Abstract
For a series of different proteins, including a structural protein, enzyme, inhibitor, protein marker, and a charge-transfer system, we have quantified the higher affinity of Na+ over K+ to the protein surface by means of molecular dynamics simulations and conductivity measurements. Both approaches show that sodium binds at least twice as strongly to the protein surface than potassium does with this effect being present in all proteins under study. Different parts of the protein exterior are responsible to a varying degree for the higher surface affinity of sodium, with the charged carboxylic groups of aspartate and glutamate playing the most important role. Therefore, local ion pairing is the key to the surface preference of sodium over potassium, which is further demonstrated and quantified by simulations of glutamate and aspartate in the form of isolated amino acids as well as short oligopeptides. As a matter of fact, the effect is already present at the level of preferential pairing of the smallest carboxylate anions, formate or acetate, with Na+ versus K+, as shown by molecular dynamics and ab initio quantum chemical calculations. By quantifying and rationalizing the higher preference of sodium over potassium to protein surfaces, the present study opens a way to molecular understanding of many ion-specific (Hofmeister) phenomena involving protein interactions in salt solutions.
Keywords: ion–protein interaction, molecular dynamics, cell environment, protein function, Hofmeister series
Sodium and potassium represent the two most abundant monovalent cations in living organisms. Despite the fact that they possess the same charge and differ only in size [Na+ having a smaller ionic radius but a larger hydrated radius than K+ (1)], sodium versus potassium ion specificity plays a crucial role in many biochemical processes. More than 100 years ago, Hofmeister discovered that Na+ destabilizes (“salts out”) hens' egg white protein more efficiently than K+ does (2), analogous behavior being later shown also for other proteins (3). In a similar way, sodium was found to be significantly more efficient than potassium, e.g., in enhancing polymerization of rat brain tubulin (4). 23Na NMR studies also were used to characterize cation-binding sites in proteins (5). The vital biological relevance of the difference between Na+ and K+ is exemplified by the low intracellular and high extracellular sodium/potassium ratio maintained in living cells by ion pumps at a considerable energy cost (6). The principal goal of this article is to quantify and rationalize the different affinities of sodium and potassium to protein surfaces by means of molecular dynamics (MD) simulations, quantum chemistry calculations, and conductivity measurements for a series of proteins and protein fragments in aqueous solutions. Our effort, which is aimed at understanding the generic, thermodynamic Na+/K+ ion specificity at protein surfaces, is thus complementary to recent computational studies of ion selectivity during active transport across the cellular membrane (7–10).
Because primitive continuum electrostatic models, which are still widely used to describe solvation of biomolecules, cannot distinguish between sodium and potassium, their ion specificity at protein surfaces has been to a large extent ignored (11). Unlike divalent cations such as Ca2+ with well recognized specific binding to proteins (6), Na+ and K+ ions often are viewed as merely defining the ionic strength of the corresponding salt solution (11). This view is also because of the fact that the specificity of ions in solutions is traditionally discussed in terms of their kosmotropic (water structure-making) versus chaotropic (water structure-breaking) behavior (12). In this respect, sodium and potassium, which are next to each other in the Hofmeister series, are roughly neutral, the former being a very weak kosmotrope and the latter a weak chaotrope (11). Moreover, recent spectroscopic measurements in electrolyte solutions showed that the effect of dissolved monovalent ions does not propagate beyond the first solvation shell (13), which underlines the importance of local ion–ion, ion–water, and water–water interactions over long-range solvent ordering effects. In this spirit, the so-called Law of Matching Water Affinities (11, 14, 15) has been proposed as an attempt to explain, among others, the Na+/K+ ion specificity. In a nutshell, based on simple electrostatic arguments it has been suggested that a cation and an anion with similar (either large or small) hydration energies tend to form contact ion pairs in aqueous solutions. Within this qualitative concept, sodium matches major intracellular anions or anionic groups, such as carboxylate, carbonate, and the phosphate monoanion (15), better than potassium does in surface charge density (and in the related hydration energy). It has been argued that Na+ thus would bind more strongly than K+ to protein surfaces containing COO− groups, which may be one of the reasons why sodium has to be pumped out of the cell (11).
Results
In the present study, we systematically investigated the ion-specific behavior of sodium and potassium at protein surfaces. To this end, we performed extensive MD simulations of a series of five very different proteins in aqueous mixtures of NaCl and KCl (containing typically 0.25 M of each salt). We chose two extracellular and three intracellular proteins. As the former, we took actin as a representative of structural proteins and bovine pancreatic trypsin inhibitor (BPTI) as an enzyme-inhibiting protein. Among the latter, ubiquitin was taken as a typical protein marker, hyperthermophilic rubredoxin as an electron-transfer system, and ribonuclease (RNase) A as a model enzyme. Our computational findings were supported by conductivity measurements of aqueous solutions of NaCl and KCl with added protein. The measured proteins were RNase A and BSA. The Na+ versus K+ ion-specific behavior was further rationalized by means of additional simulations of solvated oligopeptides and individual amino acids containing at pH = 7 a carboxylate group in the side chain [i.e., aspartate (Asp) and glutamate (Glu)] as well as by MD and ab initio calculations of ion pairing of alkali cations with small carboxylate anions (formate and acetate). All of the studied systems together with the computational and experimental methods are described in detail in Materials and Methods.
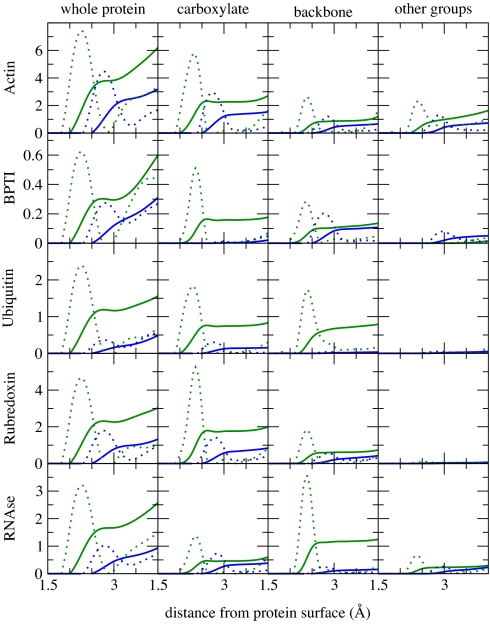
Fig. 1 summarizes the principle results of MD simulations for the five proteins under study. It shows the distributions of sodium and potassium cations in the vicinity of the protein and the integrals thereof, i.e., cumulative sums providing the number of Na+ and K+ ions within a certain distance from the protein surface. Both distributions exhibit a peak (Na+ at 2.3 Å and K+ at 2.8 Å from the protein surface), and, correspondingly, there is a plateau region for the cumulative sums that defines the number of ions in the immediate vicinity of the protein. The crucial result is that in all cases there are approximately twice as many (or even more) sodium as potassium cations near the protein surface. In absolute numbers, there are on average one to four Na+ ions at the protein surface, except for in BPTI where this number is several times smaller (Fig. 1).
Fig. 1.
Distribution functions (dotted lines) and cumulative sums (i.e., integrated distribution functions) (solid lines) of cations in the vicinity of the five investigated proteins, obtained from MD simulations. The cumulative sums provide the average number of sodium (green) or potassium (blue) ions within a given distance from the protein surface. Results are depicted for the whole protein, as well as detailed for the carboxylate groups, other side-chain functional groups, and the protein backbone. Note the larger affinity of Na+ over K+ to the protein surface and the dominant role of the COO− groups for this effect.
From the left three columns in Fig. 1, we see that the preference of Na+ over K+ for the protein surface comes mainly from cation-specific interactions with the side-chain carboxylate groups, with contributions also from the backbone and, to a lesser extent, from other side-chain groups. The ion specificity at the backbone is primarily attributable to interactions with the carbonyl oxygen of the amide group, whereas the selectivity at other side-chain groups also can have some contribution from cation–aromatic ring interactions in phenylalanine. The latter interactions were shown to be stronger for Na+ than for K+ as well (16); however, they cannot be described accurately within standard force fields. The very different nature and amino acid content of the proteins under study is reflected in varying total numbers of ions at the protein surface and relative contributions from different protein parts. However, the higher surface affinity of Na+ over K+ is revealed as a generic property for all investigated proteins.
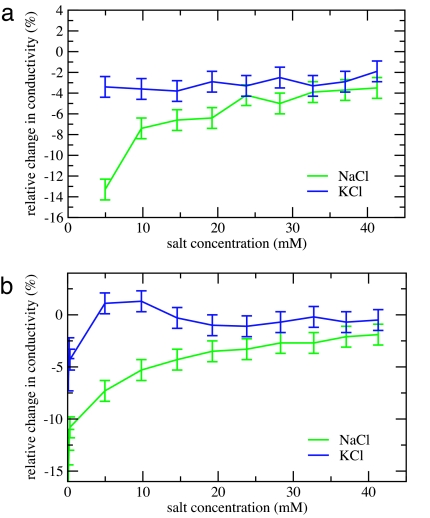
The above computational results are supported by conductivity measurements in NaCl and KCl solutions of varying concentrations with added RNase A (as one of the simulated proteins) or BSA (which is easily available but too large and without well determined structure to be effectively simulated). For both proteins, the relative decrease in conductivity is significantly larger in NaCl compared with KCl solutions (Fig. 2), which can be directly interpreted in terms of sodium being more efficiently removed from the solution to the protein surface than potassium ions are. Assuming that the decrease in conductivity is solely attributable to ions adsorbed at the protein surface, we can deduce that, e.g., for RNase A one protein molecule effectively binds approximately one to two sodium cations, whereas it attracts fewer than half the number of potassium ions. These numbers, which are only weakly dependent on salt concentration, agree well with the above results from MD simulations in solutions with a higher ionic strength. The results of conductivity measurements for BSA (the small undulation on the KCl curve in Fig. 2b being within the statistical error) can be interpreted in a quantitatively similar way.
Fig. 2.
Relative changes in measured conductivity upon adding 10 mg/ml of RNase A (a) or BSA (b) to a solution of NaCl (green) or KCl (blue) of varying concentrations. Note the much stronger conductivity decrease for NaCl compared with KCl for both proteins, which is interpreted in terms of stronger affinity of Na+ over K+ to the protein surface.
Specific ion effects at protein surfaces, particularly for higher salt concentrations, are often rationalized in terms of surface tension changes invoked by the presence of a given salt (14, 17, 18). The analogy between air–water and protein–water interfaces can be good for the exposed hydrophobic patches of the protein with low polarity and dielectric constant; however, it is much less operative for the polar or charged hydrophilic parts of protein surface. Moreover, unlike for anions, ion-specific effects for cations at the air–water interface are negligible, with sodium and potassium salts having almost the same surface tension versus concentration curves (19). This finding is consistent with the present picture indicating that the ion specificity originates to a large extent from local interactions with charged and polar groups at the protein surface (20). In particular, the higher affinity of sodium over potassium for the protein surface comes mainly from specific interactions with the carboxylate groups within the amino acid side chains.
To further verify and quantify the above conclusion, we have focused on the sodium and potassium affinity to the relevant protein fragments by simulating the two amino acids containing at pH = 7 the COO− group in the side chain. Glu and Asp were studied in a mixed NaCl and KCl solution as isolated amino acids, as well as short oligopeptides, all capped by using the acetyl and methylamide groups at the N and C termini, respectively. The principal results of these simulations are summarized in Table 1. Consistent with the results for proteins (Fig. 1), the Na+/K+ ratios reveal a two to four times stronger preference of sodium over potassium for the oligopeptide or amino acid surface (Table 1). This relative preference of Na+ is found both at the side chains and near the backbone. The former play a more important role in absolute terms because they represent ion–ion interactions, whereas the latter represent only weaker ion–dipole interactions involving the polar C O group of the amide.
O group of the amide.
Table 1.
Na+/K+ ratio in the vicinity of solvated oligopeptides, single aspartate or glutamate, and acetate anion
| System | Na+/K+ ratio (side chain) | Na+/K+ ratio (backbone) |
|---|---|---|
| Ac-(Glu)10-NMe | 2.3 | 4.0 |
| Ac-(Asp)10-NMe | 3.5 | 4.5 |
| Ac-Asp-Gly-Ser-NMe | 2.9 | 2.5 |
| Ac-Asp-Ser-NMe | 2.4 | 2.0 |
| Ac-Glu-NMe | 4.8 | 1.1 |
| Ac-Asp-NMe | 2.3 | 1.8 |
| Acetate anion | 3.4 | — |
The oligopeptides and single amino acids are terminated by acetyl (Ac) and methylamide (NMe) groups.
We further reduced the problem of the alkali cation–side-chain carboxylate interaction to a question concerning the relative strength of pairing of Na+ and K+ with acetate anion. To this end, we simulated an aqueous mixture of acetate, sodium, potassium, and chloride, following the acetate–alkali ion-pairing patterns. There is a clear preference of Na+ over K+ for the vicinity of acetate, reaching a value of 3.4 in relative terms (Table 1). For the two smallest carboxylate anions, formate and acetate, we also were able to perform ab initio quantum chemical calculations of pairing with sodium or potassium, employing a polarizable continuum model for water (see Materials and Methods for further computational details). Within these calculations, pairing with Na+ is favored over that with K+ by >2 kcal/mol, both for formate and acetate (Table 2). The ab initio results thus support the conclusions from classical MD simulations, indicating an even stronger relative affinity of Na+ over K+ to the COO− group. In a similar spirit, analogous ab initio calculations of pairing of alkali cations with formaldehyde reveal a free-energy preference of ≈1 kcal/mol in favor of sodium over potassium. This finding supports the MD results concerning ion selectivity at the protein backbone region attributable to specific interactions with the carbonyl oxygen of the amide group.
Table 2.
Difference between association free energies of aqueous sodium-carboxylate and potassium-carboxylate ion pairs, where carboxylate is a formate or acetate anion
| System | ΔΔGK+-carboxylate →Na+-carboxylate, kcal/mol |
|---|---|
| Formate | −2.58 |
| Acetate | −2.22 |
The results clearly show the preference of Na+ over K+ to pair in water with carboxylate anions.
Discussion and Conclusions
Present results provide a clear and quantitative picture of the preference of sodium over potassium for protein surfaces, rationalizing this effect primarily in terms of local cation-specific interactions with the anionic carboxylate group in Glu and Asp side chains. Additional contributions to the Na+/K+ selectivity come from interactions with carbonyl oxygens of the amide constituting the protein backbone and, to a much smaller extent, from other side-chain groups. We do not observe a qualitative dependence of the effect on salt concentration, which can be related to the local character of the Na+ versus K+ selectivity. This result is different from specific effects of anions at protein surfaces, which may exhibit even a reversal of Hofmeister ordering upon increasing salt concentration (17, 21). The importance of alkali cation–protein interactions for in vitro experiments often can be hidden behind experimental conditions optimized for a particular practical task. Nevertheless, in vivo conditions directly point to the importance of these effects, which can play a very special or even crucial role in processes where protein–protein intermolecular as well as intramolecular interactions are important, such as protein folding or build up of protein functional networks. There is ample experimental evidence concerning alkali salt effects on protein folding (22, 23). It is also well known that mostly hydrophilic surfaces of nonmembrane proteins are involved in protein–protein interaction within various intracellular and extracellular processes (24, 25). The number of charged groups on the protein surface that mediate these interactions is high, and these sites also are accessible to interactions with small monovalent ions, such as Na+ and K+. Increasing the concentration of an ion with a higher affinity to the protein effectively modulates its interaction strength and even functionality (20, 26). The present simulation and experimental results, quantifying the stronger binding of Na+ to protein surfaces over that of K+, indicate why at physiological ionic strengths the former but not the latter ion can impair the function of proteins in the cell. The energetically expensively maintained concentration differences of sodium and potassium between the interior and exterior of a living cell possibly represent a direct consequence of this fact (11).
Materials and Methods
Computational Details.
Structures of all of the studied proteins, actin, BPTI, ubiquitin, hyperthermophilic rubredoxin, and RNase A, were downloaded from the Protein Data Bank (PDB) (27). Oligopeptides [Asp-Ser, Asp-Gly-Ser, (Asp)10, and (Glu)10] were built from scratch by using amino acid templates and capped with the acetyl and methylamide groups at the N and C termini, respectively. Isolated amino acids (Asp and Glu) were terminated in the same way.
Dissociable groups in the amino acid side chains of the proteins were protonated according to target pH = 7, and the resulting structures were solvated in periodic rectangular water boxes, employing the SPC/E water model (28). Initially, a sufficiently large cavity was carved in the water box to accommodate the protein molecule. The net charge of the systems was neutralized by chloride or by sodium and potassium ions. A similar approach also was applied to the oligopeptides under study.
As a next step, equal amounts of NaCl and KCl were introduced into the simulation cell to form an ≈0.25 M NaCl + 0.25 M KCl solution. This hyperphysiological concentration was adopted to improve the statistics of the ion–protein contacts. For comparison, proteins in solutions with reduced or increased salt concentrations, as well as those containing only a single salt, also were simulated, showing very similar results concerning the Na+/K+ ion specificity at the protein surface. Solvated acetate anion with one Cl−, Na+, and K+ ion in the unit cell was created as well. The detailed parameters for individual systems were as follows.
Actin.
PDB code 1J6Z (monomeric structure), MW41920.2. The resulting net charge of −2 e was compensated by one Na+ ion and one K+ ion. The unit cell of approximate initial dimensions of 91 × 67 × 75 Å contained in addition 12,759 water molecules and 58 Na+, 58 K+, and 116 Cl− ions.
BPTI.
PDB code 6PTI, MW6531.1. The resulting net charge of +6 e was compensated by six Cl− ions. The unit cell of approximate initial dimensions of 41 × 45 × 45 Å contained in addition 2,184 water molecules and 10 Na+, 10 K+, and 20 Cl− ions.
RNase A.
PDB code 1FS3, MW13712.4. The resulting net charge of +4 e was compensated by four Cl− ions. The unit cell of approximate initial dimensions of 51 × 50 × 60 Å contained in addition 4,329 water molecules and 19 Na+, 19 K+, and 38 Cl− ions.
Hyperthermophilic Rubredoxin.
PDB code 1BRF, MW5905.6. The resulting net charge of −6 e was compensated by three Na+ and three K+ ions. The unit cell of approximate initial dimensions of 43 × 40 × 45 Å contained in addition 2,229 water molecules and 10 Na+, 10 K+, and 20 Cl− ions.
Ubiquitin.
PDB code 1UBQ, MW8579.9. The resulting net charge was 0. The unit cell of approximate initial dimensions of 43 × 40 × 45 Å contained in addition 3,688 water molecules and 17 Na+, 17 K+, and 34 Cl− ions.
Ac-(Glu)10-NMe.
The resulting net charge of −10 e was compensated by five Na+ and five K+ ions. The unit cell of approximate initial dimensions of 55 × 55 × 55 Å contained in addition 5,200 water molecules and 23 Na+, 23 K+, and 46 Cl− ions.
Ac-(Asp)10-NMe.
The resulting net charge of −10 e was compensated by five Na+ and five K+ ions. The unit cell of approximate initial dimensions of 55 × 55 × 55 Å contained in addition 5,200 water molecules and 23 Na+, 23 K+, and 46 Cl− ions.
Ac-Asp-Gly-Ser-NMe.
The resulting net charge was −1 e. The unit cell of approximate initial dimensions of 34 × 34 × 34 Å contained in addition 825 water molecules and 2 Na+, 2 K+, and 3 Cl− ions.
Ac-Asp-Ser-NMe.
The resulting net charge was −1 e. The unit cell of approximate initial dimensions of 30 × 30 × 30 Å contained in addition 540 water molecules and 2 Na+, 2 K+, and 3 Cl− ions.
Ac-Glu-NMe.
The resulting net charge was −1 e. The unit cell of approximate initial dimensions of 28 × 28 × 28 Å contained in addition 450 water molecules and 1 Na+, 1 K+, and 1 Cl− ions.
Ac-Asp-NMe.
The resulting net charge was −1 e. The unit cell of approximate initial dimensions of 28 × 28 × 28 Å contained in addition 450 water molecules and 1 Na+, 1 K+, and 1 Cl− ions.
Acetate Anion.
The resulting net charge was −1 e. The unit cell of approximate initial dimensions of 27 × 27 × 27 Å contained in addition 375 water molecules and 1 Na+, 1 K+, and 1 Cl− ions.
The MD simulations of proteins consisted of minimization, heating to 300 K, 0.5-ns equilibration at constant pressure, and 1-ns production runs in the NpT ensemble. We used the same simulation protocol as in our recent simulations on salt effects on solvated horseradish peroxidase and BPTI (20), which provided converged results for the distribution of ions around proteins. Oligopeptides, isolated amino acids, and the acetate ion were simulated in a similar way with simulation times extended to 10 ns in the former and 100 ns in the latter two cases. All calculations were carried out by using the AMBER8 package (29), employing the parm99 force field (30). An interaction cutoff of 12 Å was applied. Long-range electrostatic interactions were accounted for in a standard way by using the smooth particle mesh Ewald method (31). The SHAKE algorithm was used for constraining bonds containing hydrogen atoms (32).
The resulting trajectories were analyzed in terms of ion distributions, the integrals of which provided the total number of ions within certain distance from the solute (protein, peptide) surface. The average times the cations spent in the vicinity of the given type of amino acid (within 3.5 Å) residue also were recorded. The individual contact times of the Na+ and K+ ions with the protein surface did not exceed 30% of the total simulation time. Frequent exchanges of surface-bound and bulk ions thus ensured good statistics, which also was confirmed by longer test runs providing the same ion distributions in the vicinity of the protein surface.
Ab initio calculations of ion pairing of Na+/K+ with acetate or formate anions were performed at the MP2/aug-cc-pvtz level. For explicit correlation of all valence and outer core (subvalence) electrons additional core/valence basis functions were added (33). The aqueous solvent was described as a polarizable continuum within the COSMO model (34, 35). All parameters were taken as the default ones except for the ionic radius of sodium, which was reduced by 1.3% to match exactly the experimental difference between hydration free energies of Na+ and K+ amounting to 17.5 kcal/mol (36). The free energy of ion pairing was evaluated as the difference between the energy of the solvated contact ion pair and the energies of solvated cation and anion. All geometries were optimized in the gas phase at the same level of theory. For comparison, we also evaluated the energy of the contact ion pair for cation–anion distance taken as the first maximum on the cation–anion radial distribution function from a classical MD simulation in water. Because the shift in distance was very small, this resulted in negligible (below 0.1 kcal/mol) changes in the relative (Na+ versus K+) free energies of ion pairing. Analogous calculations also were performed for pairing of sodium or potassium with formaldehyde.
Experimental Details.
Conductivity measurements were performed by using a table conductometer Jenway 4330 (Barloworld Scientific, Dunmow, U.K.) with a vendor-supplied electrode immersed in 10 ml of solution. Single measurements were performed with three-digit accuracy. Each of the plotted values (Fig. 2) is an average over three measurements, and the estimated statistical error of the relative changes of conductivity is ≈1%. The small residual conductivity of the pure protein solution (of the order of 100 μS) was subtracted before evaluating the relative changes.
Samples.
BSA, initial fraction by heat shock, fraction V, was from Sigma-Aldrich, catalog no. A7906. RNase A, from bovine pancreas, was from Sigma-Aldrich, catalog no. R6513. Initial experimental protein concentrations were 10 mg/ml. NaCl and KCl (p. a. grade) were obtained from Lachema (Brno, Czech Republic). Proteins were dissolved in doubly distilled, deionized water, and defined amounts of 1 M stock salt (NaCl or KCl) solutions were added to reach a range of salt concentrations from 0.001 to 0.045 M.
Acknowledgments
We thank Dr. Pavel Kotrba, Dr. Lucie Marešová, and Prof. František Opekar for technical assistance with the samples and measurements, and Prof. Jan Konvalinka, Prof. Kim Collins, and Prof. Jacques Dubochet for valuable discussions. We are grateful to the METACentrum in Brno, Czech Republic, for generous allocation of computer time, and the Advanced Biomedical Computing Center, National Cancer Institute, Frederick, MD, for the use of their facilities for part of this work. Support from Czech Ministry of Education Grant LC512, Granting Agency of the Academy of Sciences Grant IAA400400503 (to P.J.), and Granting Agency of the Czech Republic Grants 203/06/1727 (to J.V.) and 203/05/H001 (to L.V.) is gratefully acknowledged.
Abbreviations
- MD
molecular dynamics
- BPTI
bovine pancreatic trypsin inhibitor
- PDB
Protein Data Bank.
Footnotes
The authors declare no conflict of interest.
References
- 1.Nightingale ER. J Phys Chem. 1959;63:1381–1387. [Google Scholar]
- 2.Hofmeister F. Arch Exp Pathol Pharmakol (Leipzig) 1888;24:247–260. [Google Scholar]
- 3.Inouye K, Kuzuya K, Tonomura B. J Biochem (Tokyo) 1998;123:847–852. doi: 10.1093/oxfordjournals.jbchem.a022014. [DOI] [PubMed] [Google Scholar]
- 4.Wolff J, Sackett DL, Knipling L. Protein Sci. 1996;5:2020–2028. doi: 10.1002/pro.5560051008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel HJ, Drakenberg T, Forsen S. In: NMR of Newly Accessible Nuclei. Laszlo P, editor. Vol 1. New York: Academic; 1983. pp. 157–197. [Google Scholar]
- 6.Darnell J, Lodish H, Baltimore D. Molecular Cell Biology. New York: Scientific American Books; 1990. [Google Scholar]
- 7.Gumbart J, Wang Y, Aksimentiev A, Tajkhorshid E, Schulten K. Curr Opin Struct Biol. 2005;15:423–431. doi: 10.1016/j.sbi.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ash WL, Zlomislic MR, Oloo EO, Tieleman DP. Biochim Biophys Acta. 2004;1666:158–189. doi: 10.1016/j.bbamem.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Wu Y, Voth GA. Biophys J. 2006;90:L73–L75. doi: 10.1529/biophysj.106.084061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen TW, Andersen OS, Roux B. Biophys J. 2006;90:3447–3468. doi: 10.1529/biophysj.105.077073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins KD. Biophys J. 1997;72:65–76. doi: 10.1016/S0006-3495(97)78647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcus Y. Ion Solvation. Chichester, UK: Wiley; 1985. [Google Scholar]
- 13.Omta AW, Kropman MJ, Woutersen S, Bakker HJ. Science. 2003;301:347–349. doi: 10.1126/science.1084801. [DOI] [PubMed] [Google Scholar]
- 14.Collins KD. Methods. 2004;34:300–311. doi: 10.1016/j.ymeth.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 15.Collins KD. Biophys Chem. 2006;119:271–281. doi: 10.1016/j.bpc.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Siu FM, Ma NL, Tsang CW. Chem Eur J. 2004;10:1966–1976. doi: 10.1002/chem.200305519. [DOI] [PubMed] [Google Scholar]
- 17.Arakawa T, Timasheff SN. Biochemistry. 1982;21:6545–6552. doi: 10.1021/bi00268a034. [DOI] [PubMed] [Google Scholar]
- 18.Lin TY, Timasheff SN. Protein Sci. 1996;5:372–381. doi: 10.1002/pro.5560050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weissenborn PK, Pugh RJ. J Colloid Interface Sci. 1996;184:550–563. doi: 10.1006/jcis.1996.0651. [DOI] [PubMed] [Google Scholar]
- 20.Vrbka L, Jungwirth P, Bauduin P, Touraud D, Kunz W. J Phys Chem B. 2006;110:7036–7043. doi: 10.1021/jp0567624. [DOI] [PubMed] [Google Scholar]
- 21.Robertson TB. J Biol Chem. 1911;9:303–326. [Google Scholar]
- 22.Maldonado S, Irun MP, Campos LA, Rubio JA, Luquita A, Lostao A, Wang RJ, Garcia-Moreno B, Sancho J. Protein Sci. 2002;11:1260–1273. doi: 10.1110/ps.2980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominy BN, Perl D, Schmid FX, Brooks CL. J Mol Biol. 2002;319:541–554. doi: 10.1016/S0022-2836(02)00259-0. [DOI] [PubMed] [Google Scholar]
- 24.Muegge I, Schweins T, Warshel A. Proteins Struct Funct Bioinfo. 1998;30:407–423. doi: 10.1002/(sici)1097-0134(19980301)30:4<407::aid-prot8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 25.Curtis RA, Ulrich J, Montaser A, Prausnitz JM, Blanch HW. Biotechnol Bioeng. 2002;79:367–380. doi: 10.1002/bit.10342. [DOI] [PubMed] [Google Scholar]
- 26.Pinna MC, Bauduin P, Tourand D, Monduzzi M, Ninham BW, Kunz W. J Phys Chem B. 2005;109:16511–16514. doi: 10.1021/jp051897v. [DOI] [PubMed] [Google Scholar]
- 27.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindaylov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berendsen HJC, Grigera JR, Straatsma TP. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 29.Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, et al. AMBER 8. San Francisco: Univ of California; 2004. [Google Scholar]
- 30.Wang JM, Cieplak P, Kollman PA. J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 31.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 32.Ryckaert J-P, Ciccotti G, Berendsen HJC. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 33.Dunning TH., Jr J Chem Phys. 1989;90:1007–1024. [Google Scholar]
- 34.Barone V, Cossi M. J Phys Chem A. 1998;102:1995–2001. [Google Scholar]
- 35.Cossi M, Rega N, Scalmani G, Barone V. J Comp Chem. 2003;24:669–681. doi: 10.1002/jcc.10189. [DOI] [PubMed] [Google Scholar]
- 36.Schmid R, Miah AM, Sapunov VN. Phys Chem Chem Phys. 2000;2:97–102. [Google Scholar]