Abstract
The catalytic subunit of PP-1 (PP-1C) is potently inhibited (IC50, ≈1 nM) by DARPP-32 (dopamine- and cAMP-regulated phosphoprotein, Mr 32,000), inhibitor-1, and inhibitor-2. The NH2-terminal 50 amino acid residues of DARPP-32 and inhibitor-1 are similar, and phosphorylation of a common threonine residue (Thr-34/Thr-35) is necessary for inhibition of PP-1C. We have characterized further the interaction between DARPP-32 and PP-1C. Using synthetic peptides derived from the NH2-terminal region of DARPP-32, residues 6–11, RKKIQF, have been shown to be required for inhibition of PP-1C. Peptides containing this motif were able to antagonize the inhibition of PP-1C by phospho-DARPP-32 and phosphoinhibitor-1. The inhibition of PP-1C by inhibitor-2, but not by okadaic acid, microcystin, or calyculin A, was also attentuated by these antagonist peptides. These results together with results from other studies support a model in which two subdomains of phospho-DARPP-32 interact with PP-1C. The region encompassing phospho-Thr-34 appears to interact with the active site of the enzyme blocking enzyme activity. The region encompassing the RKKIQF motif binds to a domain of PP-1C removed from the active site. Amino acid sequence analysis indicates that basic and hydrophobic features of the RKKIQF motif are conserved in the binding domains of certain PP-1C targeting proteins, suggesting that interaction of inhibitor proteins and targeting proteins may be mutually exclusive.
Serine/threonine protein phosphatases have been classified into four major types (PP-1, -2A, -2B, and -2C), based on their substrate specificities and sensitivities to divalent cations and protein inhibitors (1–3). A variety of naturally occurring compounds, including okadaic acid, calyculin A, microcystin, cyclosporin, and FK506, also differentially inhibit these enzymes (3, 4). PP-1 is a major eukaryotic protein phosphatase that has been shown to regulate a variety of cellular processes including cell cycle progression, cell metabolism, transcriptional regulation, and neuronal function (1–3). The catalytic subunit of PP-1 (PP-1C) is regulated by a number of heat-stable protein inhibitors. These are inhibitor-1, its homolog DARPP-32 (dopamine- and cAMP-regulated phosphoprotein, Mr 32,000), and inhibitor-2 (1, 2). Phosphorylation of Thr-35 of inhibitor-1 or Thr-34 of DARPP-32 by cAMP-dependent protein kinase (PKA) converts either protein into a potent inhibitor of PP-1C. Unphosphorylated inhibitor-2 forms a complex with PP-1C leading to enzyme inhibition and this can be reversed in a complex mechanism involving phosphorylation of inhibitor-2 by glycogen synthase kinase 3 (5). Inhibitor-1 and inhibitor-2 exhibit a relatively widespread tissue distribution and are likely, through control of the activity of PP-1, to regulate a variety of processes including the cell cycle and synaptic plasticity (6, 7). DARPP-32 is highly localized to neurons receiving D1-dopamine input, where its phosphorylation links the actions of dopamine to the regulation of neuronal excitability (2, 8, 9).
PP-1C is also regulated by its interaction with a variety of proteins, termed targeting subunits, that serve to localize the enzyme to specific subcellular compartments and to influence its substrate specificity (1–3, 10, 11). These include: the glycogen binding proteins, GM and GL; the myofibril binding protein, M110 (12); the retinoblastoma gene product, p110Rb (13); the ribosomal protein, L5 (14); the p53-binding protein, p53BP2 (15); and the nuclear proteins, NIPP-1 (16) and sds22 (17). In neurons, PP-1 is highly localized to dendritic spines (18), presumably via a novel targeting protein. Recent studies using microcystin affinity chromatography (19) and the yeast two-hybrid method (15) suggest that many additional PP-1 targeting proteins, perhaps as many as 30 or 40, remain to be identified and characterized.
The precise molecular details of the interactions of PP-1 with the various inhibitor and targeting proteins remain to be elucidated. To date there is no evidence to indicate that PP-1C can interact with more than one binding protein at the same time, suggesting that the interactions with the various binding proteins may be mutually exclusive. Recent studies of the interaction of GM, M110, and inhibitor-2 with PP-1 support this idea (12, 20). Our previous studies have suggested that two subdomains within the first 50 amino acids of DARPP-32 are necessary for its high potency as an inhibitor (21, 22). DARPP-32 and inhibitor-1 share a high degree of amino acid sequence identity within residues 1–50 and analogous studies of inhibitor-1 support the two domain model (23, 24). Subdomain 1 includes the region surrounding Thr-34 and subdomain 2 includes the region surrounding Ile-9. Furthermore, we have suggested that subdomains 1 and 2 represent, respectively, distinct inhibitory and binding sites, and that it is the conjugation of these two relatively low affinity interactions that results in the high affinity interaction observed between DARPP-32 or inhibitor-1 and PP-1 (22). In the present study, we have further examined the interaction of DARPP-32 and PP-1 and identified important residues in subdomain 2 in addition to Ile-9. Based on the information obtained, we have identified short peptide antagonists that compete with phospho-DARPP-32 or phosphoinhibitor-1. In addition, these peptide antagonists compete with the ability of inhibitor-2 to interact with PP-1. Finally, the critical residues that comprise subdomain 2 appear to be functionally conserved in certain other PP-1-binding proteins, supporting the idea that binding of the protein inhibitors and some of the targeting proteins is mutually exclusive.
MATERIALS AND METHODS
Purification of PP-1Cα from Sf9 cells.
Recombinant PP-1Cα was expressed in Sf9 cells using the baculovirus system (unpublished data). Expressed PP-1C was purified using serial column chromatography on DEAE cellulose, heparin Sepharose, Sephacryl S-200, and Mono-Q (H.-B.H., unpublished data). The purity of PP-1C was >90%. The properties of Sf9 PP-1C are the same as those of PP-1C purified from rabbit muscle (E. F. da Cruz e Silva and H.-B.H., unpublished data), and identical results were obtained for Sf9 cell or rabbit muscle PP-1C in studies of the antagonist peptides.
Peptide Synthesis.
D32[1–38c], D32[5–38], D32[6–38c], D32[8–38] (c indicates COOH-terminal cysteine) were prepared by the W. M. Keck Biotechnology Resource Center, Yale University School of Medicine (New Haven, CT). Other peptides were prepared on Rink resin (Peptides International) with fluorenylmethoxycarbonyl amino acids (Advanced ChemTech) using an automated peptide synthesizer (Advanced ChemTech Act 90). All peptides were synthesized with free NH2 termini and amidated COOH termini. Peptides were cleaved using a modified reagent K (87% trifluoroacetic acid/3.75% phenol/3.75% H2O/3.75% thioanisole/1.75% 1,2-ethanedithiol) and then purified by CM-ion-exchange HPLC (10 × 150 mm) using a gradient of NaCl in 50 mM NaOAc, pH 4.5, followed by reversed-phase HPLC on a Vydac C18 column (10 × 250 mm) with a gradient of acetonitrile in 0.05% trifluoroacetic acid. All peptides had the expected amino acid compositions and mass spectra. Peptide concentrations were determined by quantitative amino acid analysis.
Phosphorylation of Peptides and Proteins.
Recombinant DARPP-32 and DARPP-32[T34A] were prepared, and DARPP-32 phosphorylated, essentially as described (22). DARPP-32 (2 mg) was dissolved in 1 ml of 50 mM Hepes (pH 7.4), 1 mM EGTA, 10 mM magnesium acetate, and 1 mM ATP. The phosphorylation reaction was started by addition of 2 μg of PKA catalytic subunit and the incubation was carried out for 60 min. Phosphorylated DARPP-32 was purified by HPLC using a C18 column as described (22). Inhibitor-1 and inhibitor-2 were purified from rabbit skeletal muscle essentially as described (25). Synthetic peptides were phosphorylated essentially as described (21), and phosphopeptides were purified by serial chromatography using CM-ion-exchange HPLC and reversed-phase HPLC (C18 column).
PP-1 Assays.
PP-1 was assayed using 32P-phosphorylase a as substrate essentially as described (26). Assays (final volume 30 μl) contained 50 mM Tris·HCl, 0.15 mM EGTA, 15 mM 2-mercaptoethanol, 0.01% (wt/wt) Brij 35, 0.3 mg/ml BSA, 5 mM caffeine, 10 μM 32P-phosphorylase a, various protein inhibitors or toxins, and PP-1C as described. With the exception of assays involving inhibitor-2, all components except the enzyme were preincubated at 30°C for 5 min. Dephosphorylation reactions were initiated by the addition of 10 μl of PP-1C and reactions were carried out for 10 min at 30°C. In the case of inhibitor-2, PP-1C and inhibitor-2 were incubated in the absence or presence of antagonist peptides for 15 min at 4°C and reactions were initiated by addition of substrate. For kinetic analyses, 2.5–20 μM substrate (final concentration) was used. Km and Vmax values were determined from Lineweaver–Burk plots. All reactions were performed in duplicate. Okadaic acid, microcystin-LR, and calyculin A were obtained from GIBCO.
RESULTS
Inhibition of PP-1 by Dephospho- and Phospho-DARPP-32 and Related Synthetic Peptide Analogs.
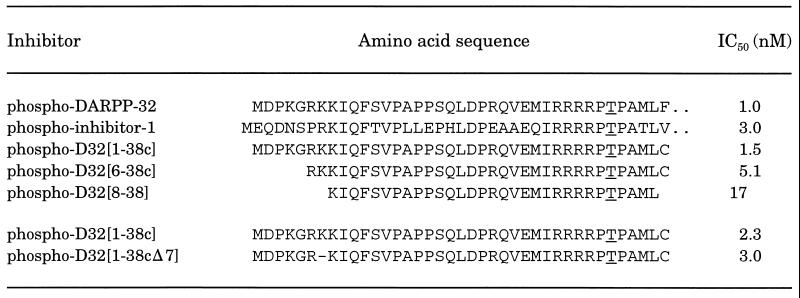
Our previous work (21) has indicated that Ile-9 within subdomain 2 of DARPP-32 was necessary for inhibition of PP-1. A phospho-peptide, D32[10–38], retained <0.5% of the inhibitory potency of intact phospho-DARPP-32, whereas phospho-D32[9–38] retained ≈20% of the activity of intact phospho-DARPP-32. To examine the contribution of NH2-terminal residues 1–8 of DARPP-32, we assessed the inhibitory potencies of a number of longer synthetic peptides (Table 1). In each case the peptides were stoichiometrically phosphorylated by PKA and their abilities to inhibit PP-1 activity were analyzed using [32P]phosphorylase a as substrate. Inclusion of amino acids 1–8 (D32[1–38c]) resulted in a phospho-peptide that had inhibitory properties indistinguishable from those of phospho-DARPP-32. Removal of residues 1–5 (D32[6–38c]) reduced inhibitory potency slightly. Removal of basic amino acids in positions 6 and 7 reduced inhibitory potency further. Residues 8–11 of DARPP-32 and 9–12 of inhibitor-1 are identical, and residue 7 of DARPP-32 and 8 of inhibitor-1 are both basic amino acids (Table 1). Although inhibitor-1 has only two basic amino acids (residues 8 and 9) whereas DARPP-32 has three (residues 6–8), phospho-inhibitor-1 has essentially identical inhibitory potency to that of phospho-DARPP-32. Accordingly, deletion of Lys-7 (resulting in a peptide that more closely resembled the inhibitor-1 sequence) had little effect on inhibitory potency.
Table 1.
Inhibition of PP-1C by phospho-DARPP-32, phospho-inhibitor-1, and related synthetic phosphopeptides
PP-1C activity was measured as described using 10 μM [32P]phosphorylase a as substrate and concentrations of phosphorylated inhibitors that bracketed the IC50 value. The phosphorylated threonine is underlined. The results shown for the last two peptides were obtained from a different series of studies. Phospho-D32[1–38c] was assayed again as an internal control.
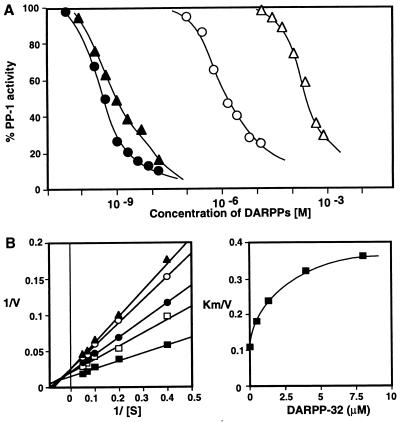
We have previously shown that dephospho-DARPP-32 binds to PP-1 and inhibits enzyme activity with an IC50 of ≈10−6 M (ref. 22 and Fig. 1A). Dephospho-D32[1–38c] was even less effective, with an IC50 value almost 6 orders of magnitude lower than the corresponding phospho-peptide (Fig. 1A). Notably, dephospho-DARPP-32 inhibited PP-1C with a mixed competitive/noncompetitive mechanism and secondary plots of KM/Vmax were nonlinear (Fig. 1B), results identical to those obtained for phospho-DARPP-32 and phospho-inhibitor-1 (21, 27). Together these results support the idea that the intact NH2 terminus of DARPP-32 is important, in particular basic amino acids in the RKKIQF motif in subdomain 2 of DARPP-32 (RKIQF in inhibitor-1). Furthermore, the results suggest that subdomain 2 interacts with a region of PP-1C that is away from the active site.
Figure 1.
Inhibition of PP-1C by dephospho- and phospho-DARPP-32. PP-1C was assayed using 10 μM [32P]phosphorylase a as substrate. (A) Phospho-DARPP-32 (•), phospho-D32[1–38c] (▴), DARPP-32[T34A] (○), and dephospho-D32[1–38c] (▵) were present in the indicated concentrations. (B). Mechanism of inhibition of PP-1C by nonphosphorylated-DARPP-32. (Left) Lineweaver–Burk analysis at various concentration of DARPP-32[T34A]. PP-1 activity was determined using a range of concentrations of [32P]phosphorylase a (2.5–20 μM), in the absence (▪) or presence of dephospho-DARPP-32 (□, 0.5 μM; •, 1.5 μM; ○, 4 μM; ▴, 8 μM). (Right) Secondary replot of Km/Vmax as a function of DARPP-32 concentration.
NH2-Terminal DARPP-32 Peptides Antagonize Inhibition of PP-1C by Phospho-DARPP-32 or Phospho-Inhibitor-1.
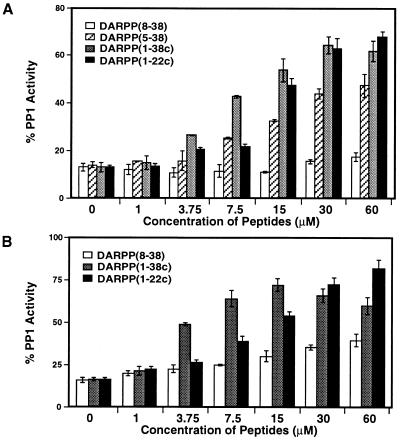
The presence of two binding sites in PP-1C for phospho-DARPP-32 and phospho-inhibitor-1 suggested that interaction of dephospho-peptides at subdomain 2 might antagonize the inhibitory actions of these two holo-proteins. Addition of 10 nM phospho-DARPP-32 or phospho-inhibitor-1 resulted in close to maximal inhibition of PP-1C (Fig. 2). Dephospho-D32[1–38c] prevented inhibition by phospho-DARPP-32 when added in concentrations (1–30 μM) (Fig. 2A) that alone had very little effect on PP-1C activity (see Fig. 1). Addition of a fixed concentration of dephospho-D32[1–38c] increased the IC50 for phospho-DARPP-32 by more than 25-fold (Fig. 3A). The antagonistic potencies of other dephospho-peptides tested were a reflection of their amino acid sequence (Fig. 2A). D32[1–22c], which does not contain subdomain 1, was as effective as D32[1–38c]. D32[5–38] was almost as effective as D32[1–38c]. However, D32[8–38] exhibited little effect in concentrations up to 60 μM. Very similar results were obtained when dephospho-peptides were used to antagonize the action of phospho-inhibitor-1, with the exception that higher concentrations of D32[8–38] were slightly effective (Fig. 2B).
Figure 2.
Effect of peptide antagonists on inhibition of PP-1 by phospho-DARPP-32 and phospho-inhibitor-1. PP-1 was assayed using 10 μM [32P]phosphorylase a as substrate in the presence of 10 nM phospho-DARPP-32 (A) or of 10 nM phospho-inhibitor-1 (B), in the presence of the indicated concentrations of peptide antagonists, D32[8–38], D32[5–38], D32[1–38c], or D32[1–22c]. In all assays, the three components, antagonist, inhibitor, and substrate, were premixed and reactions were initiated by addition of enzyme. D32[1–22c] (up to 500 μM) and D32[5–38] (<60 μM) had no effect on PP-1C activity. Error bars = SD from four independent measurements.
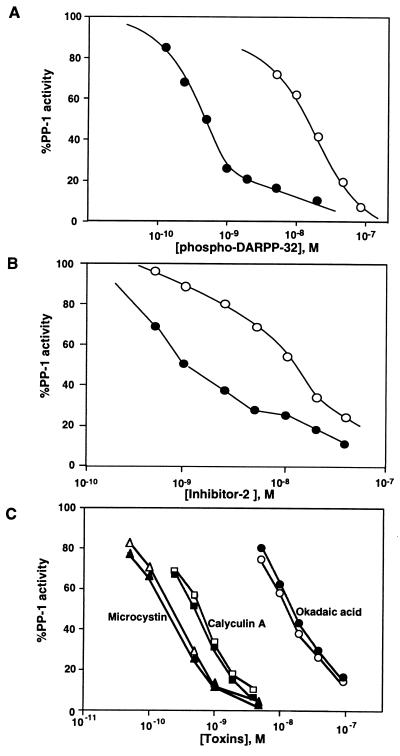
Figure 3.
Effect of peptide antagonist D32[1–38c] on the inhibition of PP-1 by phospho-DARPP-32, inhibitor-2 and various toxins. PP-1 was assayed using 10 μM [32P]phosphorylase a as substrate, in the absence (filled symbols) or presence (open symbols) of 25 μM D32[1–38c], and the indicated concentrations of (A) phospho-DARPP-32, (B) inhibitor-2, or (C) microcystin (triangles), calyculin A (squares), and okadaic acid (circles). For inhibitor-2 assays, PP-1C was preincubated with inhibitor-2 in the absence or presence of D32[1–38c] and reactions were initiated with substrate. In all other assays, antagonist, inhibitor, and substrate, were premixed and reactions were initiated by addition of enzyme.
Effect of Dephospho-D32[1–38c] on the Inhibition of PP-1 by Inhibitor-2 and Various Toxins.
Inhibitor-2 in its nonphosphorylated form is a potent inhibitor of PP-1 (1, 2). However, there is no obvious amino acid sequence homology between inhibitor-2 and DARPP-32 or inhibitor-1. In addition, no subdomain of inhibitor-2 has clearly been identified that mediates inhibition of PP-1C (5). Addition of D32[1–38c] resulted in a ≈10-fold increase in the IC50 for inhibitor-2 (Fig. 3B). PP-1 and PP-2A are potently inhibited by several naturally occurring toxins, including okadaic acid, microcystin, and calyculin A (4). Determination of the structure of PP-1 in a complex with microcystin has shown that the toxin binds at the active site of the phosphatase via several types of interaction including covalent binding to Cys-273 (28). Furthermore, recent site-directed mutagenesis studies of PP-1 have indicated that these toxins, as well as the phospho-threonine of DARPP-32, bind at the active site of PP-1 in a similar fashion despite their structural diversity (29, 30). Addition of D32[1–38c] did not affect the inhibition of PP-1 by either microcystin, calyculin A, or okadaic acid (Fig. 3C). Together, these studies further support the idea that subdomain 2 of DARPP-32 and inhibitor-1 interacts with a site in PP-1C removed from the active site and that this binding site may overlap to some extent with part of the binding site for inhibitor-2.
DISCUSSION
The results obtained in the present study, together with our previous results (21, 22), provide support for a model in which two distinct subdomains in DARPP-32 interact with PP-1C. Subdomain 1 includes the phosphorylated threonine residue (Thr-34; Table 1); notably a peptide analog containing phospho-serine in place of phospho-threonine is an ineffective inhibitor (21). Elucidation of the crystal structure of PP-1 has revealed that several acidic amino acids present in a surface groove close to the active site of PP-1 may be important in binding to four arginine residues that precede phospho-Thr-34 in DARPP-32 (28). This possibility is supported by our recent site-directed mutagenesis studies of PP-1 that suggest that acidic groove residues appear to influence the interaction of phospho-DARPP-32 with PP-1C (30). Subdomain 2 consists of a short stretch of residues NH2-terminal to the phosphorylation site. This domain includes both basic and hydrophobic amino acids within the sequence RKKIQF (residues 6–11 of DARPP-32) and it is likely that the domain interacts with a region of PP-1C away from the active site.
Dephospho-DARPP-32 inhibits PP-1C with an IC50 which is ≈1000-fold higher than that observed for phospho-DARPP-32 (22). The present study indicates that inhibition by dephospho-DARPP-32 occurs via a mixed competitive/noncompetitive mechanism, which is similar to that observed for phospho-DARPP-32 (21). Direct binding studies of either DARPP-32 (22) or inhibitor-1 (24) have also shown that the dephosphorylated proteins interact with PP-1C. Together these previous results have raised the possibility that part of the inhibitory mechanism of dephospho- or phospho-DARPP-32 might involve an allosteric contribution from binding of subdomain 2 to PP-1C. However, the present studies support the idea that subdomain 2 represents only a binding site for dephospho- or phospho-DARPP-32. Dephospho-D32[1–38c] was found to be a very poor inhibitor of PP-1C. In contrast, the phosphorylated form of this peptide was as efficient an inhibitor as phospho-DARPP-32. Moreover, dephospho-D32[1–38c] or other peptides encompassing subdomain 2 acted as antagonists of phospho-DARPP-32 at concentrations in which they had no effect by themselves on PP-1C activity.
DARPP-32 and inhibitor-1 exhibit a high percentage of amino acid identity within their NH2 termini and the inhibitory potency of the two phospho-proteins is very similar. Within subdomain 2 there are slight differences (RKKIQF for DARPP-32 and PRKIQF for inhibitor-1). The results obtained suggest that the variable basic amino acids do not significantly influence the ability of the two proteins in their phosphorylated forms to inhibit PP-1C. However, we consistently observed that the antagonist peptides more readily blocked the actions of phospho-inhibitor-1. For example, D32[8–38] did not affect inhibition by phospho-DARPP-32 but did antagonize phospho-inhibitor-1. It has also been observed that, in contrast to dephospho-DARPP-32, dephospho-inhibitor-1 does not appear to inhibit PP-1C (24). This suggests that subdomain 2 of DARPP-32 may interact more strongly with PP-1C than does subdomain 2 of inhibitor-1. Alternatively, subdomain 1 of dephospho-DARPP-32, but not of dephospho-D32[1–38c] or dephospho-inhibitor-1, may interact with PP-1C in such a way to inhibit enzyme activity. Other studies indicate that both DARPP-32 and inhibitor-1 have limited secondary structure and that this is unaffected by phosphorylation of Thr-34/Thr-35 by PKA (24, 31). Phosphorylation of the COOH terminus of DARPP-32 by protein kinases, CK1 or CK2, does influence the dephosphorylation or phosphorylation, respectively, of Thr-34 of DARPP-32 (32, 33), suggesting that the NH2 and COOH termini of DARPP-32 interact. Stabilization of the NH2-terminal inhibitory domain of DARPP-32, but not of inhibitor-1, may explain in part the ability of only dephospho-DARPP-32 to inhibit PP-1C.
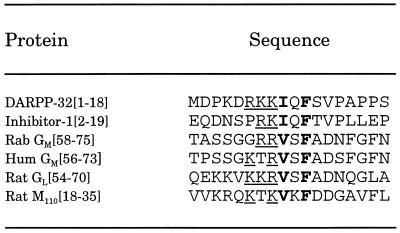
Recent studies have shown that the muscle and liver glycogen-targeting proteins, GM and GL, and the myofibrillar-targeting protein, M110, bind to a common site in PP-1C (ref. 12, but see ref. 34 for conflicting results). Short domains of GM (residues 63–75; GL has a related sequence between residues 59–72) and of M110 (residues 1–38) were able to either modulate the activities of PP-1C/targeting protein complexes or to dissociate the complexes (12). Notably, despite there being little overall amino acid sequence homology between the various proteins, these domains of GM, GL, and M110 contain basic and hydrophobic amino acids in a sequence that is very similar to that identified in subdomain 2 of DARPP-32 and inhibitor-1 (Table 2). Interestingly, the serine residue in this region of GM and GL is phosphorylated by PKA resulting in dissociation of PP-1C from either glycogen-binding protein (12). The effect of the short peptides derived from GM or M110 on inhibition of PP-1C by inhibitor-1 or inhibitor-2 have not yet been examined. However, recent studies have identified two novel PP-1C binding proteins in brain using the yeast two-hybrid method (P. Allen, unpublished data), and addition of D32[1–38c] has been found to antagonize the inhibition of PP-1C by the novel proteins (Y.-G.K. and P. Allen, unpublished data).
Table 2.
Comparison of binding domains of PP-1C inhibitors and targeting proteins.
The conserved phenylalanine residue was used to align the sequences. Conserved features of each sequence are one or more basic amino acids (underlined), a hydrophobic amino acid (boldface), a variable amino acid, and the phenylalanine residue (boldface). In GM and GL, the serine residue in the variable position is phosphorylated by PKA leading to dissociation of PP-1C from the targeting protein. Amino acid sequences were from (35, 36). Rab, rabbit; Hum, human.
The interaction of inhibitor-2 with PP-1C is believed to involve multiple sites that are related to inhibition and inactivation of the phosphatase (5). Kinetic analysis of inhibition of PP-1C by inhibitor-1 and inhibitor-2 (27), fluorescence binding studies of inhibitor-2 and okadaic acid (37), and site-directed mutagenesis analysis of PP-1C (38), suggest that part of inhibitor-2, perhaps involving phosphorylated Thr-72, may interact with the active site of PP-1C (5). The present studies have indicated that dephospho-DARPP-32 peptides can antagonize the inhibitory actions of inhibitor-2. This suggests that binding of phospho-DARPP-32 or inhibitor-2 to PP-1C involves both the active site as well as one or more additional sites removed from the active site. Other studies have suggested that binding of inhibitor-2 and targeting proteins such as the G or M proteins may be mutually exclusive (20). Our examination of the amino acid sequence of inhibitor-2 (39) has revealed that residues 134–138 (KKRQF) resemble the subdomain 2 sequence. However, the role of this domain is speculative since introduction of a fluorescent probe at residue 128 of inhibitor-2 had no effect on binding of the modified protein to PP-1C (37). Our examination of the amino acid sequences of other PP-1C binding proteins, for example, p53BP2, ribosomal protein L5, or sds22, have identified short stretches of amino acids similar to the RKKIQF motif of DARPP-32 (not shown) that could potentially be involved in binding to PP-1C. Therefore, it will be of interest to determine if all PP-1C binding proteins share this common domain, or if several completely distinct classes of PP-1C binding proteins exist.
The RKKIQF motif is common to DARPP-32, inhibitor-1, and certain other PP-1C binding proteins. However, additional domains of the inhibitor proteins and the targeting proteins bind to other distinct regions of PP-1C (see refs. 12 and 34 for discussion of the G proteins). Moreover, there must clearly be domains of targeting proteins that bind to, for example, glycogen or myofibrils. In contrast to DARPP-32, inhibitor-1, and inhibitor-2, which inhibit PP-1C by binding to the active site of the enzyme, targeting of PP-1C by other proteins does not necessarily occlude the active site, but frequently leads to changes in substrate specificity (1, 2, 12, 34). In the case of PP-1G, the complex of PP-1C and GM, the activity toward phosphorylase a is suppressed but the activity toward glycogen synthase is increased (12). Little is known about the interaction of substrate with PP-1C. The structure of the enzyme has revealed multiple surface grooves emanating from the active site (28). Site-directed mutagenesis studies suggest that amino acids surrounding the phosphorylation site of phosphorylase a might possibly occupy the COOH-terminal groove of PP-1C (30, 40). Thus, certain targeting proteins may occupy the phosphorylase a binding site while leaving alternative binding modes available in which other PP-1C surface grooves could accommodate specific substrates brought into immediate proximity by binding to the targeting proteins. In this respect it is of interest that in yeast PP-1C, mutation of a residue close to the COOH-terminal groove (equivalent to Arg-75 of mammalian PP-1C) reduces binding to GAC1, a G protein homolog (41).
In the central nervous system, DARPP-32 is highly expressed in striatum where it is localized to medium spiny neurons (2, 9). Within medium spiny neurons, DARPP-32 mediates many of the cellular responses to dopamine including regulation of the Na+, K+ ATPase, Na+ channels, and Ca2+ channels (9). Electrophysiological studies of the regulation of N- and P-type Ca2+ channels by dopamine have suggested that PP-1 is first activated and then subsequently inhibited by phospho-DARPP-32 (8). Immunoelectron microscopy studies have indicated that PP-1C is highly concentrated in dendritic spines in medium spiny neurons as well as in other types of neurons (18), presumably by a novel targeting protein. Regulation of the interaction of PP-1C with its targeting protein may therefore be important in mediating the control of N- and P-type Ca2+ channels by dopamine. The concentration of DARPP-32 in medium spiny neurons has been estimated to be 20–50 μM, and we have previously suggested (22) that DARPP-32 might interact with PP-1C even in its dephospho-form. However, the fact that PP-1C is highly localized to dendritic spines while DARPP-32 is found in the cytosol throughout the neuron suggests that the interaction of PP-1 with its targeting protein may preclude binding to dephospho-DARPP-32. It will be of interest to determine if the dendritic spine targeting protein shares the RKKIQF binding motif with DARPP-32.
In conclusion, the present studies support a model for the interaction of phospho-DARPP-32 with PP-1C that involves distinct binding and inhibitory domains. The interaction of subdomain 2 with a binding site removed from the active site of PP-1C potentially allows the development of nonpeptide molecules that could interact with this binding site in a manner in which the activity of PP-1C would not be affected. The determination of detailed structural information for the DARPP-32/PP-1C complex, as well as understanding the overlap of inhibitor and targeting protein interactions with PP-1C, will be useful in the development of such compounds, which might have value for the treatment of the dopamine disorders, schizophrenia, and Parkinson disease.
Acknowledgments
We thank Edgar da Cruz e Silva for help in analyzing the inhibitor-2 sequence. Supported by U.S. Public Health Service Grant MH 40899 (to A.C.N. and P.G.).
ABBREVIATIONS
- PP-1
-2A, -2B, and -2C, protein phosphatase 1, 2A, 2B, and 2C, respectively
- PP-1C
catalytic subunit of PP-1
- PKA
cAMP-dependent protein kinase
References
- 1.Cohen P. Annu Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 2.Shenolikar S, Nairn A C. Adv Second Messenger Phosphoprotein Res. 1991;23:1–121. [PubMed] [Google Scholar]
- 3.Shenolikar S. Annu Rev Cell Biol. 1994;10:55–86. doi: 10.1146/annurev.cb.10.110194.000415. [DOI] [PubMed] [Google Scholar]
- 4.MacKintosh C, MacKintosh R W. Trends Biochem Sci. 1994;19:444–448. doi: 10.1016/0968-0004(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 5.Park I-K, DePaoli-Roach A A. J Biol Chem. 1994;269:28919–28928. [PubMed] [Google Scholar]
- 6.Brautigan D L, Sunwoo J, Labbé J-C, Fernandez A, Lamb N J C. Nature (London) 1990;344:74–78. doi: 10.1038/344074a0. [DOI] [PubMed] [Google Scholar]
- 7.Mulkey R M, Endo S, Shenolikar S, Malenka R C. Nature (London) 1994;369:486–488. doi: 10.1038/369486a0. [DOI] [PubMed] [Google Scholar]
- 8.Surmeier D J, Bargas J, Hemmings H C, Jr, Nairn A C, Greengard P. Neuron. 1995;14:385–397. doi: 10.1016/0896-6273(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 9.Hemmings H C, Jr, Nairn A C, Bibb J A, Greengard P. In: Molecular and Cellular Mechanisms of Neostriatal Function. Ariano M A, Surmeier D J, editors. Austin, TX: Landes; 1995. pp. 279–293. [Google Scholar]
- 10.Hubbard M J, Cohen P. Trends Biochem Sci. 1993;18:172–177. doi: 10.1016/0968-0004(93)90109-z. [DOI] [PubMed] [Google Scholar]
- 11.Faux M C, Scott J D. Trends Biochem Sci. 1996;21:312–315. [PubMed] [Google Scholar]
- 12.Johnson D F, Moorhead G, Caudwell F B, Cohen P, Chen Y H, Chen M X, Cohen P T W. Eur J Biochem. 1996;239:317–325. doi: 10.1111/j.1432-1033.1996.0317u.x. [DOI] [PubMed] [Google Scholar]
- 13.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 14.Hirano K, Ito M, Hartshorne D J. J Biol Chem. 1995;270:19786–19790. doi: 10.1074/jbc.270.34.19786. [DOI] [PubMed] [Google Scholar]
- 15.Helps N R, Barker H M, Elledge S J, Cohen P T W. FEBS Lett. 1995;377:295–300. doi: 10.1016/0014-5793(95)01347-4. [DOI] [PubMed] [Google Scholar]
- 16.Van Eynde A, Wera S, Beullens M, Torrekens S, Van Leuven F, Stalmans W, Bollen M. J Biol Chem. 1995;270:28068–28074. doi: 10.1074/jbc.270.47.28068. [DOI] [PubMed] [Google Scholar]
- 17.Ohkura H, Yanagida M. Cell. 1991;64:149–157. doi: 10.1016/0092-8674(91)90216-l. [DOI] [PubMed] [Google Scholar]
- 18.Ouimet C C, da Cruz e Silva E F, Greengard P. Proc Natl Acad Sci USA. 1995;92:3396–3400. doi: 10.1073/pnas.92.8.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campos M, Fadden P, Alms G, Qian Z, Haystead T A J. J Biol Chem. 1996;271:28478–28484. doi: 10.1074/jbc.271.45.28478. [DOI] [PubMed] [Google Scholar]
- 20.Alessi D R, Street A J, Cohen P, Cohen P T W. Eur J Biochem. 1993;213:1055–1066. doi: 10.1111/j.1432-1033.1993.tb17853.x. [DOI] [PubMed] [Google Scholar]
- 21.Hemmings H C, Jr, Nairn A C, Elliott J I, Greengard P. J Biol Chem. 1990;265:20369–20376. [PubMed] [Google Scholar]
- 22.Desdouits F, Cheetham J J, Huang H-B, Kwon Y-G, da Cruz e Silva E F, Denefle P, Ehrlich M E, Nairn A C, Greengard P, Girault J-A. Biochem Biophys Res Commun. 1995;206:652–658. doi: 10.1006/bbrc.1995.1092. [DOI] [PubMed] [Google Scholar]
- 23.Aitken A, Cohen P. FEBS Lett. 1982;147:54–58. doi: 10.1016/0014-5793(82)81010-7. [DOI] [PubMed] [Google Scholar]
- 24.Endo S, Zhou X, Connor J, Wang B, Shenolikar S. Biochemistry. 1996;35:5220–5228. doi: 10.1021/bi952940f. [DOI] [PubMed] [Google Scholar]
- 25.Cohen P, Foulkes J G, Holmes C F, Nimmo G A, Tonks N K. Methods Enzymol. 1988;159:427–437. doi: 10.1016/0076-6879(88)59042-0. [DOI] [PubMed] [Google Scholar]
- 26.Cohen P, Alemany S, Hemmings B A, Resink T J, Stralfors P, Tung H Y. Methods Enzymol. 1988;159:390–408. doi: 10.1016/0076-6879(88)59039-0. [DOI] [PubMed] [Google Scholar]
- 27.Foulkes J G, Strada S J, Henderson P J, Cohen P. Eur J Biochem. 1983;132:309–313. doi: 10.1111/j.1432-1033.1983.tb07363.x. [DOI] [PubMed] [Google Scholar]
- 28.Goldberg J, Huang H-B, Kwon Y-G, Greengard P, Nairn A C, Kuriyan J. Nature (London) 1995;376:745–753. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L F, Zhang Z J, Long F X, Lee E Y C. Biochemistry. 1996;35:1606–1611. doi: 10.1021/bi9521396. [DOI] [PubMed] [Google Scholar]
- 30.Huang H-B, Horiuchi A, Goldberg J, Greengard P, Nairn A C. Proc Natl Acad Sci USA. 1997;94:3530–3535. doi: 10.1073/pnas.94.8.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neyroz P, Desdouits F, Benfenati F, Knutson J R, Greengard P, Girault J-A. J Biol Chem. 1993;268:24022–24031. [PubMed] [Google Scholar]
- 32.Girault J, Hemmings H C, Jr, Williams K R, Nairn A C, Greengard P. J Biol Chem. 1989;264:21748–21759. [PubMed] [Google Scholar]
- 33.Desdouits F, Siciliano J C, Greengard P, Girault J-A. Proc Natl Acad Sci USA. 1995;92:2682–2685. doi: 10.1073/pnas.92.7.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Kleiner U, Brautigan D L. Biochemistry. 1996;35:13858–13864. doi: 10.1021/bi961669e. [DOI] [PubMed] [Google Scholar]
- 35.Picking W D, Kudlicki W, Kramer G, Hardesty B, Vandenheede J R, Merlevede W, Park I-K, DePaoli-Roach A. Biochemistry. 1991;30:10280–10287. doi: 10.1021/bi00106a028. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, Zhao S, Long F, Zhang L, Bai G, Shima H, Nagao M, Lee E Y C. J Biol Chem. 1994;269:16997–17000. [PubMed] [Google Scholar]
- 37.Holmes C F, Campbell D G, Caudwell F B, Aitken A, Cohen P. Eur J Biochem. 1986;155:173–182. doi: 10.1111/j.1432-1033.1986.tb09473.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Zhang Z J, Brew K, Lee E Y C. Biochemistry. 1996;35:6276–6282. doi: 10.1021/bi952954l. [DOI] [PubMed] [Google Scholar]
- 39.Stuart J S, Frederick D L, Varner C M, Tatchell K. Mol Cell Biol. 1994;14:896–905. doi: 10.1128/mcb.14.2.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doherty M J, Moorhead G, Morrice N, Cohen P, Cohen P T W. FEBS Lett. 1995;375:294–298. doi: 10.1016/0014-5793(95)01184-g. [DOI] [PubMed] [Google Scholar]
- 41.Haystead C M M, Gailly P, Somlyo A P, Somlyo A V, Haystead T A J. FEBS Lett. 1995;377:123–127. doi: 10.1016/0014-5793(95)01318-0. [DOI] [PubMed] [Google Scholar]