Abstract
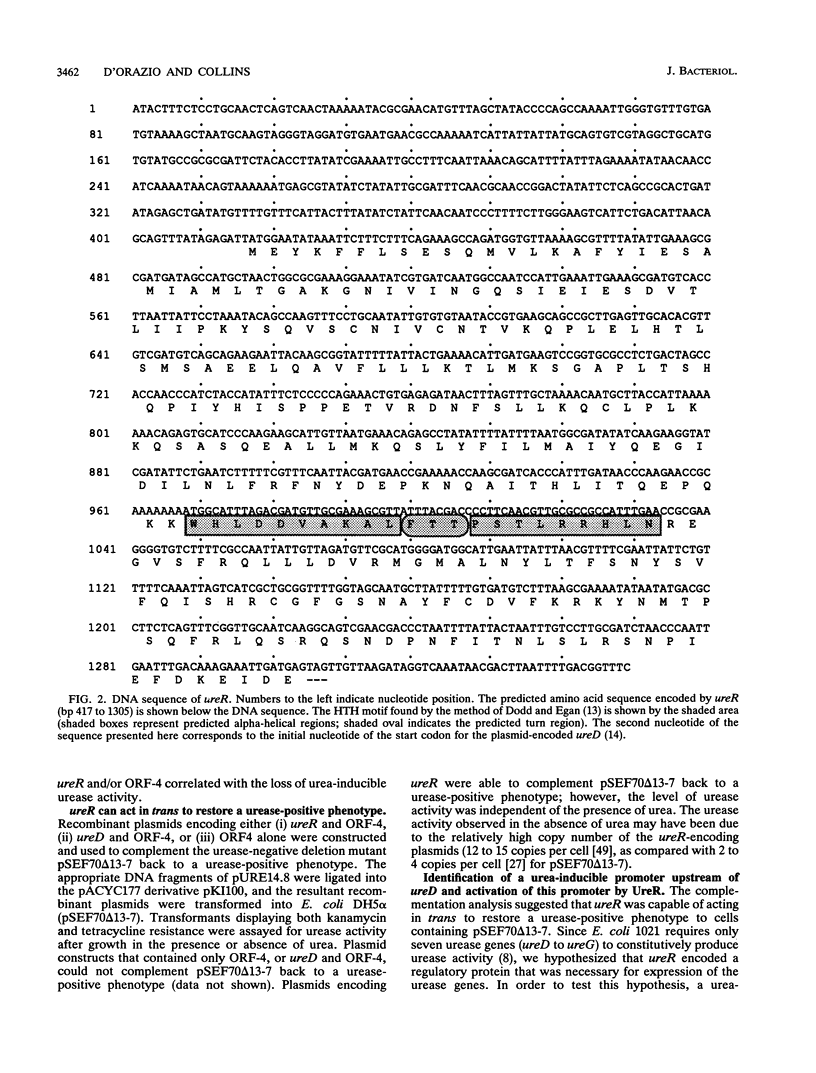
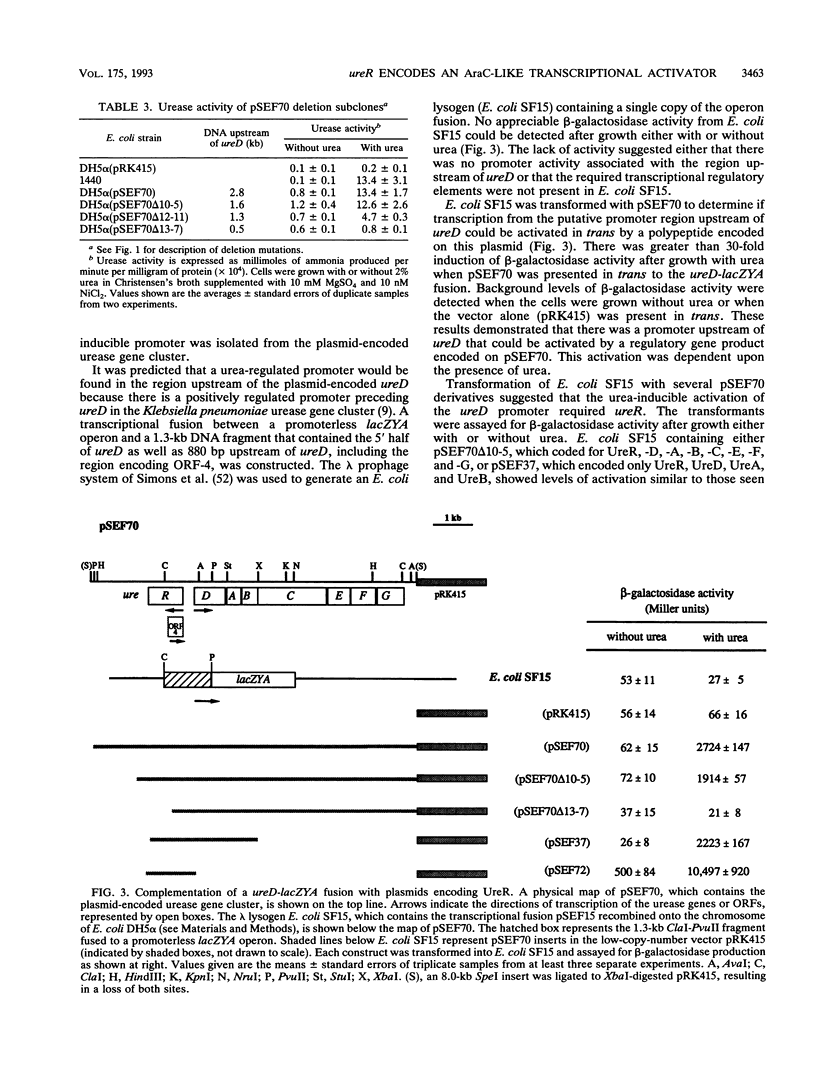
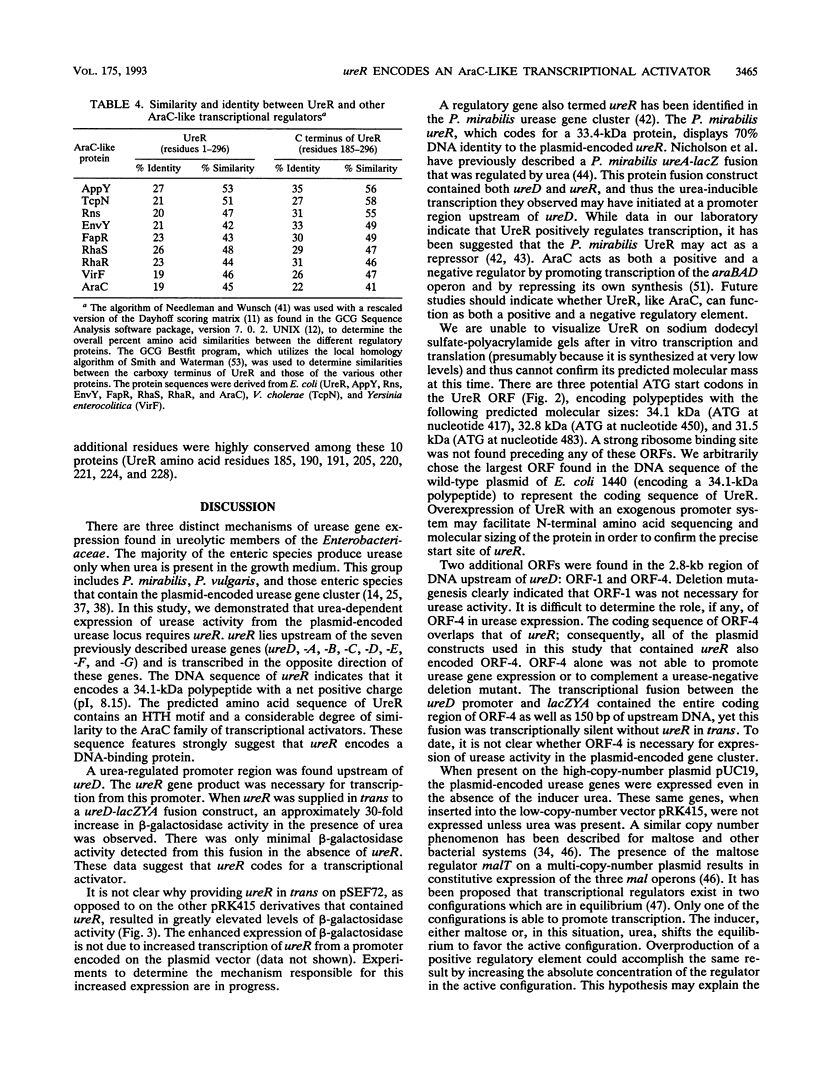
Ureolytic clinical isolates of Providencia stuartii, Salmonella spp., and some Escherichia coli strains contain large urease-encoding plasmids. Expression of urease activity from these isolates is induced at least 20-fold by urea. In order to facilitate studies on the regulatory mechanism controlling this urea-inducible expression, the plasmid-encoded urease genes were inserted into the low-copy-number vector pRK415, to form pSEF70. Deletion mutagenesis of pSEF70 demonstrated that between 1.3 and 1.6 kb of DNA upstream of ureD (the first of seven urease genes clustered in an operon-like fashion) was required for a urease-positive phenotype. An open reading frame coding for a 34.1-kDa polypeptide was found in the DNA sequence of this upstream region. This open reading frame has been designated ureR, for urease regulator. A urea-inducible promoter region was identified upstream of ureD. Transcription from this promoter was activated only when ureR was present in trans. The predicted ureR gene product contains a helix-turn-helix motif and shows significant amino acid similarity to the AraC family of transcriptional activators. We conclude that urea-dependent expression from the plasmid-encoded urease gene cluster requires ureR and that ureR codes for a positive regulatory element controlling transcription of at least one essential urease gene, ureD.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981 Apr 30;290(5809):754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Atlung T., Nielsen A., Hansen F. G. Isolation, characterization, and nucleotide sequence of appY, a regulatory gene for growth-phase-dependent gene expression in Escherichia coli. J Bacteriol. 1989 Mar;171(3):1683–1691. doi: 10.1128/jb.171.3.1683-1691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilofsky H. S., Burks C. The GenBank genetic sequence data bank. Nucleic Acids Res. 1988 Mar 11;16(5):1861–1863. doi: 10.1093/nar/16.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron J., Coffield L. M., Scott J. R. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc Natl Acad Sci U S A. 1989 Feb;86(3):963–967. doi: 10.1073/pnas.86.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen W. B. Urea Decomposition as a Means of Differentiating Proteus and Paracolon Cultures from Each Other and from Salmonella and Shigella Types. J Bacteriol. 1946 Oct;52(4):461–466. doi: 10.1128/jb.52.4.461-466.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C. M., Falkow S. Genetic analysis of Escherichia coli urease genes: evidence for two distinct loci. J Bacteriol. 1990 Dec;172(12):7138–7144. doi: 10.1128/jb.172.12.7138-7144.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C. M., Gutman D. M. Insertional inactivation of an Escherichia coli urease gene by IS3411. J Bacteriol. 1992 Feb;174(3):883–888. doi: 10.1128/jb.174.3.883-888.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C. M., Gutman D. M., Laman H. Identification of a nitrogen-regulated promoter controlling expression of Klebsiella pneumoniae urease genes. Mol Microbiol. 1993 Apr;8(1):187–198. doi: 10.1111/j.1365-2958.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Cornelis G., Sluiters C., de Rouvroit C. L., Michiels T. Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J Bacteriol. 1989 Jan;171(1):254–262. doi: 10.1128/jb.171.1.254-262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orazio S. E., Collins C. M. Characterization of a plasmid-encoded urease gene cluster found in members of the family Enterobacteriaceae. J Bacteriol. 1993 Mar;175(6):1860–1864. doi: 10.1128/jb.175.6.1860-1864.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990 Sep 11;18(17):5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich B., Magasanik B. Urease of Klebsiella aerogenes: control of its synthesis by glutamine synthetase. J Bacteriol. 1977 Aug;131(2):446–452. doi: 10.1128/jb.131.2.446-452.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatermann S., John J., Marre R. Staphylococcus saprophyticus urease: characterization and contribution to uropathogenicity in unobstructed urinary tract infection of rats. Infect Immun. 1989 Jan;57(1):110–116. doi: 10.1128/iai.57.1.110-116.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R. B., Penner J. L., Hennessy J. N., Jackowski B. J. Transferable urease activity in Providencia stuartii. J Clin Microbiol. 1981 Mar;13(3):561–565. doi: 10.1128/jcm.13.3.561-565.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Wallace J. C., Brown J. P. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 1990;183:111–132. doi: 10.1016/0076-6879(90)83009-x. [DOI] [PubMed] [Google Scholar]
- Higgins D. E., Nazareno E., DiRita V. J. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J Bacteriol. 1992 Nov;174(21):6974–6980. doi: 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L. T., Nicholson E. B., Jones B. D., Lynch M. J., Mobley H. L. Morganella morganii urease: purification, characterization, and isolation of gene sequences. J Bacteriol. 1990 Jun;172(6):3073–3080. doi: 10.1128/jb.172.6.3073-3080.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto K. S., Lory S. Formation of pilin in Pseudomonas aeruginosa requires the alternative sigma factor (RpoN) of RNA polymerase. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1954–1957. doi: 10.1073/pnas.86.6.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. D., Lockatell C. V., Johnson D. E., Warren J. W., Mobley H. L. Construction of a urease-negative mutant of Proteus mirabilis: analysis of virulence in a mouse model of ascending urinary tract infection. Infect Immun. 1990 Apr;58(4):1120–1123. doi: 10.1128/iai.58.4.1120-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. D., Mobley H. L. Proteus mirabilis urease: genetic organization, regulation, and expression of structural genes. J Bacteriol. 1988 Aug;170(8):3342–3349. doi: 10.1128/jb.170.8.3342-3349.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. D., Mobley H. L. Proteus mirabilis urease: nucleotide sequence determination and comparison with jack bean urease. J Bacteriol. 1989 Dec;171(12):6414–6422. doi: 10.1128/jb.171.12.6414-6422.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen N. T., Tamaki S., Kobayashi D., Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988 Oct 15;70(1):191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- Klaasen P., de Graaf F. K. Characterization of FapR, a positive regulator of expression of the 987P operon in enterotoxigenic Escherichia coli. Mol Microbiol. 1990 Oct;4(10):1779–1783. doi: 10.1111/j.1365-2958.1990.tb00556.x. [DOI] [PubMed] [Google Scholar]
- Lee M. H., Mulrooney S. B., Renner M. J., Markowicz Y., Hausinger R. P. Klebsiella aerogenes urease gene cluster: sequence of ureD and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. J Bacteriol. 1992 Jul;174(13):4324–4330. doi: 10.1128/jb.174.13.4324-4330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundrigan M. D., Friedrich M. J., Kadner R. J. Nucleotide sequence of the Escherichia coli porin thermoregulatory gene envY. Nucleic Acids Res. 1989 Jan 25;17(2):800–800. doi: 10.1093/nar/17.2.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- McLean R. J., Nickel J. C., Cheng K. J., Costerton J. W. The ecology and pathogenicity of urease-producing bacteria in the urinary tract. Crit Rev Microbiol. 1988;16(1):37–79. doi: 10.3109/10408418809104467. [DOI] [PubMed] [Google Scholar]
- Mermod N., Ramos J. L., Bairoch A., Timmis K. N. The xylS gene positive regulator of TOL plasmid pWWO: identification, sequence analysis and overproduction leading to constitutive expression of meta cleavage operon. Mol Gen Genet. 1987 May;207(2-3):349–354. doi: 10.1007/BF00331600. [DOI] [PubMed] [Google Scholar]
- Mobley H. L., Chippendale G. R., Fraiman M. H., Tenney J. H., Warren J. W. Variable phenotypes of Providencia stuartii due to plasmid-encoded traits. J Clin Microbiol. 1985 Nov;22(5):851–853. doi: 10.1128/jcm.22.5.851-853.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley H. L., Hausinger R. P. Microbial ureases: significance, regulation, and molecular characterization. Microbiol Rev. 1989 Mar;53(1):85–108. doi: 10.1128/mr.53.1.85-108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley H. L., Jones B. D., Jerse A. E. Cloning of urease gene sequences from Providencia stuartii. Infect Immun. 1986 Oct;54(1):161–169. doi: 10.1128/iai.54.1.161-169.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulrooney S. B., Hausinger R. P. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol. 1990 Oct;172(10):5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulrooney S. B., Pankratz H. S., Hausinger R. P. Regulation of gene expression and cellular localization of cloned Klebsiella aerogenes (K. pneumoniae) urease. J Gen Microbiol. 1989 Jun;135(6):1769–1776. doi: 10.1099/00221287-135-6-1769. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Nicholson E. B., Concaugh E. A., Foxall P. A., Island M. D., Mobley H. L. Proteus mirabilis urease: transcriptional regulation by UreR. J Bacteriol. 1993 Jan;175(2):465–473. doi: 10.1128/jb.175.2.465-473.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson E. B., Concaugh E. A., Mobley H. L. Proteus mirabilis urease: use of a ureA-lacZ fusion demonstrates that induction is highly specific for urea. Infect Immun. 1991 Oct;59(10):3360–3365. doi: 10.1128/iai.59.10.3360-3365.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogierman M. A., Manning P. A. Homology of TcpN, a putative regulatory protein of Vibrio cholerae, to the AraC family of transcriptional activators. Gene. 1992 Jul 1;116(1):93–97. doi: 10.1016/0378-1119(92)90634-2. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Restriction map of the Escherichia coli malA region and identification of the malT product. J Bacteriol. 1980 Aug;143(2):761–771. doi: 10.1128/jb.143.2.761-771.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J. L., Rojo F., Zhou L., Timmis K. N. A family of positive regulators related to the Pseudomonas putida TOL plasmid XylS and the Escherichia coli AraC activators. Nucleic Acids Res. 1990 Apr 25;18(8):2149–2152. doi: 10.1093/nar/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Tobin J. F., Schleif R. F. Positive regulation of the Escherichia coli L-rhamnose operon is mediated by the products of tandemly repeated regulatory genes. J Mol Biol. 1987 Aug 20;196(4):789–799. doi: 10.1016/0022-2836(87)90405-0. [DOI] [PubMed] [Google Scholar]
- Wallace R. G., Lee N., Fowler A. V. The araC gene of Escherichia coli: transcriptional and translational start-points and complete nucleotide sequence. Gene. 1980 Dec;12(3-4):179–190. doi: 10.1016/0378-1119(80)90100-6. [DOI] [PubMed] [Google Scholar]





