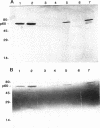
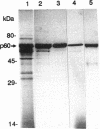
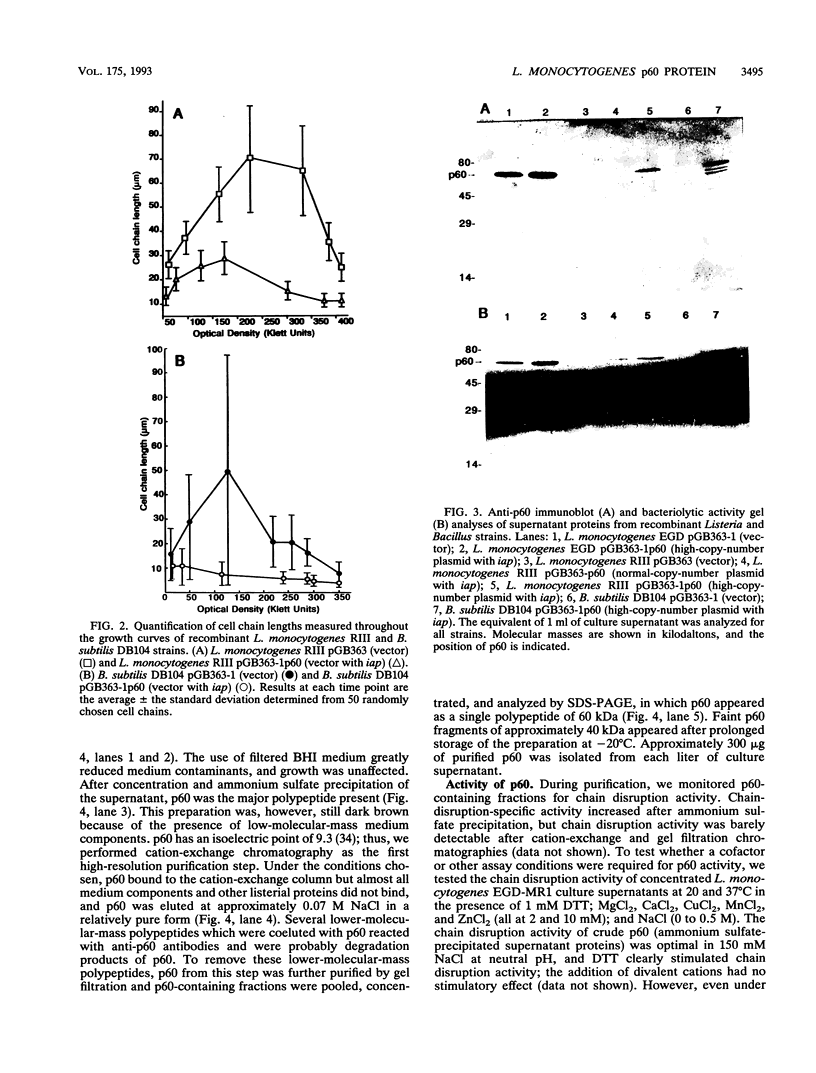
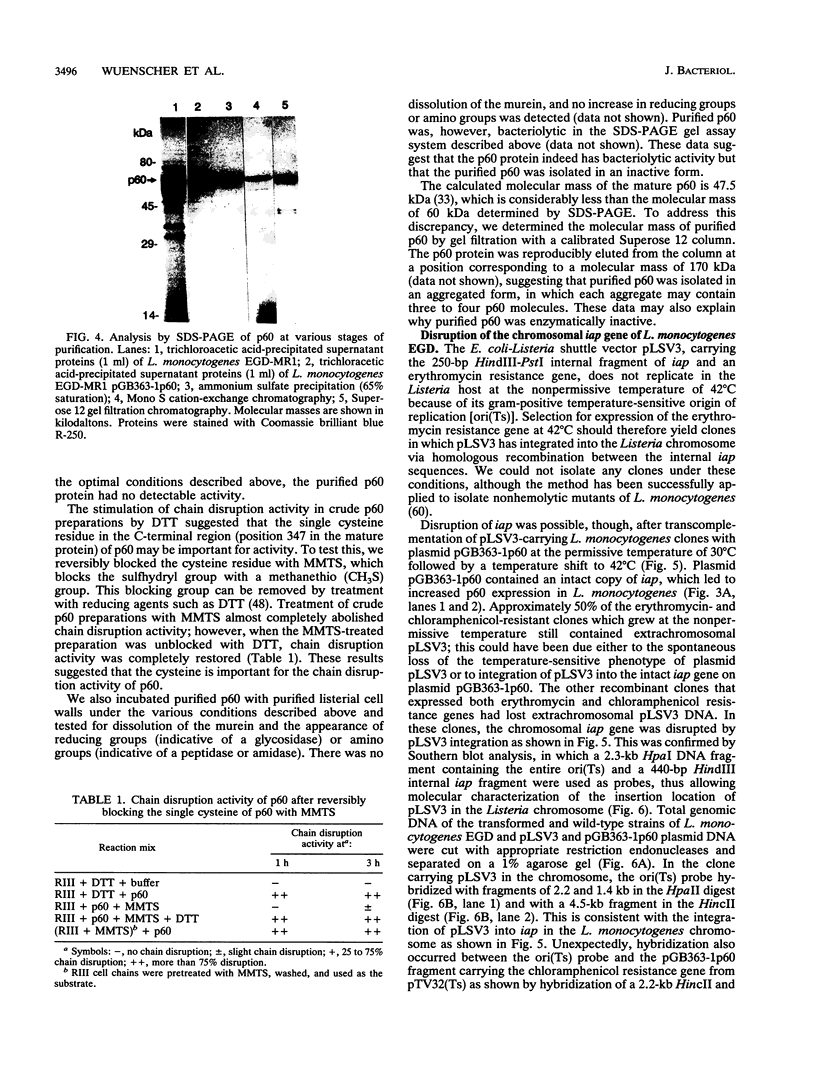
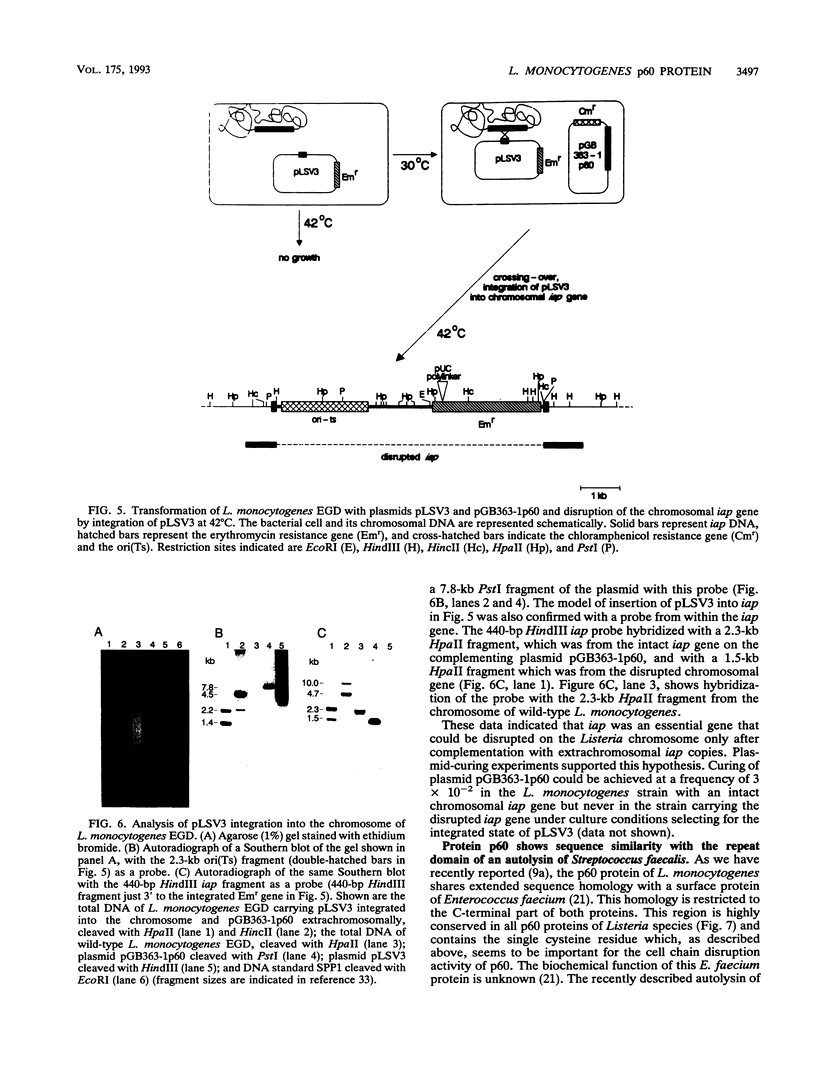
Abstract
Expression of the iap gene of Listeria monocytogenes in the L. monocytogenes rough mutant RIII and in Bacillus subtilis DB104 caused the disruption of the cell chains which these two strains normally form under exponential growth conditions. The p60 protein produced by L. monocytogenes and B. subtilis DB104 also exhibited bacteriolytic activity detected in denaturing polyacrylamide gels containing heat-killed Micrococcus lysodeikticus. Purification of the p60 protein led to aggregation of p60 and loss of the cell chain disruption and bacteriolytic activities. A cysteine residue in the C-terminal part of p60 which is conserved in all p60-like proteins from the other Listeria species seems to be essential for both activities. The iap gene could not be inactivated without a loss of cell viability, indicating that p60 is an essential housekeeping protein for L. monocytogenes and probably also for other Listeria species. These data suggest that p60 possesses a murein hydrolase activity required for a late step in cell division.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker L. A., Campbell P. A., Hollister J. R. Chemotaxigenesis and complement fixation by Listeria monocytogenes cell wall fractions. J Immunol. 1977 Nov;119(5):1723–1726. [PubMed] [Google Scholar]
- Beall B., Lutkenhaus J. Sequence analysis, transcriptional organization, and insertional mutagenesis of the envA gene of Escherichia coli. J Bacteriol. 1987 Dec;169(12):5408–5415. doi: 10.1128/jb.169.12.5408-5415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke D., Gilmore M. S. Location of antibiotic resistance determinants, copy control, and replication functions on the double-selective streptococcal cloning vector pGB301. Mol Gen Genet. 1981;184(1):115–120. doi: 10.1007/BF00271206. [DOI] [PubMed] [Google Scholar]
- Berry A. M., Lock R. A., Hansman D., Paton J. C. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect Immun. 1989 Aug;57(8):2324–2330. doi: 10.1128/iai.57.8.2324-2330.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouanchaud D. H., Scavizzi M. R., Chabbert Y. A. Elimination by ethidium bromide of antibiotic resistance in enterobacteria and staphylococci. J Gen Microbiol. 1968 Dec;54(3):417–425. doi: 10.1099/00221287-54-3-417. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bubert A., Kuhn M., Goebel W., Köhler S. Structural and functional properties of the p60 proteins from different Listeria species. J Bacteriol. 1992 Dec;174(24):8166–8171. doi: 10.1128/jb.174.24.8166-8171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubert A., Köhler S., Goebel W. The homologous and heterologous regions within the iap gene allow genus- and species-specific identification of Listeria spp. by polymerase chain reaction. Appl Environ Microbiol. 1992 Aug;58(8):2625–2632. doi: 10.1128/aem.58.8.2625-2632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béliveau C., Potvin C., Trudel J., Asselin A., Bellemare G. Cloning, sequencing, and expression in Escherichia coli of a Streptococcus faecalis autolysin. J Bacteriol. 1991 Sep;173(18):5619–5623. doi: 10.1128/jb.173.18.5619-5623.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domann E., Leimeister-Wächter M., Goebel W., Chakraborty T. Molecular cloning, sequencing, and identification of a metalloprotease gene from Listeria monocytogenes that is species specific and physically linked to the listeriolysin gene. Infect Immun. 1991 Jan;59(1):65–72. doi: 10.1128/iai.59.1.65-72.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domann E., Wehland J., Rohde M., Pistor S., Hartl M., Goebel W., Leimeister-Wächter M., Wuenscher M., Chakraborty T. A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 1992 May;11(5):1981–1990. doi: 10.1002/j.1460-2075.1992.tb05252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle R. J., Motley M. A., Carstens P. H. Turnover of cell wall in Listeria monocytogenes. Carbohydr Res. 1982 Jun 1;104(1):147–152. doi: 10.1016/s0008-6215(00)82227-2. [DOI] [PubMed] [Google Scholar]
- Fan D. P. Autolysin(s) of Bacillus subtilis as dechaining enzyme. J Bacteriol. 1970 Aug;103(2):494–499. doi: 10.1128/jb.103.2.494-499.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber J. M., Peterkin P. I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991 Sep;55(3):476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fein J. E., Rogers H. J. Autolytic enzyme-deficient mutants of Bacillus subtilis 168. J Bacteriol. 1976 Sep;127(3):1427–1442. doi: 10.1128/jb.127.3.1427-1442.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Flamm R. K., Hinrichs D. J., Thomashow M. F. Introduction of pAM beta 1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect Immun. 1984 Apr;44(1):157–161. doi: 10.1128/iai.44.1.157-161.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg C., Rogers H. J. Autolytic enzymes in growth of bacteria. Nature. 1971 Jan 22;229(5282):272–273. doi: 10.1038/229272a0. [DOI] [PubMed] [Google Scholar]
- Fürst P., Mösch H. U., Solioz M. A protein of unusual composition from Enterococcus faecium. Nucleic Acids Res. 1989 Aug 25;17(16):6724–6724. doi: 10.1093/nar/17.16.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard J. L., Berche P., Frehel C., Gouin E., Cossart P. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell. 1991 Jun 28;65(7):1127–1141. doi: 10.1016/0092-8674(91)90009-n. [DOI] [PubMed] [Google Scholar]
- Gaillard J. L., Berche P., Mounier J., Richard S., Sansonetti P. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect Immun. 1987 Nov;55(11):2822–2829. doi: 10.1128/iai.55.11.2822-2829.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltje J. V., Tomasz A. Lipoteichoic acid: a specific inhibitor of autolysin activity in Pneumococcus. Proc Natl Acad Sci U S A. 1975 May;72(5):1690–1694. doi: 10.1073/pnas.72.5.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltje J. V., Tuomanen E. I. The murein hydrolases of Escherichia coli: properties, functions and impact on the course of infections in vivo. J Gen Microbiol. 1991 Mar;137(3):441–454. doi: 10.1099/00221287-137-3-441. [DOI] [PubMed] [Google Scholar]
- Jolliffe L. K., Doyle R. J., Streips U. N. The energized membrane and cellular autolysis in Bacillus subtilis. Cell. 1981 Sep;25(3):753–763. doi: 10.1016/0092-8674(81)90183-5. [DOI] [PubMed] [Google Scholar]
- Kawamura F., Doi R. H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol. 1984 Oct;160(1):442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C., Gouin E., Tabouret M., Berche P., Ohayon H., Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992 Feb 7;68(3):521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- Kuhn M., Goebel W. Identification of an extracellular protein of Listeria monocytogenes possibly involved in intracellular uptake by mammalian cells. Infect Immun. 1989 Jan;57(1):55–61. doi: 10.1128/iai.57.1.55-61.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M., Kathariou S., Goebel W. Hemolysin supports survival but not entry of the intracellular bacterium Listeria monocytogenes. Infect Immun. 1988 Jan;56(1):79–82. doi: 10.1128/iai.56.1.79-82.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984 Dec;10(3-4):203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Köhler S., Bubert A., Vogel M., Goebel W. Expression of the iap gene coding for protein p60 of Listeria monocytogenes is controlled on the posttranscriptional level. J Bacteriol. 1991 Aug;173(15):4668–4674. doi: 10.1128/jb.173.15.4668-4674.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S., Leimeister-Wächter M., Chakraborty T., Lottspeich F., Goebel W. The gene coding for protein p60 of Listeria monocytogenes and its use as a specific probe for Listeria monocytogenes. Infect Immun. 1990 Jun;58(6):1943–1950. doi: 10.1128/iai.58.6.1943-1950.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leimeister-Wächter M., Domann E., Chakraborty T. Detection of a gene encoding a phosphatidylinositol-specific phospholipase C that is co-ordinately expressed with listeriolysin in Listeria monocytogenes. Mol Microbiol. 1991 Feb;5(2):361–366. doi: 10.1111/j.1365-2958.1991.tb02117.x. [DOI] [PubMed] [Google Scholar]
- Leimeister-Wächter M., Haffner C., Domann E., Goebel W., Chakraborty T. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of listeria monocytogenes. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8336–8340. doi: 10.1073/pnas.87.21.8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier J., Ryter A., Coquis-Rondon M., Sansonetti P. J. Intracellular and cell-to-cell spread of Listeria monocytogenes involves interaction with F-actin in the enterocytelike cell line Caco-2. Infect Immun. 1990 Apr;58(4):1048–1058. doi: 10.1128/iai.58.4.1048-1058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet A., Jr, Raines K. M., Brownback P. C. Immunopotentiating activities of cell walls, peptidoglycans, and teichoic acids from two strains of Listeria monocytogenes. Infect Immun. 1986 Oct;54(1):170–176. doi: 10.1128/iai.54.1.170-176.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy D. A., Chakraborty T., Goebel W., Cossart P. Molecular determinants of Listeria monocytogenes pathogenesis. Infect Immun. 1992 Apr;60(4):1263–1267. doi: 10.1128/iai.60.4.1263-1267.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy D. A., Jacks P. S., Hinrichs D. J. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med. 1988 Apr 1;167(4):1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potel J., Schulze-Lammers J. Listeria monocytogenes-vaccine: production and control. Zentralbl Bakteriol Mikrobiol Hyg A. 1985 May;259(3):331–340. doi: 10.1016/s0176-6724(85)80035-3. [DOI] [PubMed] [Google Scholar]
- Potvin C., Leclerc D., Tremblay G., Asselin A., Bellemare G. Cloning, sequencing and expression of a Bacillus bacteriolytic enzyme in Escherichia coli. Mol Gen Genet. 1988 Oct;214(2):241–248. doi: 10.1007/BF00337717. [DOI] [PubMed] [Google Scholar]
- Sanchez-Puelles J. M., Ronda C., Garcia J. L., Garcia P., Lopez R., Garcia E. Searching for autolysin functions. Characterization of a pneumococcal mutant deleted in the lytA gene. Eur J Biochem. 1986 Jul 15;158(2):289–293. doi: 10.1111/j.1432-1033.1986.tb09749.x. [DOI] [PubMed] [Google Scholar]
- Smith D. J., Maggio E. T., Kenyon G. L. Simple alkanethiol groups for temporary blocking of sulfhydryl groups of enzymes. Biochemistry. 1975 Feb 25;14(4):766–771. doi: 10.1021/bi00675a019. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sugai M., Akiyama T., Komatsuzawa H., Miyake Y., Suginaka H. Characterization of sodium dodecyl sulfate-stable Staphylococcus aureus bacteriolytic enzymes by polyacrylamide gel electrophoresis. J Bacteriol. 1990 Nov;172(11):6494–6498. doi: 10.1128/jb.172.11.6494-6498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugai M., Koike H., Hong Y. M., Miyake Y., Nogami R., Suginaka H. Purification of a 51 kDa endo-beta-N-acetylglucosaminidase from Staphylococcus aureus. FEMS Microbiol Lett. 1989 Oct 15;52(3):267–272. doi: 10.1016/0378-1097(89)90209-7. [DOI] [PubMed] [Google Scholar]
- Tilney L. G., Connelly P. S., Portnoy D. A. Actin filament nucleation by the bacterial pathogen, Listeria monocytogenes. J Cell Biol. 1990 Dec;111(6 Pt 2):2979–2988. doi: 10.1083/jcb.111.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney L. G., Portnoy D. A. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol. 1989 Oct;109(4 Pt 1):1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A., Moreillon P., Pozzi G. Insertional inactivation of the major autolysin gene of Streptococcus pneumoniae. J Bacteriol. 1988 Dec;170(12):5931–5934. doi: 10.1128/jb.170.12.5931-5934.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell E. A. Autolysis of listeria monocytogenes. J Bacteriol. 1973 Feb;113(2):1046–1048. doi: 10.1128/jb.113.2.1046-1048.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valisena S., Varaldo P. E., Satta G. Purification and characterization of three separate bacteriolytic enzymes excreted by Staphylococcus aureus, Staphylococcus simulans, and Staphylococcus saprophyticus. J Bacteriol. 1982 Aug;151(2):636–647. doi: 10.1128/jb.151.2.636-647.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Boland J. A., Kocks C., Dramsi S., Ohayon H., Geoffroy C., Mengaud J., Cossart P. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect Immun. 1992 Jan;60(1):219–230. doi: 10.1128/iai.60.1.219-230.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf-Watz H., Normark S. Evidence for a role of N-acetylmuramyl-L-alanine amidase in septum separation in Escherichia coli. J Bacteriol. 1976 Nov;128(2):580–586. doi: 10.1128/jb.128.2.580-586.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuenscher M. D., Köhler S., Goebel W., Chakraborty T. Gene disruption by plasmid integration in Listeria monocytogenes: insertional inactivation of the listeriolysin determinant lisA. Mol Gen Genet. 1991 Aug;228(1-2):177–182. doi: 10.1007/BF00282463. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]