Abstract
Antisense RNA may regulate the expression of a number of eukaryotic genes, but little is known about its prevalence or mechanism of action. We have used a model system in which antisense control can be studied both genetically and biochemically. Late in polyoma virus infection, early-strand mRNA levels are down-regulated by nuclear antisense RNA from the late strand. Analysis of early-strand transcripts isolated late in infection revealed extensive base modifications. In many transcripts almost half of the adenosines were altered to inosines or guanosines. These results suggest modification of RNA duplexes by double-stranded RNA adenosine deaminase or a related enzyme. Probes that detect only modified RNAs revealed that these molecules are not highly unstable, but accumulate within the nucleus and are thus inert for gene expression. Antisense-induced modifications can account for most or all of the observed regulation, with the lowered levels of early-strand RNAs commonly observed late in infection resulting from the fact that many transcripts are invisible to standard hybridization probes. This work suggests that similar antisense-mediated control mechanisms may also operate under physiological conditions in uninfected eukaryotic cells, and leads to the proposal that there is a novel pool of nuclear RNAs that cannot be seen with many molecular probes heretofore used.
Naturally occurring antisense RNA has been found both in prokaryotes and in eukaryotes. In prokaryotes, where it was first discovered, numerous examples of antisense-mediated regulation of gene expression have been reported (1). The mechanism of action of this regulation is well understood, and is usually at the translational level (2). In eukaryotes, relatively few examples of antisense RNA-mediated gene regulation have been reported (3–8). The regulation is thought to occur primarily in the nucleus (9, 10) and rarely, if at all, at the translational level (9, 11).
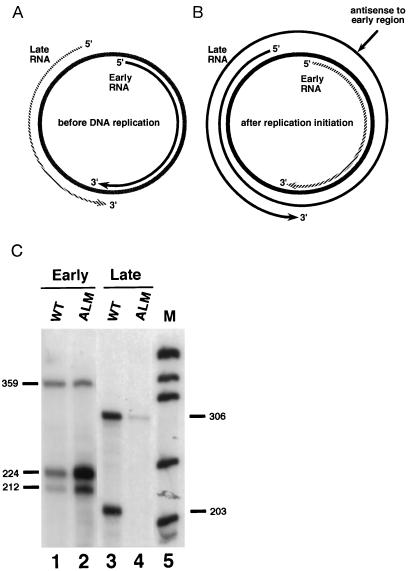
The mouse polyoma virus serves as a useful and general model system to study naturally occurring mammalian antisense-induced regulation of transcript levels, as well as the fate of double-stranded RNA (dsRNA) molecules within the nucleus. This virus is small and depends heavily on the host for its gene expression. The double stranded, circular, polyoma genome is divided into early and late transcription units that are expressed from opposite strands of the viral genome. The life cycle of this virus is divided into two phases: the early phase, which occurs immediately after infection and before DNA replication, and the late phase, which begins after the onset of DNA replication. There exists striking temporal regulation of early- and late-strand transcript levels in polyoma. During the early phase of productive infection, early-strand transcripts accumulate preferentially over late-strand transcripts (12–21). The late-strand transcripts are synthesized during this period, but are presumably degraded rapidly in the nucleus (21). However, at late times the late-strand transcripts are much more abundant than the early-strand transcripts (12–21). As expected, a DNA replication-dependent increase in the levels of the late-strand transcripts is observed. Interestingly, a corresponding replication-dependent increase in the levels of the early-strand transcripts is not seen; instead, down-regulation of early-strand RNA levels is observed. Previous studies have shown that the efficiency of both the early as well as the late promoters is constant throughout infection and that regulation of both early- and late-strand RNA levels is posttranscriptional (16, 17). The stability of the late-strand transcripts increases at the onset of the late phase. This has been reported to be linked to the inefficiency of late-strand transcription termination at late times. During the late phase of infection, RNA polymerase II encircles the genome multiple times (refs. 16 and 21–27; Fig. 1B). These giant multigenomic transcripts have multiple copies of the small, 57-base noncoding “late leader” exon, which is expressed immediately downstream of the late promoter. Leader-to-leader splicing occurs, and this event appears to stabilize late-strand transcripts (21). Thus, late in infection, late-strand transcripts are much more abundant than those from the early strand. Further, these multigenomic RNAs have the potential to form sense–antisense hybrids with early-strand transcripts (Fig. 1B). Importantly, the region of the late strands that can form antisense hybrids with early-strand RNAs is entirely within intronic sequences that do not appear in late-strand messages.
Figure 1.
Temporal regulation of polyoma virus transcript levels. (A) During the early phase of viral infection, early-strand transcripts accumulate preferentially over late-strand transcripts. Late-strand transcripts are processed inefficiently and are relatively unstable. Prior to DNA replication the ratio of late-strand to early-strand RNAs is less than 1:10. (B) During the late phase of infection, after the onset of DNA replication, late-strand transcripts are more abundant than early-strand transcripts. Transcription termination is inefficient during this period, allowing RNA polymerase II to encircle the genome multiple times. The resulting multigenomic transcripts contain sequences complementary to early-strand transcripts and thus have the potential to act as natural antisense regulators within the nucleus. Hatched lines denote unstable transcripts. (C) The antisense effect. Total cell RNA was harvested 48 hr after transfection with either wild-type genomes (WT) or mutant ALM (which contains a 51-bp deletion in the viral late region, which leads to unstable late-strand primary transcripts) genomes, and analyzed for early-strand (lanes 1 and 2) and late-strand (lanes 3 and 4) transcripts using specific riboprobes that span splicing junctions. Positions of expected early-strand and late-strand bands from wild-type and marked constructs are indicated. M, molecular weight markers produced by digesting pUC18 with MspI. Quantitation using a Packard InstantImager revealed that mutant ALM produced less than 10% as much late-strand RNA as wild type, and overexpressed early-strand RNAs by a factor of 5.
We previously presented two lines of evidence demonstrating that early-strand RNA levels are, in fact, down-regulated by these antisense late-strand RNAs in the nucleus (28). First, expression of nuclear-targeted antisense RNA in trans resulted in substantially reduced levels of early-strand RNAs in a dose-dependent manner (28). Second, numerous mutations affecting the nuclear stability of antisense transcripts indicated that sense and antisense levels appear to be reciprocally regulated: low levels of antisense transcripts always correlate with high levels of early-strand RNAs, and vice versa (28). However, the exact mechanism of down-regulation of early-strand transcripts by the antisense transcripts was not known.
In the present work, we have examined the fate of these hypothetical sense–antisense hybrids in the nucleus. More specifically, we have asked whether the reduced levels of early-strand RNAs at late times in infection result from antisense-induced RNA degradation or rather, are early-strand transcripts, as a result of base modifications, merely invisible (noncomplementary) to the molecular probes used in RNase protection assays? This latter possibility was considered because mammalian nuclei contain a well-described enzymatic activity that is capable of modifying dsRNA molecules. The enzyme dsRAD/DRADA (double-stranded RNA-specific adenosine deaminase), originally discovered in the nucleus of Xenopus laevis (29, 30), was subsequently found to be ubiquitous in the animal kingdom (31). This activity is largely confined to the nucleus [although it was reported recently that a cytoplasmic form of this deaminase may be induced by interferon (32)] and catalyzes the conversion of adenosines to inosines within dsRNA (33, 34) by the mechanism of hydrolytic deamination (35). The resultant RNA contains I–U base pairs, which may lead to partial unwinding of the RNA duplex (34). In vitro studies suggest that the only substrate for this enzyme is dsRNA. The activity cannot be competitively inhibited by single-stranded RNA (ssRNA), dsDNA, or ssDNA (36). It has been demonstrated that the mammalian glutamate receptor subunit mRNA (37–39) and hepatitis delta virus RNA (40, 41) are in vivo substrates for modification by dsRAD/DRADA. Other cases where such modifications have been detected include RNAs from the defective measles virus, other negative-strand RNA viruses and retroviruses (refs. 42–46 and references therein), as well as in the 4f-rnp transcripts in Drosophila (47). Finally, dsRAD/DRADA has been postulated to be involved in antisense regulation (48, 49), although no in vivo evidence has been demonstrated so far.
To elucidate the fate of dsRNAs in the nucleus, we set out to determine whether early-strand transcripts are modified at late times in infection. Adenosine modifications would suggest that early-strand RNA hybridizes to complementary late-strand RNA, resulting in deamination by dsRAD/DRADA or a related enzyme such as RED1 (38). Our results demonstrate that about half of all detectable early-strand RNAs in the nucleus at late times contain modified adenosines (which appear as guanosines when sequenced), and in these modified transcripts, about 50% of the adenosines are modified. The base preferences of the modified adenosines are consistent with the possibility that the modifying activity is dsRAD/DRADA or a related enzyme. Since the only substrate for dsRAD/DRADA and related enzymes is dsRNA, adenosine-to-guanosine (or inosine) modifications suggest that sense–antisense duplexes are formed within the nuclei of polyoma-infected cells, and these are subsequently modified. Furthermore, using probes that specifically detect modified transcripts, the fate of these transcripts was followed. Our results indicate that these extensively modified transcripts are retained in the nucleus; they are not transported to the cytoplasm and hence become dead-end transcripts that are not translated.
MATERIALS AND METHODS
Cell Culture, Infections, and Transfections.
Mouse NIH 3T3 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, l-glutamine, penicillin, and streptomycin at 37°C in 5% CO2. Cells were infected at 35–40% confluency with polyoma strain 59RA (multiplicity of infection = 5). Transfections were carried out with a modified calcium phosphate DNA coprecipitation method (50). In experiments to measure RNA decay rates, actinomycin D was added to transfected cultures at a final concentration of 20 μg/ml.
To block DNA replication, the cells were treated with aphidicolin 1 hr posttransfection, and 48 hr later total RNA was harvested using the single-step guanidinium isothiocyanate method (51, 52). RNase protection analysis revealed the presence of sufficient amounts of early RNAs, while late-strand RNAs were barely detectable (data not shown). These cells express very low levels of late-strand transcripts.
RNA Isolation.
Nuclear and cytoplasmic RNAs were isolated at various time points postinfection by a modification of the guanidine isothiocyanate method (51, 52). Total RNA was harvested using the single-step guanidinium isothiocyanate method (51, 52).
RNase Protection Assays.
Internally labeled RNA probes were made by in vitro transcription by T3 or T7 RNA polymerase in the presence of [α-32P]UTP. DNA templates were removed by RQ1 DNase digestion followed by phenol/chloroform extraction. The riboprobes were hybridized to target RNAs at 60°C overnight, as described (53). The hybridization products were digested with a T1/T2 mixture (54) at 37°C for 1.5 hr, and the resulting samples were resolved on 6% denaturing polyacrylamide gels. Routinely, 40 μg of RNA samples were used for RNase protection. The protected bands were quantitated using a Packard InstantImager. Background was subtracted using regions of identical size located immediately below each of the experimental bands.
Reverse Transcription–PCR (RT-PCR).
RNAs were reverse transcribed and amplified using the Gene Amp RNA PCR Kit (Perkin–Elmer/Cetus) according to the manufacturer’s instructions, using primers that would hybridize to the RNA irrespective of adenosine modifications. The primer used for RT was 5′-AGAAAGAACAGCA-3′. The second primer used for PCR amplification was 5′-TCCCCCTGCTCCT-3′.
Cloning and Sequencing.
The desired fragments were cloned into pBluescript SK(+) by standard cloning protocols (51, 52). Sequencing was carried out by the dideoxy chain termination method using the Sequenase Version 2.0 DNA sequencing kit (United States Biochemical), according to the manufacturer’s protocol.
RESULTS
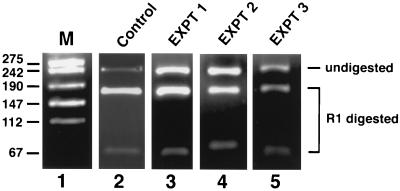
During the polyoma virus life cycle, early-strand RNA levels are down-regulated by antisense transcripts from the late strand. Previous results from our laboratory have shown that mutations that destabilize the late-strand transcripts result in increased expression of the early-strand RNAs (28, 55, 56). An example of this antisense-induced regulation is shown in Fig. 1C. Mutant ALM contains a 51-bp deletion in the late region, which leads to unstable late-strand primary transcripts and consequently only very low levels of antisense RNA to the early strand (Fig. 1C). This mutant overexpresses early-strand RNAs about 5-fold. This 5-fold difference in early-strand RNA levels represents the typical antisense effect in the polyoma system (refs. 28, 55 and 56; data not shown). We set out to investigate whether the mechanism of antisense-mediated repression of early-strand RNA levels is via the formation of sense–antisense hybrids that are subsequently modified by dsRAD or a related enzyme. To this end, nuclear RNA was harvested 30 hr postinfection, a time point when both sense and antisense transcripts coexist within the nucleus. Early region RNA was amplified by RT-PCR using primers that were insensitive to adenosine modifications. Thus, the RT primer lacked T residues, and the second PCR primer lacked A residues. The amplified DNA fragment harbors a unique EcoRI site and will become EcoRI-resistant should any A residue within this site be modified to any other residue. As expected, a 220-bp PCR product was obtained that corresponded to the correct fragment size. In three independent experiments we observed that 40–50% of the 220-bp amplified fragment was resistant to EcoRI (Fig. 2, lanes 3–5). In the control experiment DNA replication was blocked by addition of aphidicolin, thus preventing accumulation of late-strand RNA and hence the antisense effect. As a result, almost all of the RT-PCR-amplified fragments were sensitive to EcoRI (Fig. 2, lane 2). In fact, in this experiment, when the small amount of EcoRI-resistant DNA from the control experiment was excised from the gel and redigested with EcoR1, almost all was cut, while the EcoRI-resistant bands from the experimental lanes remained mostly resistant (data not shown). Primers specific for a different part of the polyoma early region spanning a DraI restriction enzyme site revealed about 50% DraI-resistant RNAs, and primers specific for the complementary, late-strand, also revealed modifications (data not shown). The preceding results suggest that a significant level of modification of dsRNA may have occurred within the sense–antisense duplex RNA. It should be noted that EcoRI-resistant fragments may represent only part of the total pool of modified fragments, because not all modified transcripts may have sustained modifications within the EcoRI site.
Figure 2.
Analysis of RT-PCR products. A region of the early sense strand was reverse-transcribed and amplified by RT-PCR using primers that would hybridize to the RNA irrespective of adenosine modifications. The primer used for reverse transcription was 5′-AGAAAGAACAGCA-3′. The second primer used for PCR amplification was 5′-TCCCCCTGCTCCT-3′. The amplified product was gel-purified and subjected to EcoRI digestion. The bands were separated by agarose gel electrophoresis. Lane 1, marker. Lane 2, control experiment where cells were treated with aphidicolin to block DNA replication. RNase protection analysis revealed the presence of sufficient amounts of early-strand RNAs, while late-strand RNAs were barely detectable (data not shown). These cells express very low levels of late-strand transcripts. Lanes 3–5, results of three independent experiments. Bands corresponding to the EcoRI-digested and resistant fragments are indicated. As seen in the figure, a considerable amount of RT-PCR amplified product was resistant to EcoRI as compared with the control. The EcoRI-resistant band was purified, cloned into pBluescript SK(+), and sequenced.
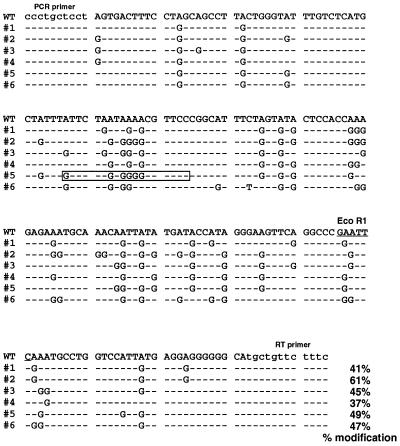
To determine whether base modifications had actually occurred, EcoRI-resistant, 220-bp fragments (Fig. 2, lanes 3–5) were purified, cloned, and sequenced. Comparison of the genomic and cDNA sequences of six different clones are shown in Fig. 3. Strikingly, about half the adenosines were mutated to guanosines on the early sense strand. This suggests, but does not prove, modification of adenosines to inosines in the sense–antisense hybrid dsRNA by the enzyme dsRAD/DRADA. Inosine base pairs with cytosine during reverse transcription, and this in turn would direct the incorporation of guanosine during the sequencing reaction, resulting in a net A-to-G conversion.
Figure 3.
Sequence comparison of six modified clones with that of genomic DNA. WT, sequence of the wild-type polyoma genome. #1–#6, sequences of the modified clones. Only the modified bases are shown in the six clones; other sequences were identical, except as noted. Lowercase letters indicate primer binding sites. The EcoR1 site is underlined, and the percentage of the modified adenosines is indicated for each sequence. A synthetic DNA oligodeoxynucleotide complementary to the sequence within the boxed region was used for the experiments described in Fig. 4.
Modified sequences were next examined to decipher the rules by which adenosines are modified in vivo. Modified adenosines exhibit a 5′ neighbor preference in the order U > A > C = G (Table 1). No strong 3′ neighbor preference was seen. These in vivo results are in strikingly close agreement with those reported from in vitro studies of dsRAD/DRADA activity (57). This is strongly indicative of the fact that dsRAD/DRADA is responsible for antisense-induced base modification in our system. However, as the base preferences for other dsRNA-specific adenosine deaminases have not yet been reported, it remains possible that a relative of dsRAD/DRADA is actually responsible for the observed modifications.
Table 1.
Analysis of 5′ and 3′ neighbor preferences of modified adenosines
| Neighbor preference | Total adenosines with indicated neighbor | Modified adenosines | % Modification |
|---|---|---|---|
| U* | 96 | 67 | 70 |
| A* | 90 | 51 | 57 |
| C* | 60 | 13 | 22 |
| G* | 48 | 8 | 17 |
| U† | 108 | 50 | 46 |
| A† | 90 | 38 | 42 |
| C† | 42 | 22 | 52 |
| G† | 66 | 32 | 48 |
5′ neighbor preference.
3′ neighbor preference.
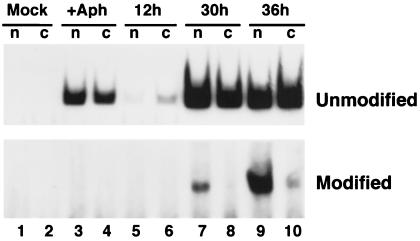
We next addressed the fate of RNAs modified as a result of sense–antisense interaction. Modified RNAs are invisible to many standard hybridization probes, owing to extensive base pair mismatching. To determine whether modified RNAs are transported to the cytoplasm, we utilized an RT-PCR strategy, similar to that described in Fig. 2, that only detects a subset of modified species. Cells were infected for various times, and nuclear and cytoplasmic RNAs were isolated and subjected to RT-PCR with primer pairs specific for either unmodified or modified RNAs and from the same region of the viral genome (see Fig. 3). Importantly, the 3′ base of the RT primer for modified RNAs was a C, which would preclude base pairing with any RNA not modified at the complementary position. Lack of a 3′ base pair prevented priming on RNAs not modified at this position. We note that a probe such as this will still be expected to detect RNAs with many diverse modifications in other regions of their sequences. However, due to the variable nature of base modifications in different transcripts, it is impossible to design a single probe that would detect all modified species without also detecting unmodified species. Results are shown in Fig. 4. As expected, unmodified RNAs were apparent throughout infection and in the absence of DNA replication. Modified RNAs, however, could only be detected at late times in infection, and were not present in the absence of viral DNA replication. The intensities of the bands using probes for modified and unmodified RNAs could not be compared, because separate PCR reactions and conditions were used. Most interestingly, however, at 30 hr postinfection, modified RNA could be seen only in the nuclear fraction (Fig. 4, lane 7), while unmodified RNAs were found in both cellular compartments, consistent with previous data. As expected, the unmodified RNA signal increased with time (lane 9). These results suggest that RNAs modified by antisense action in the nucleus become dead-end transcripts that may serve no functional role in gene expression. Alternatively, modified RNAs could be transported to the cytoplasm but would be degraded very rapidly in that compartment.
Figure 4.
Determination of the intracellular fate of modified RNAs. Mouse NIH 3T3 cells were mock-infected, or infected with wild-type polyoma virus for the times indicated. At the indicated times, nuclear and cytoplasmic RNAs were isolated, and 5 μg of each fraction used for RT-PCR amplification, as in Fig. 2. Reverse transcription was performed using a primer containing the sequence complementary to the modified or unmodified sequence shown in the boxed area in Fig. 3 (5′-GGAACGCCCCACTAGAAC-3′). Subsequent PCR amplification used, in addition, the “PCR primer” noted in lowercase letters in Fig. 3. The PCR products corresponding to amplification of modified or unmodified RNAs are 74 bp in length, and are indicated in the figure.
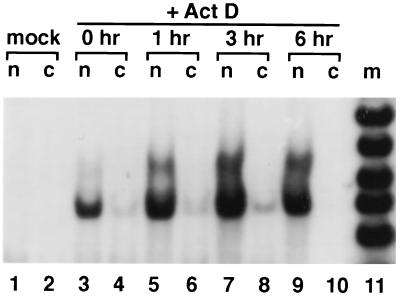
To determine the stability of the modified transcripts, the cells were treated with actinomycin D to block transcription (Fig. 5). The signal from the modified transcripts increased with time, and even 6 hr after actinomycin D treatment, a strong signal was detected. This suggests that modified RNAs are rather stable within the nucleus, or that the process of modification is rather slow. The conclusion that modified RNAs are relatively stable is supported by the results of Fig. 2, where modified RNAs were readily detected (almost 50% of the RT-PCR product). If modified RNAs were rapidly degraded, we would have expected to find far fewer of them in this sort of experiment. Alternatively, it is possible that the turnover of modified transcripts is mediated by an activity that is itself sensitive to inhibition by actinomycin D.
Figure 5.
Actinomycin D time course measurements of RNA stabilities. Nuclear and cytoplasmic RNAs were isolated at various times after treatment of cells with actinomycin D, and 5 μg of each fraction was used for RT-PCR amplification, as in Fig. 2, using the same primers as in Fig. 4.
DISCUSSION
Numerous examples of naturally existing antisense-mediated regulation of gene expression have been documented in prokaryotes (1). In most cases the regulation occurs at the translational level (1, 2). The antisense transcript hybridizes to the sense transcript and blocks access of the translational machinery to the 5′ end of the sense transcript. This in turn leads to reduced levels of protein synthesis. On the other hand, of the few examples of natural antisense that are known in eukaryotes (3–8), most, if not all, are thought to act within the nucleus (9, 10). In fact, it is likely that many complementary RNAs are expressed within the nucleus, either by design (antisense regulation) or by accident (symmetrical transcription or transcriptional read-through events). The polyoma virus system described here provides a powerful experimental system to study mammalian antisense regulation and the fate of naturally occurring double-stranded RNAs within the nucleus.
Why does polyoma regulate the levels of its early-strand RNAs? During the late phase of a productive infection, intronic regions of late-strand transcripts form antisense to early-strand transcripts, and these antisense RNAs lead to lower than expected levels of early-strand messages. During the early phase of infection, viral regulatory proteins (the T antigens) are needed to serve as inducers of cell proliferation and facilitators of viral DNA replication. The late gene products, virion capsid proteins, are not needed before the late phase, which occurs after the onset of DNA replication. During the late phase, however, there is little need for continued expression of early gene products. To balance the efficiencies of both replication and packaging it is quite possible that the virus has evolved a simple antisense strategy to down-regulate early-strand mRNA levels.
Using the polyoma model system, we have provided insight into the likely mechanism of natural mammalian antisense regulation. Using RT-PCR to specifically amplify the early-strand transcripts, we analyzed them to see if they were subject to RNA editing by dsRAD/DRADA or a related enzyme. Resistance of the RT-PCR amplified fragments to the enzyme EcoR1 showed that about 50% of the early-strand RNAs had been modified. It is important to note that these EcoR1-resistant fragments represent only a subset of all the modified fragments, since others may have sustained modifications in regions outside of this particular enzyme site. Sequence analysis of the EcoR1-resistant fragments confirmed that 40–60% of the adenosines within the early-strand RNAs had been modified to guanosines. These A-to-G changes in the cDNAs are suggestive of A-to-I changes within the dsRNAs, consistent with modifications induced by dsRAD/DRADA or a related enzyme like RED1 (38). However, our studies on the neighbor preferences of the modified adenosines are most consistent with in vitro studies on dsRAD/DRADA activity (57). Thus, we consider it highly likely that dsRAD/DRADA is responsible for antisense-induced base modifications in the polyoma system. Since the modifications caused by dsRAD/DRADA can be detected only in dsRNA, the polyoma sense–antisense transcripts almost certainly form duplexes within the nucleus.
We note that A-to-G (or to I) changes alter the genetic coding potential of mRNAs but can never create stop codons. Therefore, if modified RNAs were processed and transported to the cytoplasm at late times in polyoma infection, hundreds or even thousands of different viral T antigens could be produced. This would dramatically complicate our understanding of the polyoma virus life cycle. Our results using probes that specifically detect modified RNAs indicate that extensive antisense-induced base modification results in nuclear accumulation of the altered transcripts. Modified messages are most likely trapped in the nucleus, either because of their base compositions (presumed inosine content), altered secondary structures, or possibly partially duplex structures (resulting from incomplete dissociation of the two RNA strands). Alternatively, they could be transported to the cytoplasm but be rapidly degraded there. In any case, they appear to be inert for gene expression, as they cannot associate with the translation machinery. To further characterize the fate of modified transcripts in the polyoma system, we are currently carrying out experiments with other probes as well as probes from other regions of the early transcription unit. We should note, however, that the probes we have used here actually are themselves detecting a large number of different modified RNA molecules.
RNA modifications can account for most, if not all, of the antisense effect in the polyoma virus system. The extent of the antisense effect (Fig. 1C) is about 5-fold, while about 50% of the early-strand RT-PCR product at late times in infection is EcoR1-resistant (Fig. 2). However, of all modified RNAs, only 17% would have the first A residue modified (based on the observed neighbor preferences; Table 1), while only 57% would have the second A residue modified. Because not all modified RNAs are EcoR1-resistant, we can therefore use the observed base preferences to estimate that 60–65% of early-strand transcripts must have been modified. Assuming modified and unmodified RNAs have the same nuclear stability, the modifications account for about two-thirds of the antisense effect. So far, we have been unable to demonstrate any large differences in stability between modified and unmodified transcripts (data not shown). However, if modified transcripts were even slightly more rapidly turned over than unmodified transcripts, then the entire antisense effect could easily be accounted for.
It is most likely that inosines, rather than double-stranded RNA, cause nuclear retention. This conclusion derives from a careful analysis of the nuclear and cytoplasmic distribution of unmodified early-strand RNAs at different times in the polyoma life cycle. At early times, unmodified RNAs are almost equally distributed between the cytoplasm and the nucleus. At late times, the distribution pattern is exactly the same (data not shown). If double-stranded RNAs were the cause of nuclear retention, then we would expect the nuclear signal to be higher at late times, because this is the only time when high levels of both sense and antisense transcripts exist in the nucleus.
One well-studied mRNA that is edited by the same or a similar mechanism is that for the glutamate receptor. However, the GluRB message only has several modified residues. If each inosine had only a modest effect on nuclear retention, and if these effects were additive, one would not notice an effect on GluRB mRNA export, whereas the more extensively modified species examined here would show a much more severe export block. Also, if inosine were a nuclear retention signal, it is possible that for GluRB these few residues would be buried within secondary structure. In other known systems where there is significant A-to-I modification, the modified RNAs are retained in the nucleus (42, 43).
Is the regulation described here specific to polyoma, or is this a general mechanism for higher eukaryotes? We think that what we have observed reflects a general quality control system by which the cell deals with double-stranded RNA molecules. Polyoma is small and almost completely dependent on the host cell machinery for its propagation and must use the host cell biochemical pathways for these modifications. Finally, the prevalence of natural antisense regulation by this mechanism is still unknown, at least partly because appropriate probes do not exist to detect modified transcripts. The studies reported here ultimately should help in elucidating the detailed mechanism of action of eukaryotic antisense RNA as well as aiding in the design of more effective artificial antisense constructs for clinical and therapeutic applications.
Acknowledgments
We are grateful to D. Batt for providing us with the control RNA and E. Carmichael for useful suggestions. We thank members of our laboratory: D. Batt, Y. Huang, P. Leahy, X. Li, L. Rapp, M. Szlachetka, and K. Wimler for helpful comments on the manuscript, and K. Wimler for technical assistance. This work was supported by Grant CA45382 from the National Cancer Institute.
ABBREVIATIONS
- dsRNA
double-stranded RNA
- RT-PCR
reverse transcription–PCR
- dsRAD/DRADA
double-stranded RNA-specific adenosine deaminase
References
- 1.Inouye M. Gene. 1988;72:25–34. doi: 10.1016/0378-1119(88)90124-2. [DOI] [PubMed] [Google Scholar]
- 2.Simons R W. Gene. 1988;72:35–44. doi: 10.1016/0378-1119(88)90125-4. [DOI] [PubMed] [Google Scholar]
- 3.Tosic M, Roach A, de Rivaz J-C, Dolivo M, Matthieu J-M. EMBO J. 1990;9:401–406. doi: 10.1002/j.1460-2075.1990.tb08124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volk R, Köster M, Pöting A, Hartmann L, Knöchel W. EMBO J. 1989;8:2983–2988. doi: 10.1002/j.1460-2075.1989.tb08448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams T, Fried M. Nature (London) 1986;322:275–279. doi: 10.1038/322275a0. [DOI] [PubMed] [Google Scholar]
- 6.Adelman J P, Bond C T, Douglass J, Herbert E. Science. 1987;235:1514–1517. doi: 10.1126/science.3547652. [DOI] [PubMed] [Google Scholar]
- 7.Spencer C A, Gietz R D, Hodgetts R B. Nature (London) 1986;322:279–281. doi: 10.1038/322279a0. [DOI] [PubMed] [Google Scholar]
- 8.Nellen W, Lichtenstein C. Trends Biochem Sci. 1993;18:419–423. doi: 10.1016/0968-0004(93)90137-c. [DOI] [PubMed] [Google Scholar]
- 9.Cornelissen M. Nucleic Acids Res. 1989;17:7203–7209. doi: 10.1093/nar/17.18.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray J A H, Crockett N. In: Antisense Techniques: An Overview. Murray J A H, editor. New York: Wiley-Liss; 1992. pp. 1–49. [Google Scholar]
- 11.Hofgen R, Axelsen K B, Kannangara C G, Schuttke I, Pohlenz H D, Willmitzer L, Grimm B, Vonwettstein D. Proc Natl Acad Sci USA. 1994;91:1726–1730. doi: 10.1073/pnas.91.5.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cogen B. Virology. 1978;85:222–230. doi: 10.1016/0042-6822(78)90426-9. [DOI] [PubMed] [Google Scholar]
- 13.Piper P. J Mol Biol. 1979;131:399–407. doi: 10.1016/0022-2836(79)90083-4. [DOI] [PubMed] [Google Scholar]
- 14.Fenton R G, Basilico C. Virology. 1982;121:384–392. doi: 10.1016/0042-6822(82)90176-3. [DOI] [PubMed] [Google Scholar]
- 15.Fenton R G, Basilico C. Proc Natl Acad Sci USA. 1982;79:7142–7146. doi: 10.1073/pnas.79.23.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farmerie W G, Folk W R. Proc Natl Acad Sci USA. 1984;81:6919–6923. doi: 10.1073/pnas.81.22.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyde-DeRuyscher R, Carmichael G G. Proc Natl Acad Sci USA. 1988;85:8993–8997. doi: 10.1073/pnas.85.23.8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Carmichael G G. Proc Natl Acad Sci USA. 1993;90:8494–8498. doi: 10.1073/pnas.90.18.8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamen R, Lindstrom D M, Shure H, Old R W. Cold Spring Harb Symp Quant Biol. 1974;39:187–198. doi: 10.1101/sqb.1974.039.01.025. [DOI] [PubMed] [Google Scholar]
- 20.Beard P, Acheson N H, Maxwell I H. J Virol. 1976;17:20–26. doi: 10.1128/jvi.17.1.20-26.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyde-DeRuyscher R P, Carmichael G G. J Virol. 1990;64:5823–5832. doi: 10.1128/jvi.64.12.5823-5832.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acheson N. Cell. 1976;8:1–12. doi: 10.1016/0092-8674(76)90179-3. [DOI] [PubMed] [Google Scholar]
- 23.Birg F, Favaloro J, Kamen R. Proc Natl Acad Sci USA. 1977;74:3138–3142. doi: 10.1073/pnas.74.8.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acheson N. J Virol. 1981;37:628–635. doi: 10.1128/jvi.37.2.628-635.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Acheson N H. Proc Natl Acad Sci USA. 1978;75:4754–4758. doi: 10.1073/pnas.75.10.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acheson N. Mol Cell Biol. 1984;4:722–729. doi: 10.1128/mcb.4.4.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treisman R, Kamen R. J Mol Biol. 1981;148:273–301. doi: 10.1016/0022-2836(81)90539-8. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Batt D B, Carmichael G G. Proc Natl Acad Sci USA. 1994;91:4258–4262. doi: 10.1073/pnas.91.10.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bass B L, Weintraub H. Cell. 1987;48:607–613. doi: 10.1016/0092-8674(87)90239-x. [DOI] [PubMed] [Google Scholar]
- 30.Rebagliati M R, Melton D A. Cell. 1987;48:599–605. doi: 10.1016/0092-8674(87)90238-8. [DOI] [PubMed] [Google Scholar]
- 31.Wagner R W, Yoo C, Wrabetz L, Kamholz J, Buchhalter J, Hassan N F, Khalili K, Kim S U, Perussia B, McMorrin F A, Nishikura K. Mol Cell Biol. 1990;10:5586–5590. doi: 10.1128/mcb.10.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patterson J B, Samuel C E. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner R W, Smith J E, Cooperman B S, Nishikura K. Proc Natl Acad Sci USA. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bass B L, Weintraub H. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 35.Polson A G, Crain P F, Pomerantz S C, McCloskey J A, Bass B L. Biochemistry. 1991;30:11507–11514. doi: 10.1021/bi00113a004. [DOI] [PubMed] [Google Scholar]
- 36.Wagner R W, Nishikura K. Mol Cell Biol. 1988;8:770–777. doi: 10.1128/mcb.8.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higuchi M, Single F N, Köhler M, Sommer B, Sprengel R, Seeburg P H. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 38.Melcher T, Maas S, Herb A, Sprengel R, Seeburg P H, Higuchi M. Nature (London) 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 39.Dabiri G A, Lai F, Drakas R A, Nishikura K. EMBO J. 1996;15:34–45. [PMC free article] [PubMed] [Google Scholar]
- 40.Luo G X, Chao M, Hsieh S Y, Sureau C, Nishikura K, Taylor J. J Virol. 1990;64:1021–1027. doi: 10.1128/jvi.64.3.1021-1027.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polson A G, Bass B L, Casey J L. Nature (London) 1996;380:454–456. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 42.Bass B L, Weintraub H, Cattaneo R, Billeter M A. Cell. 1989;56:331. doi: 10.1016/0092-8674(89)90234-1. [DOI] [PubMed] [Google Scholar]
- 43.Cattaneo R, Billeter M A. Curr Top Microbiol Immunol. 1992;176:63–74. doi: 10.1007/978-3-642-77011-1_5. [DOI] [PubMed] [Google Scholar]
- 44.Billeter M A, Cattaneo R, Spielhofer P, Kaelin K, Huber M, Schmid A, Baczko K, ter Meulen V. Ann NY Acad Sci. 1994;724:367–377. doi: 10.1111/j.1749-6632.1994.tb38934.x. [DOI] [PubMed] [Google Scholar]
- 45.Cattaneo R. Curr Biol. 1994;4:134–136. doi: 10.1016/s0960-9822(94)00030-8. [DOI] [PubMed] [Google Scholar]
- 46.Hajjar A M, Linial M L. J Virol. 1995;69:5878–5882. doi: 10.1128/jvi.69.9.5878-5882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petschek J P, Mermer M J, Scheckelhoff M R, Simone A A, Vaughn J C. J Mol Biol. 1996;259:885–890. doi: 10.1006/jmbi.1996.0365. [DOI] [PubMed] [Google Scholar]
- 48.Bass B L. Semin Dev Biol. 1992;3:425–433. [Google Scholar]
- 49.Kim U, Nishikura K. Semin Cell Biol. 1993;4:285–293. doi: 10.1006/scel.1993.1034. [DOI] [PubMed] [Google Scholar]
- 50.Cahill K B, Roome A J, Carmichael G G. J Virol. 1990;64:992–1001. doi: 10.1128/jvi.64.3.992-1001.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current Protocols in Molecular Biology. New York: Greene and Wiley; 1989. [Google Scholar]
- 52.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 53.Adami G R, Marlor C W, Barrett N L, Carmichael G G. J Virol. 1989;63:85–93. doi: 10.1128/jvi.63.1.85-93.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lichtler A, Barrett N L, Carmichael G G. Biotechniques. 1992;12:231–232. [PubMed] [Google Scholar]
- 55.Barrett N L, Carmichael G G, Luo Y. Nucleic Acids Res. 1991;19:3011–3017. doi: 10.1093/nar/19.11.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adami G R, Carmichael G G. Nucleic Acids Res. 1987;15:2593–2610. doi: 10.1093/nar/15.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polson A G, Bass B L. EMBO J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]