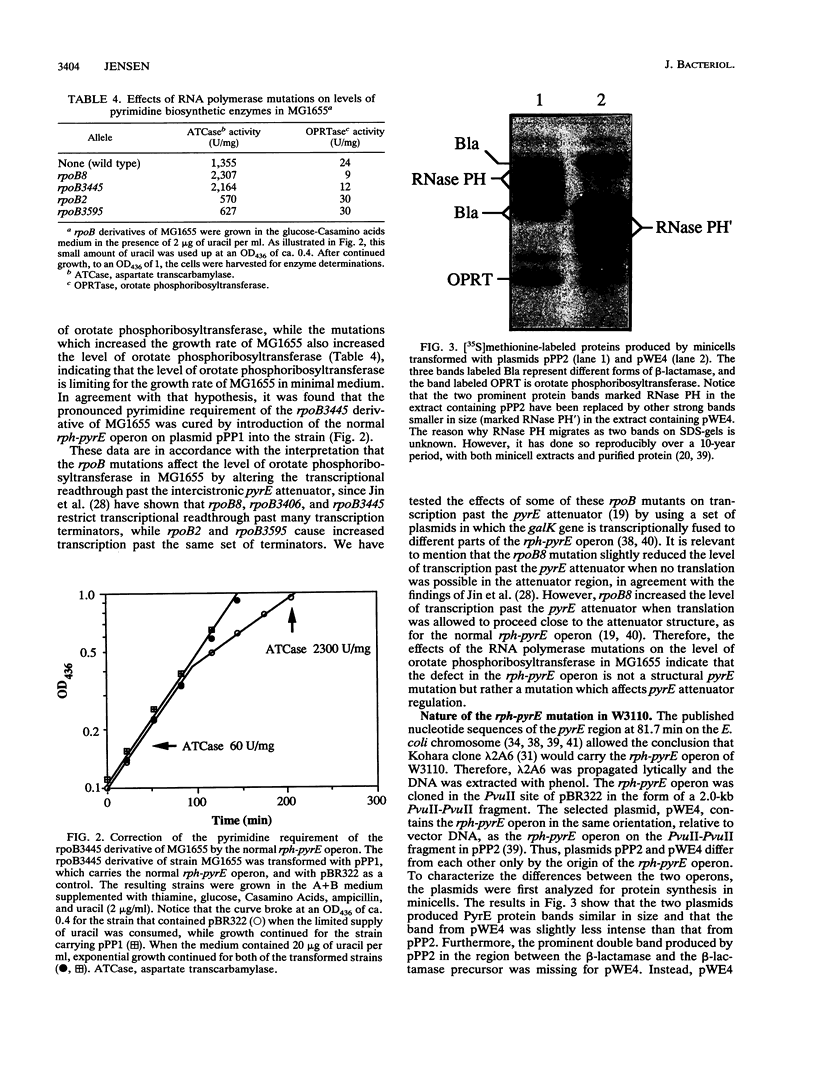
The widely used and closely related Escherichia coli "wild types" W3110 and MG1655, as well as their common ancestor W1485, starve for pyrimidine in minimal medium because of a suboptimal content of orotate phosphoribosyltransferase, which is encoded by the pyrE gene. This conclusion was based on the findings that (i) the strains grew 10 to 15% more slowly in pyrimidine-free medium than in medium containing uracil; (ii) their levels of aspartate transcarbamylase were highly derepressed, as is characteristic for pyrimidine starvation conditions; and (iii) their levels of orotate phosphoribosyltransferase were low. After introduction of a plasmid carrying the rph-pyrE operon from strain HfrH, the growth rates were no longer stimulated by uracil and the levels of aspartate transcarbamylase were low and similar to the levels observed for other strains of E. coli K-12, E. coli B, and Salmonella typhimurium. To identify the mutation responsible for these phenotypes, the rph-pyrE operon of W3110 was cloned in pBR322 from Kohara bacteriophage lambda 2A6. DNA sequencing revealed that a GC base pair was missing near the end of the rph gene of W3110. This one-base-pair deletion results in a frame shift of translation over the last 15 codons and reduces the size of the rph gene product by 10 amino acid residues relative to the size of RNase PH of other E. coli strains, as confirmed by analysis of protein synthesis in minicells. The truncated protein lacks RNase PH activity, and the premature translation stop in the rph cistron explains the low levels of orotate phosphoribosyltransferase in W3110, since close coupling between transcription and translation is needed to support optimal levels of transcription past the intercistronic pyrE attenuator.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- An G., Justesen J., Watson R. J., Friesen J. D. Cloning the spoT gene of Escherichia coli: identification of the spoT gene product. J Bacteriol. 1979 Mar;137(3):1100–1110. doi: 10.1128/jb.137.3.1100-1110.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J. T., Poulsen P., Jensen K. F. Attenuation in the rph-pyrE operon of Escherichia coli and processing of the dicistronic mRNA. Eur J Biochem. 1992 Jun 1;206(2):381–390. doi: 10.1111/j.1432-1033.1992.tb16938.x. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bonekamp F., Andersen H. D., Christensen T., Jensen K. F. Codon-defined ribosomal pausing in Escherichia coli detected by using the pyrE attenuator to probe the coupling between transcription and translation. Nucleic Acids Res. 1985 Jun 11;13(11):4113–4123. doi: 10.1093/nar/13.11.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonekamp F., Clemmesen K., Karlström O., Jensen K. F. Mechanism of UTP-modulated attenuation at the pyrE gene of Escherichia coli: an example of operon polarity control through the coupling of translation to transcription. EMBO J. 1984 Dec 1;3(12):2857–2861. doi: 10.1002/j.1460-2075.1984.tb02220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmesen K., Bonekamp F., Karlström O., Jensen K. F. Role of translation in the UTP-modulated attenuation at the pyrBI operon of Escherichia coli. Mol Gen Genet. 1985;201(2):247–251. doi: 10.1007/BF00425666. [DOI] [PubMed] [Google Scholar]
- Craven M. G., Henner D. J., Alessi D., Schauer A. T., Ost K. A., Deutscher M. P., Friedman D. I. Identification of the rph (RNase PH) gene of Bacillus subtilis: evidence for suppression of cold-sensitive mutations in Escherichia coli. J Bacteriol. 1992 Jul;174(14):4727–4735. doi: 10.1128/jb.174.14.4727-4735.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandanell G., Hammer K. Two operator sites separated by 599 base pairs are required for deoR repression of the deo operon of Escherichia coli. EMBO J. 1985 Dec 1;4(12):3333–3338. doi: 10.1002/j.1460-2075.1985.tb04085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. L., Plunkett G., 3rd, Burland V., Blattner F. R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992 Aug 7;257(5071):771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- Deutscher M. P., Marshall G. T., Cudny H. RNase PH: an Escherichia coli phosphate-dependent nuclease distinct from polynucleotide phosphorylase. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4710–4714. doi: 10.1073/pnas.85.13.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GERHART J. C., PARDEE A. B. The enzymology of control by feedback inhibition. J Biol Chem. 1962 Mar;237:891–896. [PubMed] [Google Scholar]
- Gaal T., Gourse R. L. Guanosine 3'-diphosphate 5'-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5533–5537. doi: 10.1073/pnas.87.14.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. F., Andersen J. T., Poulsen P. Overexpression and rapid purification of the orfE/rph gene product, RNase PH of Escherichia coli. J Biol Chem. 1992 Aug 25;267(24):17147–17152. [PubMed] [Google Scholar]
- Jensen K. F. Apparent involvement of purines in the control of expression of Salmonella typhimurium pyr genes: analysis of a leaky guaB mutant resistant to pyrimidine analogs. J Bacteriol. 1979 Jun;138(3):731–738. doi: 10.1128/jb.138.3.731-738.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. F. Hyper-regulation of pyr gene expression in Escherichia coli cells with slow ribosomes. Evidence for RNA polymerase pausing in vivo? Eur J Biochem. 1988 Aug 15;175(3):587–593. doi: 10.1111/j.1432-1033.1988.tb14232.x. [DOI] [PubMed] [Google Scholar]
- Jensen K. F., Larsen J. N., Schack L., Sivertsen A. Studies on the structure and expression of Escherichia coli pyrC, pyrD, and pyrF using the cloned genes. Eur J Biochem. 1984 Apr 16;140(2):343–352. doi: 10.1111/j.1432-1033.1984.tb08107.x. [DOI] [PubMed] [Google Scholar]
- Jensen K. F., Neuhard J., Schack L. RNA polymerase involvement in the regulation of expression of Salmonella typhimurium pyr genes. Isolation and characterization of a fluorouracil-resistant mutant with high, constitutive expression of the pyrB and pyrE genes due to a mutation in rpoBC. EMBO J. 1982;1(1):69–74. doi: 10.1002/j.1460-2075.1982.tb01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D. J., Gross C. A. Characterization of the pleiotropic phenotypes of rifampin-resistant rpoB mutants of Escherichia coli. J Bacteriol. 1989 Sep;171(9):5229–5231. doi: 10.1128/jb.171.9.5229-5231.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D. J., Gross C. A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988 Jul 5;202(1):45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- Jin D. J., Gross C. A. Three rpoBC mutations that suppress the termination defects of rho mutants also affect the functions of nusA mutants. Mol Gen Genet. 1989 Apr;216(2-3):269–275. doi: 10.1007/BF00334365. [DOI] [PubMed] [Google Scholar]
- Jin D. J., Walter W. A., Gross C. A. Characterization of the termination phenotypes of rifampicin-resistant mutants. J Mol Biol. 1988 Jul 20;202(2):245–253. doi: 10.1016/0022-2836(88)90455-x. [DOI] [PubMed] [Google Scholar]
- Kelly K. O., Deutscher M. P. The presence of only one of five exoribonucleases is sufficient to support the growth of Escherichia coli. J Bacteriol. 1992 Oct;174(20):6682–6684. doi: 10.1128/jb.174.20.6682-6684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K. O., Reuven N. B., Li Z., Deutscher M. P. RNase PH is essential for tRNA processing and viability in RNase-deficient Escherichia coli cells. J Biol Chem. 1992 Aug 15;267(23):16015–16018. [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lundberg L. G., Thoresson H. O., Karlström O. H., Nyman P. O. Nucleotide sequence of the structural gene for dUTPase of Escherichia coli K-12. EMBO J. 1983;2(6):967–971. doi: 10.1002/j.1460-2075.1983.tb01529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhard J., Stauning E., Kelln R. A. Cloning and characterization of the pyrE gene and of PyrE::Mud1 (Ap lac) fusions from Salmonella typhimurium. Eur J Biochem. 1985 Feb 1;146(3):597–603. doi: 10.1111/j.1432-1033.1985.tb08693.x. [DOI] [PubMed] [Google Scholar]
- O'Callaghan C. H., Morris A., Kirby S. M., Shingler A. H. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972 Apr;1(4):283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen P., Andersen J. T., Jensen K. F. Molecular and mutational analysis of three genes preceding pyrE on the Escherichia coli chromosome. Mol Microbiol. 1989 Mar;3(3):393–404. doi: 10.1111/j.1365-2958.1989.tb00184.x. [DOI] [PubMed] [Google Scholar]
- Poulsen P., Bonekamp F., Jensen K. F. Structure of the Escherichia coli pyrE operon and control of pyrE expression by a UTP modulated intercistronic attentuation. EMBO J. 1984 Aug;3(8):1783–1790. doi: 10.1002/j.1460-2075.1984.tb02046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen P., Jensen K. F. Effect of UTP and GTP pools on attenuation at the pyrE gene of Escherichia coli. Mol Gen Genet. 1987 Jun;208(1-2):152–158. doi: 10.1007/BF00330436. [DOI] [PubMed] [Google Scholar]
- Poulsen P., Jensen K. F., Valentin-Hansen P., Carlsson P., Lundberg L. G. Nucleotide sequence of the Escherichia coli pyrE gene and of the DNA in front of the protein-coding region. Eur J Biochem. 1983 Sep 15;135(2):223–229. doi: 10.1111/j.1432-1033.1983.tb07641.x. [DOI] [PubMed] [Google Scholar]
- Roland K. L., Liu C. G., Turnbough C. L., Jr Role of the ribosome in suppressing transcriptional termination at the pyrBI attenuator of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7149–7153. doi: 10.1073/pnas.85.19.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrenk W. J., Weisberg R. A. A simple method for making new transducing lines of coliphage lambda. Mol Gen Genet. 1975;137(2):101–107. doi: 10.1007/BF00341676. [DOI] [PubMed] [Google Scholar]
- Schwartz M., Neuhard J. Control of expression of the pyr genes in Salmonella typhimurium: effects of variations in uridine and cytidine nucleotide pools. J Bacteriol. 1975 Mar;121(3):814–822. doi: 10.1128/jb.121.3.814-822.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy T. J., Beckwith J. Isolation and characterization of mutants of Escherichia coli K12 affected in protein localization. Methods Enzymol. 1983;97:11–40. doi: 10.1016/0076-6879(83)97115-x. [DOI] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H. C., Arps P. J., Connolly D. M., Winkler M. E. Absence of hisT-mediated tRNA pseudouridylation results in a uracil requirement that interferes with Escherichia coli K-12 cell division. J Bacteriol. 1991 Nov;173(22):7395–7400. doi: 10.1128/jb.173.22.7395-7400.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Nordström K. A runaway-replication mutant of plasmid R1drd-19: temperature-dependent loss of copy number control. Mol Gen Genet. 1978 Oct 4;165(2):167–179. doi: 10.1007/BF00269904. [DOI] [PubMed] [Google Scholar]
- VanBogelen R. A., Neidhardt F. C. The gene-protein database of Escherichia coli: edition 4. Electrophoresis. 1991 Nov;12(11):955–994. doi: 10.1002/elps.1150121114. [DOI] [PubMed] [Google Scholar]
- Yura T., Mori H., Nagai H., Nagata T., Ishihama A., Fujita N., Isono K., Mizobuchi K., Nakata A. Systematic sequencing of the Escherichia coli genome: analysis of the 0-2.4 min region. Nucleic Acids Res. 1992 Jul 11;20(13):3305–3308. doi: 10.1093/nar/20.13.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]



