Abstract
Meningitis and sepsis caused by serogroup B meningococcus are two severe diseases that still cause significant mortality. To date there is no universal vaccine that prevents these diseases. In this work, five antigens discovered by reverse vaccinology were expressed in a form suitable for large-scale manufacturing and formulated with adjuvants suitable for human use. The vaccine adjuvanted by aluminum hydroxide induced bactericidal antibodies in mice against 78% of a panel of 85 meningococcal strains representative of the global population diversity. The strain coverage could be increased to 90% and above by the addition of CpG oligonucleotides or by using MF59 as adjuvant. The vaccine has the potential to conquer one of the most devastating diseases of childhood.
Keywords: meningococcus B, reverse vaccinology
Contributed by Rino Rappuoli, May 12, 2006
Neisseria meningitidis is an encapsulated, Gram-negative bacterium that colonizes the upper respiratory tract of ≈10% of humans. With a frequency of one to three cases per 100,000 of the population, the bacterium enters the bloodstream, where it multiplies to high density and causes a form of sepsis characterized by the dramatic disruption of the endothelium and microvasculature. From the bloodstream the bacterium can cross the blood–brain barrier and cause meningitis. The invasive infection is very dramatic, affecting mostly infants, children, and adolescents who do not have bactericidal antibodies to the infecting strain. Within a few hours otherwise healthy subjects become severely sick, and, despite the sophistication of modern medicine and availability of effective antibiotics, 5–15% die, and up to 25% of the survivors have lifelong sequelae. During epidemics the frequency of disease increases to >10/100,000 of the population and rises to as high as 400/100,000. Based on the chemical composition of the polysaccharide capsule, N. meningitidis strains can be classified into 13 different serogroups. Strains representative of five serogroups (A, B, C, Y, and W135) cause nearly all disease in humans (1, 2).
Most of the strains isolated from invasive meningococcal disease have also been classified by multilocus enzyme electrophoresis (3) into several hypervirulent lineages (electrophoretic types: ET37, ET5, cluster A4, lineage 3, and subgroups I, III, and IV-1) or by multilocus sequence typing (4) into sequence type (ST) complexes (cpx) ST11, ST32, ST8, ST41/44, ST1, ST5, and ST4.
N. meningitidis causes ≈1.2 million cases per year globally, of which 3,000 occur in the United States and 7,000 occur in Europe, where the organism remains the most common cause of bacterial meningitis in children and young adults (5). Serogroup B meningococcus (MenB) is responsible for ≈32% of meningococcal disease reported in the United States, and from 45% to >80% of meningococcal disease reported in Europe (6, 7). Immunity against the disease can be acquired naturally or induced by vaccination and correlates with the presence of bactericidal antibodies, which kill the bacterium in the presence of complement (8). Tetravalent vaccines composed of the purified capsular polysaccharides of serogroups A, C, Y, and W135 have been available for three decades for use in adults. Conjugate vaccines, effective in all age groups, were developed a decade ago (9). Large-scale vaccination against serogroup C meningococcus in the United Kingdom in the year 2000 showed that these vaccines are very safe and effective in eliminating the disease (10) and led to the licensure of tetravalent conjugate vaccines against serotypes A, C, Y, and W-135. Once universally deployed, these vaccines will likely eliminate forever the disease caused by strains with these serogroups.
MenB differs from the others because it is decorated by a capsular polysaccharide identical to the polysialic acid [α(2–8)N-acetylneuraminic acid] present in many human glycoproteins. This mimicry rules out the use of polysaccharide–protein conjugate vaccines as an effective strategy to combat MenB disease. Therefore, currently there is no universal vaccine available against this bacterium. Protein-based vaccines composed of outer membrane vesicles (OMV) purified from the bacterium have been produced; however, they induce immunity mostly against the highly variable PorA membrane protein, and efficacy trials have shown serosubtype-specific protection (11). To develop a universal vaccine against MenB, a few years ago we determined the sequence of the genome of the bacterium and used it to discover novel antigens (12, 13). Thus, MenB became the prototype for the use of genomics for vaccine development, a process termed reverse vaccinology (14).
Twenty-eight novel antigens that were able to induce antibodies with bactericidal activity (BCA) in mice when used for immunization using Freund’s complete adjuvant (FCA) were identified. Some of the antigens had a high degree of sequence conservation using a historical panel of MenB strains and were recognized by sera of patients convalescent from meningococcal disease (15, 16). Detailed studies on some of the most promising antigens confirmed that some of these antigens were good candidates for a universal vaccine (16, 17), whereas others turned out to be less interesting. For instance, GNA33 was found to be a mimotope of PorA and thus unlikely to be useful as a vaccine because of serosubtype specificity (18).
However, although the in vitro antibody data obtained in mice immunized with the individual antigens in FCA were encouraging, it was not clear whether the responses were sufficient for the development of a vaccine suitable for human use and able to induce universal protection against recent, genetically diverse, and epidemiologically relevant clinical isolates.
Results
Antigen Selection.
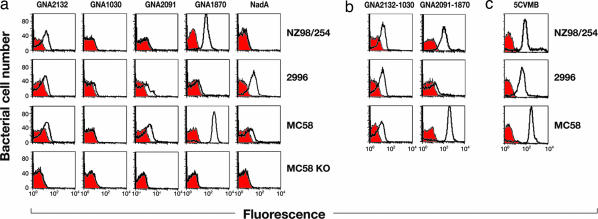
The antigens selected by reverse vaccinology were prioritized based on their ability to induce broad protection as inferred by BCA or observed in passive protection in the infant rat or mouse protection assays (19, 20). The three top antigens that met the prioritization criteria were GNA2132 (21), GNA1870 (16, 22, 23), and NadA (17, 24). Two additional antigens, GNA1030 and GNA2091 (13), were selected because they also induced protective immunity but only in some of the assays. FACS analysis (Fig. 1a) showed that the sera obtained from mice immunized with each of the five antigens formulated with aluminum hydroxide stained the surface of one or more of the MenB strains NZ98/254, 2996, and MC58, which were selected as representative of the most common patterns observed among a variety of different strains. The serum against GNA1030, which appears only slightly positive in the figure, showed positive binding to 2996 when the serum used had been obtained by immunizing with complete FCA instead of aluminum (data not shown). The figure also shows that none of the antisera prepared against the individual antigens stained all of the strains well. To confirm the specificity of the analysis, none of the sera stained the MC58 knockout mutants where the corresponding gene had been deleted (Fig. 1a, MC58 KO). Binding activity observed in FACS paralleled antibody functional activity observed in the BCA assay (Table 1). The presence of BCA was tested in sera obtained by immunizing mice with each of the antigens formulated with aluminum hydroxide or FCA (Table 1). With the exception of GNA2091, which, although unable to induce BCA, had been selected for its ability to induce protection in the mouse protection assay (19, 20) (data not shown), each of the other individual antigens induced BCA against one or more of the three strains. The proteins GNA2132, GNA1030, and GNA1870 induced better titers when formulated with FCA than when formulated with aluminum. In the case of NadA, the two adjuvants gave a similar titer.
Fig. 1.
FACS analysis of N. meningitidis strains using sera prepared against vaccine antigens. (a) Binding of polyclonal antiserum raised against the individual antigens GNA2132, GNA1030, GNA2091, GNA1870, and NadA (left to right) to live encapsulated MenB strains NZ98/254 (first row), 2996 (second row), and MC58 (third row). The fourth row contains the MC58 knockout mutants (KO) of each antigen probed with the homologous antiserum. Red profiles represent the negative control (preimmune sera). White profiles show reaction with immune sera. (b and c) Binding of polyclonal antibodies against fusion proteins (b) and 5VCMB vaccine (c). First row, NZ98/254 (B:4:P1.4), ST cpx 41/44, ET lin.3, nadA−. Second row, 2996 (B:2b:P1.5,2), ST cpx 8, ET A4. Third row, MC58 (B:15:P1.7,16b), ST cpx 32, ET ET5.
Table 1.
Bactericidal activity of mice polyclonal antibodies
| Immunogens | Strains | |||||
|---|---|---|---|---|---|---|
| MC58 | 2996 | NZ98/254 | ||||
| FCA | Al(OH)3 | FCA | Al(OH)3 | FCA | Al(OH)3 | |
| Antigens | ||||||
| GNA2132 | 16,384* | 512 | 16,384 | 1,024 | 16,384 | 512 |
| GNA1030 | <4 | <4 | 2,048 | <4 | <4 | 8 |
| GNA2091 | <4 | <4 | <4 | <4 | <4 | <4 |
| GNA1870 | 65,536 | 16,384 | 64 | <4 | 16,384 | 64 |
| NadA | 512 | 1,024 | 4,096 | 4,096 | <4 | <4 |
| Fusions | ||||||
| GNA2132–1030 | n.d. | 2,048 | n.d. | 2,048 | n.d. | 4,096 |
| GNA2091–1870 | n.d. | 32,768 | n.d. | <4 | n.d. | 1,024 |
| Vaccine | ||||||
| 5CVMB | 65,536 | 32,768 | 8,192 | 4,096 | 16,384 | 4,096 |
*Bactericidal titers were expressed as the reciprocal of the serum dilution yelding ≥50% bactericidal killing. n.d., not determined.
Although the five antigens had been selected based on their individual behavior, none of them induced protective antibody responses that were sufficiently broad enough to cover all strains tested.
A Combination of Antigens to Develop an Effective Serogroup B Vaccine.
Given that vaccines containing several antigens have been shown to confer better protection than those containing only one or two antigens (25), we combined the five antigens into one multicomponent vaccine, trusting that this would increase the breadth of the vaccine coverage and avoid selection of escape mutants. To facilitate large-scale industrial manufacturing of the vaccine, four of the five antigens were expressed as fusion proteins. Among >30 protein–protein fusions generated, the most promising were the combinations of GNA2132 with GNA1030 and of GNA2091 with GNA1870. NadA did not perform well when fused to other proteins, probably because of the loss of the trimeric organization (data not shown) (24, 26). The two fusion proteins formulated with aluminum hydroxide induced immune responses in both FACS (Fig. 1b) and BCA assays (Table 1) that were generally more potent than those induced by the individual antigens. Twenty micrograms of each of the two fusion proteins and of the NadA antigen was adsorbed to aluminum hydroxide to make the vaccine formulation that was used in subsequent studies. We named the multicomponent vaccine 5CVMB (5 component vaccine against MenB). In FACS analysis (Fig. 1c) the antibodies obtained by immunizing with this vaccine stained the three representative MenB strains very well. Similarly, the antibodies had BCA titers against each of the three strains, ranging from 4,096 to 32,768 (Table 1). The respective titers were only slightly lower than those obtained by immunizing with FCA, which is the most potent adjuvant known for mouse immunization but is unsuitable for human use.
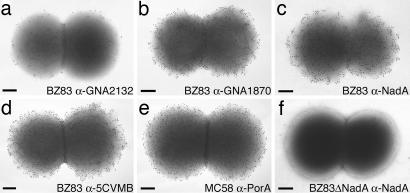
The potency of the multicomponent vaccine was also demonstrated by ImmunoGold staining of the bacterial surface using the three vaccine antigens that elicited the best BCA titers (Table 1). The antibodies against GNA2132, GNA1870, and NadA stained the surface of MenB strain BZ83 with different density (Fig. 2a–c). However, the staining performed with antisera raised against the vaccine (Fig. 2d) greatly increased the density of gold particles to a level that was comparable to the one obtained using antibodies against MC58 PorA (Fig. 2e), which is known to confer protection in humans.
Fig. 2.
ImmunoGold labeling and transmission electron microscopy. Analysis of BZ83 strain was performed with antisera raised against vaccine antigens: anti-GNA2132 (a), anti-GNA1870 (b), anti-NadA (c), and anti-5CVMB (d) vaccine. (e) Anti-PorA staining of strain MC58 performed by using a mAb raised against PorA 1.7. (f) Representative control showing anti-NadA staining of strain BZ83 mutant knockout for the nadA gene (BZ83ΔNadA). Similar results were obtained when knockout strains in gna2132 and gna1870 genes were analyzed with the corresponding antisera (data not shown). (Scale bars: 200 nm.)
Coverage of the MenB Population Diversity.
To properly test whether the 5CVMB vaccine formulation was able to induce protection against the majority of the MenB strains, we assembled and characterized a very large collection of clinical isolates representing as closely as possible the global population diversity. In total, 214 N. meningitidis strains were collected: 56 from the U.S. and Canada, 44 from the United Kingdom, 98 from other European countries, 2 from New Zealand, 6 from Australia, and 8 from other regions of the world. The collection contained mostly recent clinical isolates, although a minority of older laboratory strains were included to bridge the data to those generated in the past with well known strains. Eighty-five of the 214 strains were found to be suitable for BCA assays using several batches of baby rabbit complement. The main properties of the 85 strains are reported (Table 2). Four meningococcal hypervirulent clusters, ST32 cpx or ET5, ST41/44 cpx or lineage 3, ST8 cpx or cluster A4, and ST11 cpx or ET37, represent 56 of 85 (65.9%) of the total strains; 29 of 85 (34.1%) of the strains represent emerging lineages of pathogenic strains that were classified as “others” because they do not belong to any known hypervirulent cluster.
Table 2.
Characteristics of 85 N. meningitidis strains used for bactericidal assays
| Hypervirulent clusters | Year of isolation | Total | ||
|---|---|---|---|---|
| Before 1980 | 1980–1995 | 1996–2002 | ||
| ST32 cpx (ET5) | 1 | 4 | 6 | 11 |
| ST41/44 cpx (lineage 3) | 0 | 5 | 16 | 21 |
| ST8 cpx (A4) | 1 | 2 | 4 | 7 |
| ST11 cpx (ET37) | 2 | 4 | 11 | 17 |
| Others* | 3 | 8 | 18 | 29 |
| Total | 7 | 23 | 55 | 85 |
*The strains that do not belong to any of the hypervirulent clusters have been classified as others.
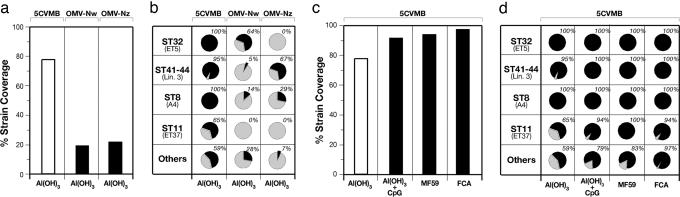
The sera obtained by immunizing mice with the 5CVMB vaccine formulated with aluminum hydroxide were tested in a bactericidal assay against this panel of 85 strains. We found that 66 of 85 (77.7%) of the strains tested were killed at serum dilutions ≥1/128 (Fig. 3a). This threshold titer correlates with a positive result using human complement (27). Killing of ≈78% of the strains by a meningococcal B vaccine is very encouraging, especially when compared with the 17 of 85 strains (20%) and with the 18 of 85 strains (21.2%) killed by the OMV vaccines made from the Norwegian strain H44/76 (Fig. 3a, center column) and the New Zealand strain NZ98/254 (Fig. 3a, right column), which represent the best vaccines available so far.
Fig. 3.
Protective effect of 5CVMB vaccine. The analysis has been performed by testing the immune sera for BCA against 85 N. meningitidis strains using baby rabbit serum as complement source (16). Results are expressed as the percentage of strains killed with titers ≥128. (a and b) Comparison of strain coverage induced by 5CVMB, OMVNw, and OMVNz vaccines (a) and the respective vaccine coverage of the 85 strains grouped into hypervirulent clusters (b). (c and d) Strain coverage induced by different formulations of 5CVMB vaccine (with aluminum hydroxide, aluminum hydroxide plus CpG 1826, MF59, or FCA) (c) and the respective vaccine coverage of hypervirulent clusters (d).
A closer observation of the strains covered showed that the Nw vaccine induced BCA titers >8,000 against all 9 strains with PorA P1.16, identical to that of the vaccine strain, titers of 512 or lower against 8 strains carrying a different PorA, and no BCA against the remaining 68 strains. Similarly, the Nz vaccine induced high BCA titers against the 11 strains with PorA P1.4 identical to the vaccine strain, low BCA titers against 7 strains with different PorA, and no BCA against the remaining 67 strains. The poor coverage of strains and hypervirulent lineages provided by OMV vaccines (Fig. 3b) validates the fragmentary observations obtained so far in preclinical and clinical studies showing that OMV vaccines induce immunity that is mostly PorA-specific.
In marked contrast to the OMV vaccines, and as expected from the antigen composition, the 5CVMB vaccine induced broader protection that was not serosubtype-specific and covered most of the population diversity. The protection induced by 5CVMB formulated with aluminum (Fig. 3b) was complete (100%) against all strains of the panel from the hypervirulent cpx ST32 and ST8, almost complete (95%) against the hypervirulent cpx ST41/44, and substantial against strains of hypervirulent cluster ST11 and those classified as others (65% and 59%, respectively). A subset of strains, for which human complement could be used for the bactericidal assay, was killed by the vaccine-induced antibodies in the presence of human complement. For instance, the BCA titers obtained against MenB strains MC58, 2996, and NZ98/254 were 1,024, 128, and 64, respectively.
To investigate whether the coverage of the 5CVMB vaccine could be increased, mice were immunized with the same vaccine antigens but adjuvanted by aluminum hydroxide combined with CpG oligonucleotides (28, 29). The results showed that the strain coverage increased to 91.7% (Fig. 3c). We also tested the vaccine using an oil-in-water emulsion, MF59, which is licensed for human use in Europe, as adjuvant (30). The presence of the MF59 increased the coverage to 94.1% (Fig. 3c). Interestingly, when FCA was used as adjuvant, coverage by BCA reached 97.6% (Fig. 3c). The addition of CpG to 5CVMB with aluminum hydroxide or the use of MF59 or FCA (Fig. 3d) significantly increased the coverage of strains belonging to the ST11 cluster and of those classified as others. These data show that in mice the antigens included in the vaccine can induce immunity against nearly all MenB strains when the immune response is optimized. The increased strain coverage could be due to the ability of the adjuvants to induce a higher immune response or to influence antigen conformation by exposing additional epitopes to the immune system. Further work will be needed to address this issue. In humans, the immune response can be optimized by the addition of CpG, by using MF59, or with a booster dose after primary immunization.
All animals treated with different formulations of 5CVMB vaccine were healthy throughout the experiments, based on daily observations, and no overt toxicity was observed.
To confirm that the antisera obtained were able to confer protection in vivo, representative sera from mice immunized with 5CVMB were also used for passive immunization of infant rats, which were subsequently challenged i.p. with MenB strain 2996. In two experiments where the animals were challenged with 6 × 104 and 5 × 103 virulent bacteria, respectively (Table 3), all infant rats that had received PBS as negative control developed a bacteremia after 18 h. In contrast, no bacteria were recovered from the blood of rats that had been passively immunized.
Table 3.
Passive protection in the infant rat meningococcal bacteremia model
| Exp. | 2996 (challenged CFU) | Treatment | Serum dilution or dose of mAb, μg, per rat | No. positive/total | CFU/ml (geometric mean) |
|---|---|---|---|---|---|
| 1 | 63,000 | Anti-PorA(Pl.2) mAb | 2 | 0/3 | <1 |
| Anti-B5CVMB mouse serum | 1:5 | 0/3 | <1 | ||
| 1:20 | 0/3 | <1 | |||
| Anti-5CVMB rabbit serum | 1:5 | 0/3 | <1 | ||
| 1:20 | 0/3 | 5 | |||
| PBS control | 3/3 | 146,590 | |||
| 2 | 5,000 | Anti-PorA(Pl.2) mAb | 1 | 0/4 | <1 |
| Anti-5CVMB mouse serum | 1:10 | 0/4 | <1 | ||
| 1:50 | 0/4 | <1 | |||
| Anti-5CVMB rabbit serum | 1:10 | 0/4 | <1 | ||
| 1:50 | 0/4 | <1 | |||
| PBS control | 4/4 | 8,250 |
CFU, colony-forming unit.
Discussion
We have described a universal vaccine against MenB, a bacterium that causes a disease against which conventional vaccinology has been ineffective and that still kills many young people in both developed and developing countries. The vaccine, developed by using novel antigens identified by reverse vaccinology, covers 78% of the strains in the basic formulation using aluminum hydroxide as adjuvant, and coverage can be increased to >95% of the strains by using other adjuvants. The progress compared with vaccines available so far is gigantic; in fact, OMV vaccines, which are the best vaccines against MenB developed during the last 30 years, are shown to cover only 20% of the MenB strains.
The work also shows that universal protein-based vaccines can be developed against those encapsulated bacteria that are usually targeted by conjugate vaccines. This finding should foster research on reverse vaccinology to identify protein-based vaccines for other pathogens, such as pneumococcus, where there are too many serotypes of capsular polysaccharides to make conjugate vaccines capable of protecting against all strains, against Staphylococcus, group A Streptococcus, and other microorganisms.
Although protein-based vaccines have also been described for group B Streptococcus (31) and other pathogens, so far most of the data in the literature were obtained by using vaccines formulated with adjuvants unsuitable for human use, such as FCA, and usually tested against a limited number of bacterial strains, so that the coverage of the population diversity could not be totally predicted. Therefore, the question of whether protein-based vaccines against encapsulated bacteria are feasible for use in humans was still pending. In this study we have used the antigens derived from the genome mining of MenB, and we have asked the critical questions necessary for extending their large-scale use to humans. To do so, we expressed them in a system suitable for industrial manufacturing; we formulated them with an adjuvant suitable for human use, such as aluminum hydroxide, and we tested the immune response against an unusually large panel of strains enriched with recent clinical isolates and designed to represent as much as possible the true diversity of the bacterial population.
If the results reported here are confirmed in clinical trials, this vaccine is likely to conquer one of the most devastating diseases that still cause morbidity and mortality in children and adolescents.
Materials and Methods
Cloning Expression and Purification of Vaccine Components.
Single recombinant proteins were expressed and purified as previously described (13, 16, 17). Genes considered in this study were from the genome of N. meningitidis NZ98/254 strain in the case of GNA2132, MC58 strain in the case of GNA1870, and 2996 strain in the case of GNA1030, GNA2091, and NadA.
All of the proteins present in the multicomponent vaccine 5CVMV were produced in Escherichia coli devoid of the predicted leader peptide using the pET24b+ as expression vector and E. coli BL21 (DE3) as expression host. NadA corresponds to the NadAΔ351–405 recombinant form (17).
To generate the fusion GNA2132–1030, the gna2132 gene was amplified by PCR from the NZ98/254 strain by using the following oligonucleotides: 2132 forward (CGCGGATCCGCTAGC-CCCGATGTTAAATCGGC) and 2132 reverse (CGGGGATCC-ATCCTGCTCTTTTTTGCCGG) containing the NheI and BamHI restriction sites, respectively (restriction sites are underlined); the gna1030 gene was amplified from the 2996 strain by using 1030 forward (CGCGGATCCGGTGGTGGTGGT-GCCACCTACAAAGTGGAC) and 1030 reverse (CCCGCTCGAG-TTATTGTTTGGCTGCCTCGAT) oligonucleotides, which include the BamHI and XhoI restriction sites, respectively. A polyglycines linker was added between the two proteins of the fusion inserting the corresponding DNA sequence as tail in the sequence of the primer forward. The insertion of NheI and BamHI sites results in alanine–serine or glycine–serine additional amino acids, respectively.
Amplified DNA fragments were digested with NheI/BamHI and with BamHI/XhoI enzymes and cloned in pET24b+ vector digested with NheI/XhoI.
To create the fusion GNA2091–1870, the gna2091 gene was amplified from the 2996 strain by using the oligonucleotides 2091 forward (CGCGGATCCCATATG-GTCAGCGCAGTAATCGGA) and 2091 reverse (CGGGGATCC-GCGTTGGACGTAGTTTTG) containing the NdeI and BamHI restriction sites, respectively; the gna 1870 gene was amplified from the MC58 strain by using 1870 forward (CGCGGATCC-GGAGGGGGTGGTGTCG) and 1870 reverse (CCCGCTCGAG-TTATTGCTTGGCGGCAAG) oligonucleotides, including the BamHI and XhoI restriction sites, respectively.
Amplified DNA fragments were digested with NdeI/XhoI and BamHI/XhoI enzymes and cloned in pET24b+ vector digested with NdeI/XhoI.
The recombinant proteins were expressed in E. coli and purified by standard methods (32).
Mouse Immunizations.
To prepare antisera, 20 μg of individual antigens GNA2132, GNA1030, GNA2091, GNA1870, NadA, GNA2132–1030, and GNA2091–1870 or a combination of 20 μg each of GNA2132–1030, GNA2091–1870, and NadA was used to immunize 6-week-old CD1 female mice (Charles River Breeding Laboratories). Five to 10 mice per group were used. The antigens were administered i.p. together with aluminum hydroxide (3 mg/ml) on days 0, 21, and 35. The same immunization schedule was performed with different adjuvants such as CpG 1826 plus aluminum hydroxide, MF59, or FCA. Blood samples for analysis were taken on day 49. To prepare antisera against OMV preparation (from H44/76 strain for NW and NZ98/254 strain for NZ), 10 μg of each adsorbed to aluminum hydroxide was used. The same immunization schedule was used to immunize New Zealand White rabbits. The rabbits were injected in several lymph nodes with 5CVMB vaccine (25 μg of each antigen). The treatments were performed in accordance with internal animal ethical committee and institutional guidelines.
FACS Analysis.
The analysis was determined by using a FACScan flow cytometer as described previously (16). Polyclonal sera were tested at a 1/200 dilution. Antibody binding was detected by using a secondary anti-mouse (whole molecule) FITC-conjugated (Sigma) antibody.
Immunoelectron Microscopy.
Whole-cell, negative-stain, ImmunoGold labeling was performed by using a modified procedure described by Fletcher et al. (22). Briefly, N. meningitidis serogroup B strains cells were fixed for 60 min at room temperature in 4% paraformaldehyde plus 0.05% glutaraldehyde in PBS (pH 7.2). After fixation, droplets of cells were placed on Formvar carbon-coated grids (Agar Scientific) for 5 min. Excess fluid was wicked off, and blocking was accomplished in two stages by using PBS plus 1% BSA for 5 min and, later, PBS containing 1% coldwater fish gelatin (Fluka) for 10 min. Excess aldehyde was quenched by using 0.02 M glycine in PBS for 5 min. Grids were inverted over polyclonal antibodies or mAbs (diluted in PBS plus 1% BSA) for 60 min in a humidified chamber. Grids were rinsed five times for 1 min in PBS plus 1% BSA. Antigen was detected by incubation for 60 min with goat anti-mouse IgG plus IgM conjugated to 10-nm colloidal gold beads (BB International, Cardiff, U.K.) diluted in PBS–BSA. Rinsing took place in PBS (four times for 1 min). Grids with cells were stabilized with 1% glutaraldehyde in PBS. Each sample was then rinsed five times in distilled water. Grids were finally treated with uranyl acetate and examined by a TEM GEOL 1200EX II transmission electron microscope.
Serum Bactericidal Assay.
Serum BCA against N. meningitidis strains was evaluated as previously described (23). The complement source was pooled baby rabbit sera (Cedarlane Laboratories or Pel-Freez Biologicals) or human serum from a healthy adult with no detectable intrinsic bactericidal activity.
Infant Rat Protection Assay.
The ability of anti-5CVMB to confer protection against N. meningitidis bacteremia was evaluated as previously described in infant rats challenged i.p. (33). Bacteria numbers in the blood cultures were obtained by plating out onto chocolate agar 100 μl of blood collected 18 h after challenge.
Acknowledgments
We thank Marco Tortoli, Giacomo Matteucci, and the animal care facility at Novartis Vaccines for technical assistance; Roberto Olivieri and Erwin Swennen for fermentations; Sandra Nuti for help with FACS analysis; Anna Rita Taddei for technical help with ImmunoGold labeling and transmission electron microscopy; Maria Scarselli for help in bioinformatics; and Cesira Galeotti, Duccio Medini, and Claudio Donati for discussion. We also thank Catherine Mallia for manuscript editing and Giorgio Corsi for artwork.
Abbreviations
- ET
electrophoretic type
- ST
sequence type
- MenB
serogroup B meningococcus
- OMV
outer membrane vesicle(s)
- BCA
bactericidal activity
- cpx
complex
- FCA
Freund’s complete adjuvant.
Footnotes
Conflict of interest statement: All authors either are employed by or have an ongoing collaboration with Novartis Vaccines.
See accompanying Profile on page 10831.
References
- 1.Gotschlich E. C., Goldschneider I., Artenstein M. S. J. Exp. Med. 1969;129:1367–1384. doi: 10.1084/jem.129.6.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gotschlich E. C., Liu T. Y., Artenstein M. S. J. Exp. Med. 1969;129:1349–1365. doi: 10.1084/jem.129.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caugant D. A., Bovre K., Gaustad P., Bryn K., Holten E., Hoiby E. A., Froholm L. O. J. Gen. Microbiol. 1986;132:641–652. doi: 10.1099/00221287-132-3-641. [DOI] [PubMed] [Google Scholar]
- 4.Maiden M. C., Bygraves J. A., Feil E., Morelli G., Russell J. E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D. A., et al. Proc. Natl. Acad. Sci. USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. Wkly. Epidemiol. Rec. 2001;37:281–288. [Google Scholar]
- 6.Rosenstein N. E., Perkins B. A., Stephens D. S., Lefkowitz L., Cartter M. L., Danila R., Cieslak P., Shutt K. A., Popovic T., Schuchat A., et al. J. Infect. Dis. 1999;180:1894–1901. doi: 10.1086/315158. [DOI] [PubMed] [Google Scholar]
- 7.Noah N., Henderson B. Surveillance of Bacterial Meningitis in Europe 1997/8. London: Communicable Disease Surveillance Centre; 2002. pp. 1–16. [Google Scholar]
- 8.Goldschneider I., Gotschlich E. C., Artenstein M. S. J. Exp. Med. 1969;129:1307–1326. doi: 10.1084/jem.129.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costantino P., Viti S., Podda A., Velmonte M. A., Nencioni L., Rappuoli R. Vaccine. 1992;10:691–698. doi: 10.1016/0264-410x(92)90091-w. [DOI] [PubMed] [Google Scholar]
- 10.Snape M. D., Pollard A. J. Lancet Infect. Dis. 2005;5:21–30. doi: 10.1016/S1473-3099(04)01251-4. [DOI] [PubMed] [Google Scholar]
- 11.Tappero J. W., Lagos R., Ballesteros A. M., Plikaytis B., Williams D., Dykes J., Gheesling L. L., Carlone G. M., Hoiby E. A., Holst J., et al. J. Am. Med. Assoc. 1999;281:1520–1527. doi: 10.1001/jama.281.16.1520. [DOI] [PubMed] [Google Scholar]
- 12.Tettelin H., Saunders N. J., Heidelberg J., Jeffries A. C., Nelson K. E., Eisen J. A., Ketchum K. A., Hood D. W., Peden J. F., Dodson R. J., et al. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 13.Pizza M., Scarlato V., Masignani V., Giuliani M. M., Arico B., Comanducci M., Jennings G. T., Baldi L., Bartolini E., Capecchi B., et al. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 14.Rappuoli R. Curr. Opin. Microbiol. 2000;3:445–450. doi: 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 15.Litt D. J., Savino S., Beddek A., Comanducci M., Sandiford C., Stevens J., Levin M., Ison C., Pizza M., Rappuoli R., Kroll J. S. J. Infect. Dis. 2004;190:1488–1497. doi: 10.1086/424464. [DOI] [PubMed] [Google Scholar]
- 16.Masignani V., Comanducci M., Giuliani M. M., Bambini S., Adu-Bobie J., Arico B., Brunelli B., Pieri A., Santini L., Savino S., et al. J. Exp. Med. 2003;197:789–799. doi: 10.1084/jem.20021911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comanducci M., Bambini S., Brunelli B., Adu-Bobie J., Arico B., Capecchi B., Giuliani M. M., Masignani V., Santini L., Savino S., et al. J. Exp. Med. 2002;195:1445–1454. doi: 10.1084/jem.20020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granoff D. M., Moe G. R., Giuliani M. M., Adu-Bobie J., Santini L., Brunelli B., Piccinetti F., Zuno-Mitchell P., Lee S. S., Neri P., et al. J. Immunol. 2001;167:6487–6496. doi: 10.4049/jimmunol.167.11.6487. [DOI] [PubMed] [Google Scholar]
- 19.West D., Reddin K., Matheson M., Heath R., Funnell S., Hudson M., Robinson A., Gorringe A. Infect. Immun. 2001;69:1561–1567. doi: 10.1128/IAI.69.3.1561-1567.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorringe A. R., Reddin K. M., Funnell S. G., Johansson L., Rytkonen A., Jonsson A. B. Vaccine. 2005;23:2214–2217. doi: 10.1016/j.vaccine.2005.01.053. [DOI] [PubMed] [Google Scholar]
- 21.Welsch J. A., Moe G. R., Rossi R., Adu-Bobie J., Rappuoli R., Granoff D. M. J. Infect. Dis. 2003;188:1730–1740. doi: 10.1086/379375. [DOI] [PubMed] [Google Scholar]
- 22.Fletcher L. D., Bernfield L., Barniak V., Farley J. E., Howell A., Knauf M., Ooi P., Smith R. P., Weise P., Wetherell M., et al. Infect. Immun. 2004;72:2088–2100. doi: 10.1128/IAI.72.4.2088-2100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giuliani M. M., Santini L., Brunelli B., Biolchi A., Arico B., Di Marcello F., Cartocci E., Comanducci M., Masignani V., Lozzi L., et al. Infect. Immun. 2005;73:1151–1160. doi: 10.1128/IAI.73.2.1151-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capecchi B., Adu-Bobie J., Di Marcello F., Ciucchi L., Masignani V., Taddei A., Rappuoli R., Pizza M., Arico B. Mol. Microbiol. 2005;55:687–698. doi: 10.1111/j.1365-2958.2004.04423.x. [DOI] [PubMed] [Google Scholar]
- 25.Olin P., Rasmussen F., Gustafsson L., Hallander H. O., Heijbel H. Lancet. 1997;350:1569–1577. doi: 10.1016/s0140-6736(97)06508-2. [DOI] [PubMed] [Google Scholar]
- 26.Barocchi M. A., Masignani V., Rappuoli R. Nat. Rev. Microbiol. 2005;3:349–358. doi: 10.1038/nrmicro1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrews N., Borrow R., Miller E. Clin. Diagn. Lab. Immunol. 2003;10:780–786. doi: 10.1128/CDLI.10.5.780-786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis H. L., Weeratna R., Waldschmidt T. J., Tygrett L., Schorr J., Krieg A. M. J. Immunol. 1998;160:870–876. [PubMed] [Google Scholar]
- 29.Weeratna R. D., McCluskie M. J., Xu Y., Davis H. L. Vaccine. 2000;18:1755–1762. doi: 10.1016/s0264-410x(99)00526-5. [DOI] [PubMed] [Google Scholar]
- 30.Podda A., Del Giudice G. Expert Rev. Vaccines. 2003;2:197–203. doi: 10.1586/14760584.2.2.197. [DOI] [PubMed] [Google Scholar]
- 31.Maione D., Margarit I., Rinaudo C. D., Masignani V., Mora M., Scarselli M., Tettelin H., Brettoni C., Iacobini E. T., Rosini R., et al. Science. 2005;309:148–150. doi: 10.1126/science.1109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coligan J. E., Dunn B. M., Ploegh H. L., Speiches D. W., Wingfield P. T., editors. Current Protocols in Protein Science. New York: Wiley; 1997. pp. 6.0.1–6.7.14.pp. 9.4.1–9.4.16. [Google Scholar]
- 33.Moe G. R., Tan S., Granoff D. M. Infect. Immun. 1999;67:5664–5675. doi: 10.1128/iai.67.11.5664-5675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]