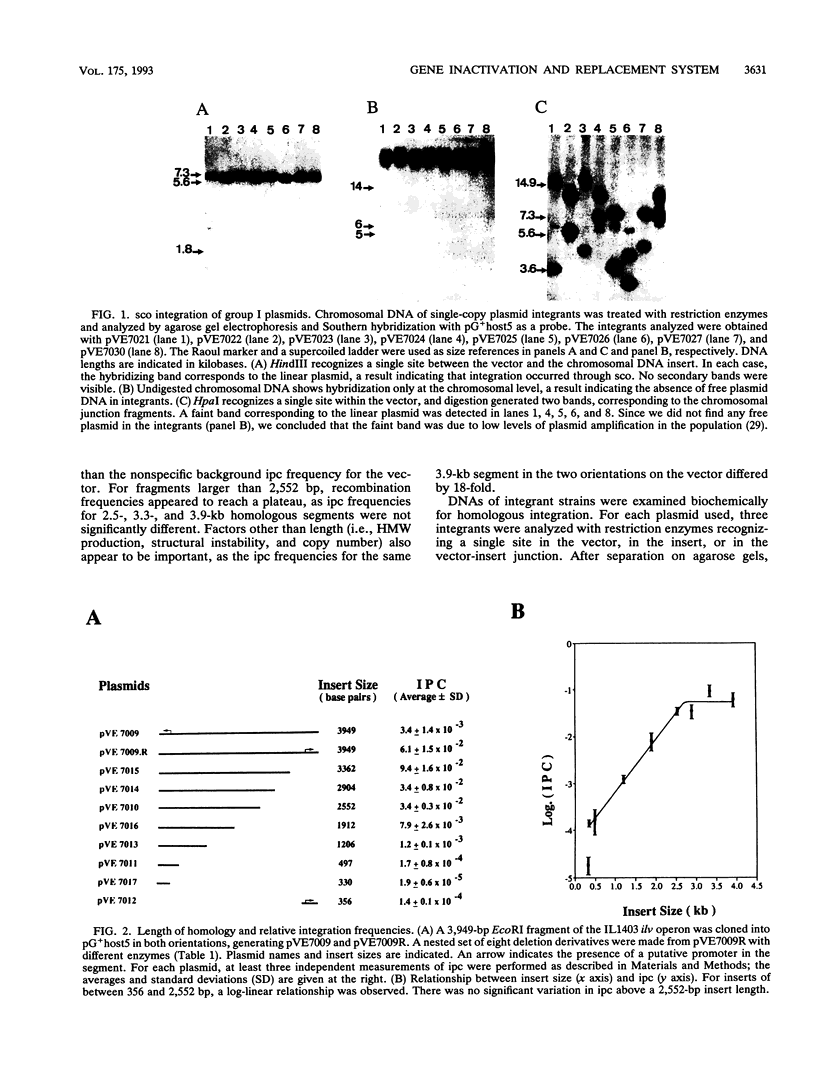
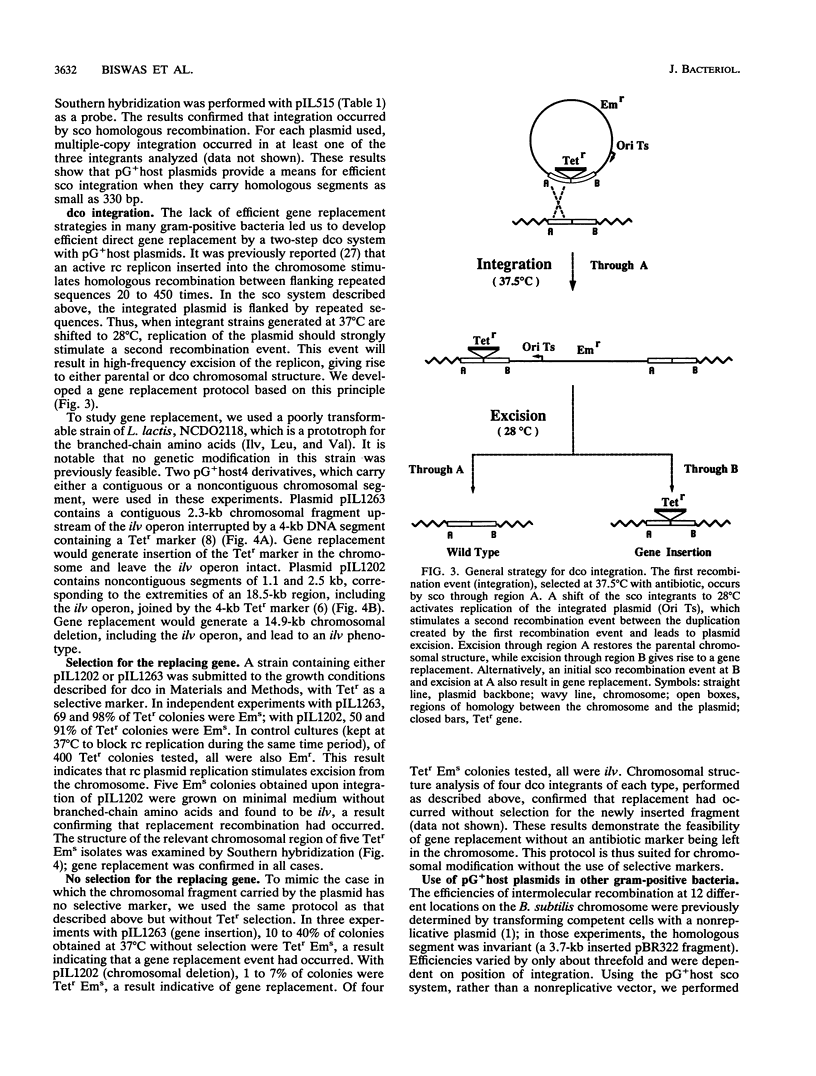
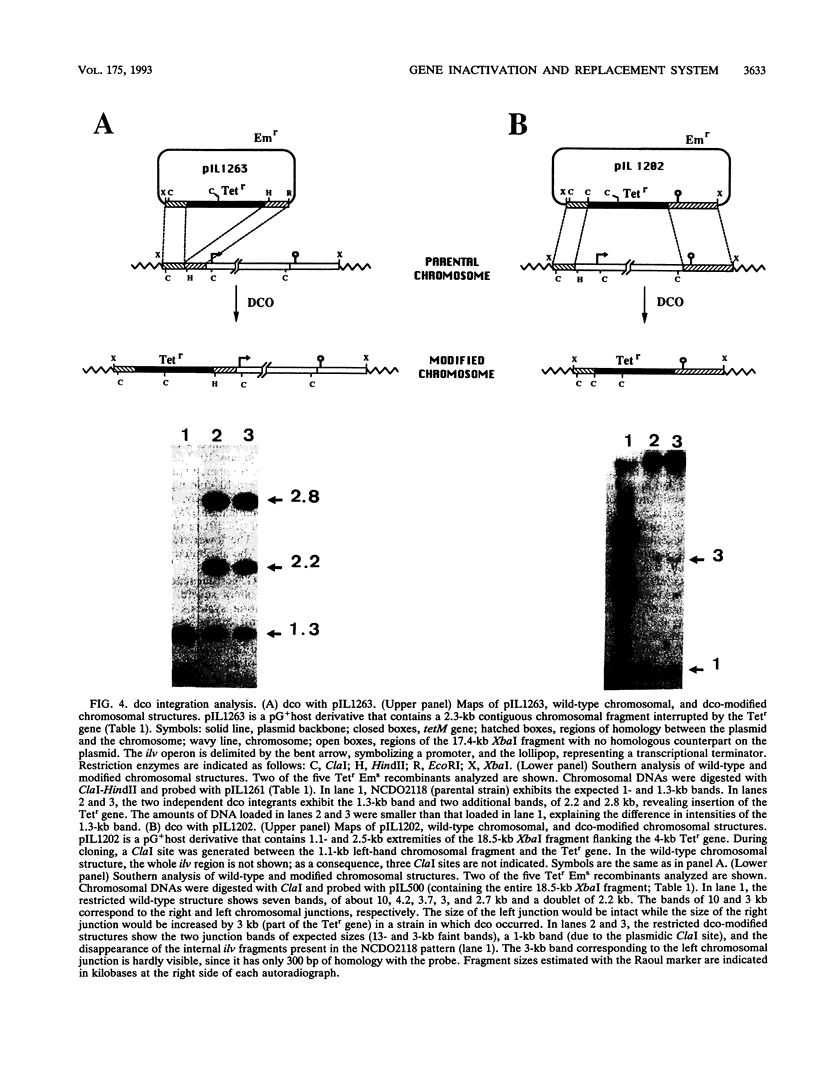
Abstract
A system for high-efficiency single- and double-crossover homologous integration in gram-positive bacteria has been developed, with Lactococcus lactis as a model system. The system is based on a thermosensitive broad-host-range rolling-circle plasmid, pG+host5, which contains a pBR322 replicon for propagation in Escherichia coli at 37 degrees C. A nested set of L. lactis chromosomal fragments cloned onto pG+host5 were used to show that the single-crossover integration frequency was logarithmically proportional to the length of homology for DNA fragments between 0.35 and 2.5 kb. Using random chromosomal 1-kb fragments, we showed that homologous integration can occur along the entire chromosome. We made use of the reported stimulatory effect of rolling-circle replication on intramolecular recombination to develop a protocol for gene replacement. Cultures were first maintained at 37 degrees C to select for a bacterial population enriched for plasmid integrants; activation of the integrated rolling-circle plasmid by a temperature shift to 28 degrees C resulted in efficient plasmid excision by homologous recombination and replacement of a chromosomal gene by the plasmid-carried modified copy. More than 50% of cells underwent replacement recombination when selection was applied for the replacing gene. Between 1 and 40% of cells underwent replacement recombination when no selection was applied. Chromosomal insertions and deletions were obtained in this way. These results show that gene replacement can be obtained at an extremely high efficiency by making use of the thermosensitive rolling-circle nature of the delivery vector. This procedure is applicable to numerous gram-positive bacteria.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biswas I., Vagner V., Ehrlich S. D. Efficiency of homologous intermolecular recombination at different locations on the Bacillus subtilis chromosome. J Bacteriol. 1992 Sep;174(17):5593–5596. doi: 10.1128/jb.174.17.5593-5596.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Duncan C. H., Wilson G. A., Young F. E. Mechanism of integrating foreign DNA during transformation of Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3664–3668. doi: 10.1073/pnas.75.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ftouhi N., Guillén N. Genetic analysis of fusion recombinants in Bacillus subtilis: function of the recE gene. Genetics. 1990 Nov;126(3):487–496. doi: 10.1093/genetics/126.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon J. J., Chopin M. C., Ehrlich S. D. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992 Oct;174(20):6580–6589. doi: 10.1128/jb.174.20.6580-6589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. Insertion of foreign DNA into plasmids from gram-positive bacteria induces formation of high-molecular-weight plasmid multimers. J Bacteriol. 1988 Mar;170(3):1183–1190. doi: 10.1128/jb.170.3.1183-1190.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton C. M., Aldea M., Washburn B. K., Babitzke P., Kushner S. R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989 Sep;171(9):4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holo H., Nes I. F. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989 Dec;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khasanov F. K., Zvingila D. J., Zainullin A. A., Prozorov A. A., Bashkirov V. I. Homologous recombination between plasmid and chromosomal DNA in Bacillus subtilis requires approximately 70 bp of homology. Mol Gen Genet. 1992 Sep;234(3):494–497. doi: 10.1007/BF00538711. [DOI] [PubMed] [Google Scholar]
- Kok J., van der Vossen J. M., Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984 Oct;48(4):726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourgeois P., Lautier M., Mata M., Ritzenthaler P. Physical and genetic map of the chromosome of Lactococcus lactis subsp. lactis IL1403. J Bacteriol. 1992 Nov;174(21):6752–6762. doi: 10.1128/jb.174.21.6752-6762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourgeois P., Mata M., Ritzenthaler P. Genome comparison of Lactococcus strains by pulsed-field gel electrophoresis. FEMS Microbiol Lett. 1989 May;50(1-2):65–69. doi: 10.1016/0378-1097(89)90460-6. [DOI] [PubMed] [Google Scholar]
- Leenhouts K. J., Kok J., Venema G. Campbell-like integration of heterologous plasmid DNA into the chromosome of Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1989 Feb;55(2):394–400. doi: 10.1128/aem.55.2.394-400.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenhouts K. J., Kok J., Venema G. Replacement recombination in Lactococcus lactis. J Bacteriol. 1991 Aug;173(15):4794–4798. doi: 10.1128/jb.173.15.4794-4798.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenhouts K. J., Tolner B., Bron S., Kok J., Venema G., Seegers J. F. Nucleotide sequence and characterization of the broad-host-range lactococcal plasmid pWVO1. Plasmid. 1991 Jul;26(1):55–66. doi: 10.1016/0147-619x(91)90036-v. [DOI] [PubMed] [Google Scholar]
- Maguin E., Duwat P., Hege T., Ehrlich D., Gruss A. New thermosensitive plasmid for gram-positive bacteria. J Bacteriol. 1992 Sep;174(17):5633–5638. doi: 10.1128/jb.174.17.5633-5638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P., Trieu-Cuot P., Courvalin P. Nucleotide sequence of the tetM tetracycline resistance determinant of the streptococcal conjugative shuttle transposon Tn1545. Nucleic Acids Res. 1986 Sep 11;14(17):7047–7058. doi: 10.1093/nar/14.17.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Recombination efficiency is a quadratic function of the length of homology during plasmid transformation of Bacillus subtilis protoplasts and Escherichia coli competent cells. EMBO J. 1984 Dec 1;3(12):2879–2884. doi: 10.1002/j.1460-2075.1984.tb02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaudet B., Goze A., Ehrlich S. D. Insertional mutagenesis in Bacillus subtilis: mechanism and use in gene cloning. Gene. 1982 Oct;19(3):277–284. doi: 10.1016/0378-1119(82)90017-8. [DOI] [PubMed] [Google Scholar]
- Noirot P., Petit M. A., Ehrlich S. D. Plasmid replication stimulates DNA recombination in Bacillus subtilis. J Mol Biol. 1987 Jul 5;196(1):39–48. doi: 10.1016/0022-2836(87)90509-2. [DOI] [PubMed] [Google Scholar]
- Petit M. A., Mesas J. M., Noirot P., Morel-Deville F., Ehrlich S. D. Induction of DNA amplification in the Bacillus subtilis chromosome. EMBO J. 1992 Apr;11(4):1317–1326. doi: 10.1002/j.1460-2075.1992.tb05176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi G., Guild W. R. Modes of integration of heterologous plasmid DNA into the chromosome of Streptococcus pneumoniae. J Bacteriol. 1985 Mar;161(3):909–912. doi: 10.1128/jb.161.3.909-912.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner V., Ehrlich S. D. Efficiency of homologous DNA recombination varies along the Bacillus subtilis chromosome. J Bacteriol. 1988 Sep;170(9):3978–3982. doi: 10.1128/jb.170.9.3978-3982.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]





