Abstract
Arabidopsis det2 mutants are small dark-green dwarfs displaying pleiotropic defects in light-regulated development during multiple stages of the plant life cycle. The DET2 gene encodes a protein that shares ≈40% sequence identity with mammalian steroid 5α-reductases and is implicated in the synthesis of a class of plant steroids, the brassinosteroids. Here we show that the DET2 protein, when expressed in human embryonic kidney 293 cells, catalyzes the 5α-reduction of several animal steroid substrates and has similar kinetic properties to the mammalian steroid 5α-reductase enzymes. Moreover, human steroid 5α-reductases expressed in det2 mutant plants can substitute for DET2 in brassinosteroid biosynthesis. These data indicate that DET2 is an ortholog of the mammalian steroid 5α-reductases and provide further evidence that brassinosteroids play an essential role in light-regulated plant development. The structural and functional conservation between DET2 and human steroid 5α-reductases raise interesting issues concerning the evolutionary origin of the steroid hormone signaling system.
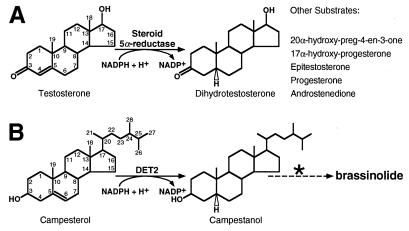
Steroid hormones play important roles in mammalian development and homeostasis (1). While many steroids have been identified in plants (2), to date only the brassinosteroids (BRs) are widely distributed throughout the plant kingdom and elicit unique growth promoting activity when applied exogenously (3). Despite these properties, the physiological role of endogenous BRs in plant development has been unclear. We recently isolated the Arabidopsis DET2 gene by positional cloning (4) and found that DET2 encodes a 262-amino acid protein that shares significant sequence identity with mammalian steroid 5α-reductases, enzymes that play crucial roles in steroid hormone metabolism and action by catalyzing an NADPH-dependent reduction of the Δ4,5 double bond in a variety of steroids (Fig. 1A) (5). Mutations in DET2, including a missense mutation at residue Glu-204, which is absolutely required for human steroid 5α-reductase (hS5R) activity (5), lead to many defects in light-dependent Arabidopsis development that can be ameliorated by application of brassinolide, an active BR (4, 6). Based on these results, we proposed that DET2 encodes a 5α-reductase in a BR biosynthetic pathway (Fig. 1B) and that BRs constitute an important class of plant hormones (4).
Figure 1.
Chemical reactions catalyzed by mammalian steroid 5α-reductases (A) and that proposed for Arabidopsis DET2 (B).
Support for the postulated importance of BRs in plant development came from the identification of a second Arabidopsis gene, CPD (constitutive photomorphogenesis and dwarfism) (7). The CPD gene encodes a protein that shares sequence identity with several mammalian cytochrome P450 proteins including steroid hydroxylases. Mutations in CPD cause phenotypic defects that are similar to those of det2 mutations. Moreover, brassinolide treatment also restores a wild-type phenotype to cpd mutants. Rescue studies with intermediates in the BR biosynthetic pathway suggest that CPD may encode a steroid 23-hydroxylase (7).
In mammals, steroid hormones are synthesized from cholesterol via pregnenolone through a series of reactions that modify the ring structure and the side chain of the sterol (8). Similarly, BRs are thought to be derived from several major phytosterols (e.g., campesterol, sitosterol, and stigmasterol) via multiple oxidation steps (9). Many mammalian steroid hormones have a Δ4,5 double bond in ring A and the reduction of this bond, catalyzed by steroid 5α-reductase, serves to either decrease or enhance hormone activity depending on the steroid and tissue in which the reaction is catalyzed (5). In contrast, all known naturally occurring and biologically active BRs lack double bonds in the A and B rings and contain a 5α-reduced stereochemistry. A steroid 5α-reductase must therefore be required for the formation of the trans A/B ring junction that is essential for the biological activity of BRs (3).
Two steroid 5α-reductase isozymes (types 1 and 2) have been identified in the human, rat, mouse, and monkey (5, 10). The isozymes share ≈50% sequence identity, show similar substrate preferences, have similar gene structures and are both integral membrane proteins of the endoplasmic reticulum; however, they differ in their affinities for steroid substrates, pH optima, sensitivities to certain 4-azasteroid inhibitors, tissue distributions, and physiological functions (5). Inherited deficiencies of steroid 5α-reductase type 2, but not type 1, result in a form of male pseudohermaphroditism in which the external genitalia fail to develop normally (11). The Arabidopsis DET2 protein shares 38–42% sequence identity with the mammalian steroid 5α-reductase isozymes and has a similar hydropathy profile. Eighty percent of the absolutely conserved residues in the mammalian enzymes are present in the predicted sequence of the DET2 protein. This extensive similarity to mammalian steroid 5α-reductases suggests that DET2 may perform a similar in vivo biochemical function as the mammalian enzymes. Here, we test this hypothesis and show that DET2 is a functional homolog of the mammalian steroid 5α-reductases and capable of acting on progesterone and testosterone. Moreover, stable transformation of det2 mutant plants with either a human type 1 or type 2 steroid 5α-reductase cDNA rescue the pleiotropic defects caused by det2 mutations.
MATERIALS AND METHODS
Plant Materials and Growth Conditions.
The standard wild-type genotype used was Arabidopsis thaliana Columbia (Col-0). The det2-1 mutant used in this study has been described (6). Seeds were surfaced sterilized by washing for 1 min in 95% ethanol, followed by 12 min in a 1:3 dilution of bleach (Clorox) containing 0.02% (vol/vol) Tween-20. The seeds were then washed three times with sterilized distilled water, and resuspended in 0.08% phytagar (GIBCO/BRL). After treatment for 2 days at 4°C to induce germination, seeds were sown in Petri plates containing 0.5× MS (12) medium (pH 5.7) supplemented with 1% sucrose, 0.8% phytagar, and 1× Gamborg’s B5 vitamins (Sigma) and maintained in growth chambers at 21°C with a 16-hr photoperiod. Plants were grown in the dark by wrapping plates with three layers of aluminum foil. For all experiments, det2 and the Col-0 wild-type plants were grown side-by-side under the same light and humidity conditions.
Expression of DET2 cDNA in Human Embryonic Kidney 293 Cells.
A 948-bp fragment from a DET2 cDNA clone containing 18 bp of 5′-untranslated sequence, an ORF of 786 bp and 154 bp of 3′-untranslated sequence was ligated into the pCMV5 expression vector (13). As a control, a mutant det2 protein was also expressed in the pCMV5 vector (pCMV5-det2-1). This construct, which contains a single nucleotide mutation (G-A) that changes Glu-204 to Lys-204, was made by replacing the wild-type DET2 sequence with the same restriction fragment derived from PCR-amplified det2-1 genomic DNA. The resulting plasmid was then digested with EcoRI/BglII and cloned into the pCMV5 expression vector. Five μg of expression plasmid containing either wild-type DET2 or mutated det2-1 cDNA was transfected into human embryonic kidney 293 cells (ATCC CRL1573) by a calcium phosphate precipitation procedure as described (14). Sixteen hours after transfection, steroid 5α-reductase activity was assayed in either intact cells or cell lysates as described (15, 16).
Transformation of det2-1 Mutants.
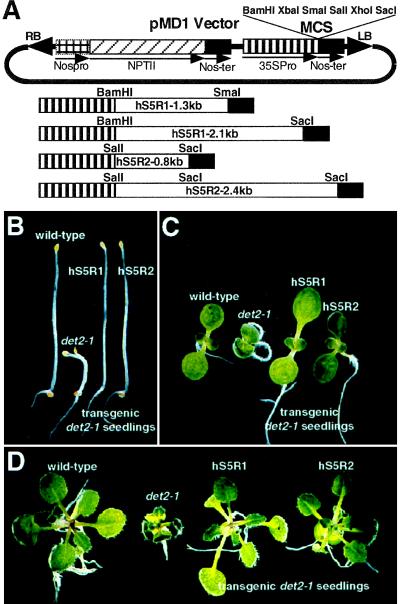
A human cDNA encoding either the type 1 or type 2 steroid 5α-reductase was cloned into the pMD1 vector, a derivative of pBI121 (CLONTECH), which contains the cauliflower mosaic virus 35S promoter (CaMV) and a nopaline synthase transcriptional terminator, to generate pMD1-hS5R plasmids (see Fig. 4A). Agrobacterium strain GV3101, transformed with a pMD1-hS5R construct, was used to transform det2-1 mutants by the vacuum infiltration method as described previously (17). The transformants (T1) were selected on 0.5× MS medium (pH 5.7) (12) supplemented with 1% sucrose, 0.8% phytagar, 1× Gamborg’s B5 vitamins, and 25 μg/ml kanamycin. Kanamycin-resistant seedlings were transferred to soil, maintained at 23°C under a 16-hr light/8-hr dark cycle, allowed to self-pollinate, and T2 seeds were collected.
Figure 4.
hS5R can complement the det2-1 mutation. (A) Schematic representations of pMD-hS5R expression plasmids used to transform det2-1 mutants. Two constructs were made for each type of hS5R cDNA: a shorter one containing a truncated 3′-untranslated region and a longer one containing a full-length human cDNA. DNA fragments indicated are hS5R, nopaline synthase promoter (NosPro) and transcriptional terminator (Nos-ter), neomycin phosphotransferase gene (NPTII), CaMV 35S promoter (35Spro), and multiple cloning sites (MCS). (B–D) Complementation of det2-1 mutation by hS5R cDNA. (B) Dark-grown 7-day-old seedlings. (C) Light-grown 7-day-old seedlings. (D) Light-grown 3-week-old seedlings. From left to right in B–D: Wild-type Col-0, det2-1, transgenic det2-1 containing the full-length hS5R type 1 cDNA, and transgenic det2-1 containing the full-length hS5R type 2 cDNA.
Treatment of Etiolated Seedlings with 17β-(N,N-diethyl)carbamoyl-4-methyl-4-aza-5α-androstan-3-one (4-MA).
Seeds were germinated on 0.5× MS (pH 5.7) medium (12) supplemented with 1% sucrose, 1× Gamborg’s B5 vitamins, 0.8% phytagar, and varying concentrations of 4-MA (gift from Merck Sharp & Dohme Research Laboratories). Following a 2-hr light treatment, plates were wrapped with three layers of aluminum foil and kept at 21°C in a growth chamber. The hypocotyl lengths of 10-day-old etiolated seedlings were measured.
DNA and RNA Analyses.
Arabidopsis DNA was isolated as described (18). For amplification of a genomic DET2 fragment, 1 μl (10–20 ng) of plant DNA was used as a template in a 50-μl reaction mixture containing 5 μl of 10× Taq polymerase buffer (Stratagene), 200 μM dNTPs, 125 ng each of forward and reverse primers, and 2.5 units of Taq polymerase. Amplification reactions were conducted in a thermocycler (Ericomp, San Diego) by denaturing the template DNA for 10 min at 95°C followed by 40 cycles of denaturation at 94°C for 45 sec, annealing at 50°C for 45 sec, extension at 72°C for 90 sec, and a final extension period of 10 minutes at 72°C. For amplifying hS5R cDNAs in transgenic det2-1 plants, 10% dimethyl sulfoxide was added in the PCR (to overcome the problem caused by the high G+C content of the cDNAs), together with primers derived from the CaMV 35S promoter and each of the cDNAs. Total RNA was isolated from 2-week-old seedlings by the method of Napoli et al. (19) and RNA gel blot hybridizations were performed as described (6).
RESULTS
DET2 Is a Functional Steroid 5α-Reductase.
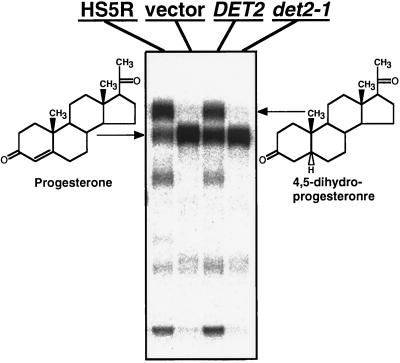
To determine whether the Arabidopsis DET2 locus encodes a functional steroid 5α-reductase, a full-length DET2 cDNA was cloned into a mammalian expression vector, pCMV5 (12), and introduced into cultured human embryonic kidney 293 cells by a CaPO4-mediated transfection protocol (13). Sixteen hours after transfection, radiolabeled steroids were added to the cell medium and their conversion to 5α-reduced forms was monitored by TLC (14). As shown in Fig. 2, cells transfected with a pCMV5 expression vector lacking a cDNA insert displayed no measurable steroid 5α-reductase activity, while introduction of a pCMV5 expression vector containing a cDNA of either type 1 hS5R or the Arabidopsis DET2 resulted in reduction of radiolabeled progesterone to 4,5-dihydroprogesterone. Consistent with our previous prediction (4), the Glu-204 → Lys mutation of det2-1 totally inactivated the steroid 5α-reductase activity of DET2, as 293 cells transfected with det2-1 cDNA failed to convert the radiolabeled progesterone to its 5α-reduced form (Fig. 2).
Figure 2.
Arabidopsis DET2 has steroid 5α-reductase activity. TLC analysis of steroids produced by incubating 64 nM [14C]progesterone with human embryonic 293 cells transfected with a pCMV5 expression plasmid containing cDNA of type 1 hS5R (lane 1), no insert (lane 2), wild-type DET2 (lane 3), or det2-1 (lane 4), as described.
Like mammalian steroid 5α-reductases (5), Arabidopsis DET2 can catalyze the 5α-reduction of several steroids with a 3-oxo,Δ4,5 structure, including testosterone and androstenedione (data not shown). Because the hypothesized substrates of Arabidopsis DET2 in BR biosynthesis is campesterol or its analogs (sitosterol or stigmasterol), which contain a 3β-hydroxyl,Δ5,6 structure, we measured the steroid 5α-reductase activity of DET2 toward several radiolabeled steroids with this structure including cholesterol, pregnenolone, and dehydroepiandrosterone. In line with the results obtained with mammalian steroid 5α-reductases (20), the 293 cells transfected with a cDNA of either the type 1 hS5R or DET2 failed to 5α-reduce these substrates (data not shown), implying that plants require the presence of an additional enzyme to convert campesterol to 3-oxo,Δ4,5-campesterol before 5α-reduction by the DET2 enzyme.
Kinetic Properties of Expressed DET2 Protein.
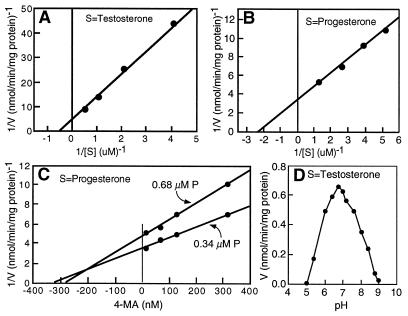
An in vitro assay (14) was used to study the enzyme kinetics of the kidney 293 cell-expressed DET2 protein. The apparent Km value for testosterone was determined to be 2.5 μM with a Vmax of 0.2 nmol/min−1·mg−1 (Fig. 3A), while the apparent Km value for progesterone was 0.4 μM with a Vmax of 0.5 nmol/min−1·mg−1 (Fig. 3B). These values compare favorably with those calculated for the human type 1 isozyme (Km = 1.7 μM for testosterone and Km = 1.3 μM for progesterone) (5). Like the mammalian enzymes (5), the steroid 5α-reductase activity of DET2 requires NADPH rather than NADH as a cofactor and the catalyzed reaction was irreversible (data not shown).
Figure 3.
Biochemical characterization of the steroid 5α-reductase activity of DET2. DET2 cDNA cloned into the pCMV5 expression plasmid was transfected into cultured 293 cells. Sixteen hours later, cell lysates were prepared and 5 μg aliquots of protein were assayed for steroid 5α-reductase activity as described (15). Analysis of enzyme kinetic data was performed with the k.cat program from BioMetallics (Princeton). (A) Lineweaver–Burk plot for [14C]testosterone. (B) Lineweaver–Burk plot for [14C]progesterone (C) Competitive inhibition of DET2 enzyme activity by 4-MA. Steroid 5α-reductase activity was assayed in the presence of the indicated concentration of 4-MA and either 0.34 μM or 0.68 μM [14C]progesterone. The intersection of the two lines defines the Ki (21). (D) pH profile of the DET2 steroid 5α-reductase activity using [14C]testosterone as a substrate. Assays were carried out at the indicated pHs in the presence of 5 μg of cell lysate protein, 1 μM [14C]testosterone, and 2.0 mM NADPH for 20 min at 37°C.
4-Azasteroids are a class of selective and potent inhibitors of mammalian steroid 5α-reductase isozymes (5). To determine if the DET2 enzyme was also inhibited by these drugs, cell lysates containing recombinant DET2 were incubated with progesterone as substrate and various amounts of the 4-azasteroid, 4-MA. Increasing the concentration of 4-MA in the reaction mixture progressively inhibited DET2 enzyme activity (Fig. 3C). Inspection of the data revealed a competitive mode of inhibition with a calculated apparent Ki of 300 nM. In experiments not shown, two other inhibitors of mammalian steroid 5α-reductase isozymes, finasteride (a 4-azasteroid) and LY191704 (a nonsteroidal benzoquinolinone), did not affect the activity of recombinant DET2.
The mammalian steroid 5α-reductase type 1 and type 2 isozymes are distinguished by their pH optima. In all species so far examined, the type 1 isozyme has an alkaline pH optimum (Vmax at about pH 8.0,) and the type 2 isozyme has an acidic pH optimum (Vmax at pH 5.0–5.5) (5). The pH optimum of the plant enzyme was measured to determine which mammalian isozyme the DET2 enzyme most closely resembled with respect to this biochemical parameter. The data of Fig. 3D show that the pH versus enzyme activity curve obtained with cell lysates containing recombinant DET2 protein was symmetrical with an optimum at pH 6.8, which is between the pH optima of the two mammalian isozymes.
hS5R cDNAs Complement the Plant det2-1 Mutation.
If DET2 is a functional homolog of hS5R, then expression of one or the other human enzyme should complement the det2 mutation in plants and rescue the mutant phenotypes. To test this hypothesis, we stably introduced cDNAs encoding either the type 1 or type 2 hS5R into det2-1 mutants using Agrobacterium-mediated transformation (17). In these experiments, two constructs with either a full-length cDNA or a cDNA with a truncated 3′-untranslated region were prepared for each type of hS5R cDNA. These were cloned into a binary vector, pMD1, which contains a CaMV 35S promoter and a nopaline synthase 3′-untranslated sequence (Fig. 4A).
In one set of transformation experiments, 64 transgenic plants were obtained. Among these, 57 displayed wild-type phenotypes and a remaining 7 lines displayed intermediate phenotypes between det2-1 and wild-type plants (Table 1). As shown in Fig. 4B-4D, the presence of either hS5R cDNA in the det2-1 mutant background rescued both the dark and light phenotypes of the mutation. All 64 transgenic lines were tested for the insertion of a human cDNA into their genomes by PCR using oligonucleotide primers derived from the CaMV 35S promoter and a hS5R cDNA. PCR products of the expected size for either type of hS5R cDNA were amplified from the genomic DNAs of all transgenic plants (summarized in Table 1), while nontransformed controls of wild-type or det2-1 heterozygous and homozygous plants did not yield positive amplification signals. Since the det2-1 mutation causes a G-A transition, leading to the elimination of an MnlI restriction site, an MnlI restriction fragment length polymorphism (RFLP) exists between det2-1 mutants and wild-type plants. A PCR-based RFLP analysis was used to confirm that each of the 64 transgenic lines still contained the det2-1 mutation (Table 1), thereby eliminating the possibility that the observed phenotypic normalization was due to an accidental introduction of a wild-type DET2 gene into their genomes or to reversion at the mutated site.
Table 1.
Summary of transgenic det2-1 plants carrying hS5R cDNAs
| Constructs | No. of plants
|
|||
|---|---|---|---|---|
| Showing kanamycin resistance | Containing hS5R cDNA | Having det2-1 mutation | Displaying wild-type phenotype | |
| hS5R1, 1.3 kb | 8 | 8 | 8 | 8 |
| hS5R1, 2.1 kb | 18 | 18 | 18 | 18 |
| hS5R2, 0.8 kb | 18 | 18 | 18 | 16 |
| hS5R2, 2.4 kb | 20 | 20 | 20 | 15 |
Transgenic plants of each construct were obtained from two independent transformation experiments. See Materials and Methods and Fig. 4A for a description of constructs.
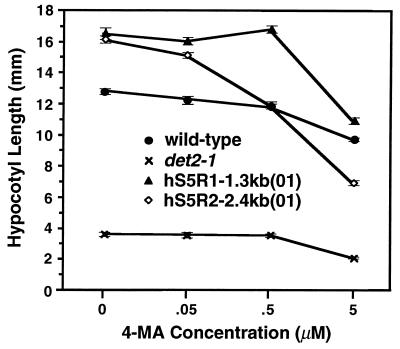
To prove that the observed phenotypic normalization was caused by expression of a hS5R cDNA in the transgenic plants, we germinated seeds collected from two representative transgenic lines, containing either a type 1 or a type 2 human cDNA, on a synthetic growth medium with different concentrations of 4-MA, an inhibitor of the mammalian steroid 5α-reductases. Fig. 5 shows the effect of increasing concentrations of 4-MA on the hypocotyl elongation of dark-grown plants. While increasing concentrations of 4-MA had little effect on the hypocotyl elongation of either wild-type or det2-1 seedlings, the drug caused a significant decrease of hypocotyl growth in the two transgenic lines. The differential effects of 4-MA on det2-1, wild-type, and the two transgenic lines observed in these experiments were consistent with the biochemical properties of the individual hS5R and DET2. The Ki for 4-MA of the hS5R type 1 (8.0 nM) is twice that of the type 2 enzyme (4.0 nM) (5), while the Ki for 4-MA of DET2 is at least 30-fold higher (300 nM) than those of the human isozymes (Fig. 3C).
Figure 5.
Effect of 4-MA on the hypocotyl lengths of det2-1, wild-type, and transgenic det2-1 plants. Data represent the mean ± SE obtained from 25 seedlings of each genotype.
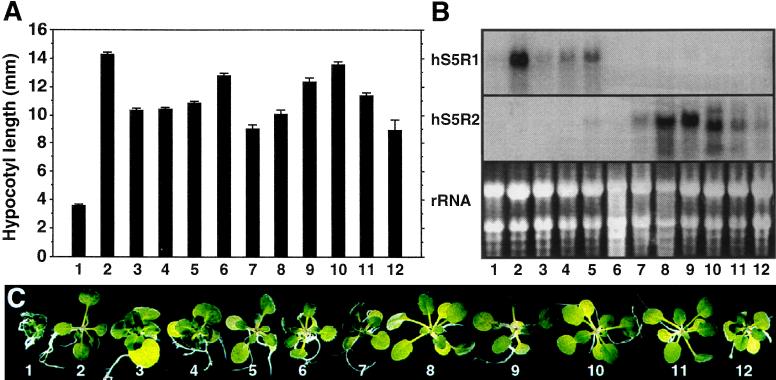
The second line of evidence that expression of hS5R cDNAs rescued det2-1 mutant phenotypes came from observations that the progeny of some of the individual transformants looked more similar to det2-1 mutants than to wild-type plants, although their parental lines exhibited a fully wild-type phenotype. We suspected that this phenomenon was due to loss of transgene expression in the T2 generation, which has been previously observed in various transgenic lines with the pMD1 binary vector (unpublished results). Therefore, the expression levels of the human cDNAs were examined in several transgenic lines by RNA blotting. The steady-state level of hS5R mRNA expressed in the transgenic det2-1 plants (Fig. 6B) was correlated with the degree of mutant rescue as indicated by both the hypocotyl length of the dark-grown seedlings (Fig. 6A) and the overall morphology of light-grown plants (Fig. 6C). Plants showing the highest level of human transcripts among all lines examined exhibited full wild-type stature in the light and their hypocotyls were even longer than those of wild-type controls in the dark. In contrast, the lines showing the lowest levels of human mRNAs looked more like det2-1 mutants than wild-type plants in the light and had intermediate hypocotyl lengths in the dark. Based on these results, we concluded that the human cDNAs can rescue det2-1 defects and that the expression of hS5R cDNAs in the det2-1 transgenic plants correlated with the degree of rescue of their mutant phenotypes.
Figure 6.
The expression level of hS5R cDNA is correlated with phenotype in transgenic det2-1 plants. (A) Hypocotyl length of dark-grown seedlings of wild-type Col-0, det2-1 and segregating progeny of primary transgenic det2-1 plants. Data represent the mean ± SE obtained from a population with an average sample size of 60 seedlings. (B) Northern blot analysis of transgene expression. Each lane contained 20 μg of total plant RNA. (C) Morphology of 14-day-old light-grown seedlings. From left to right: 1, det2-1; 2, hS5R1 1.3 kb (04); 3, hS5R1 1.3 kb (08); 4, hS5R1 2.1 kb (04); 5, hS5R1 2.1 kb (17); 6, wild-type; 7, hS5R2 0.8 kb (01); 8, hS5R2 0.8 kb (12); 9, hS5R2 0.8 kb (15); 10, hS5R2 2.4 kb (04); 11, hS5R2 2.4 kb (05); 12, hS5R2 2.4 kb (12).
DISCUSSION
In this paper we show that the Arabidopsis DET2 gene encodes an ortholog of mammalian steroid 5α-reductases. First, DET2 expressed in human embryonic kidney 293 cells is capable of reducing several mammalian steroids with a 3-oxo,Δ4,5 structure including testosterone, androstenedione, and progesterone. Second, hS5R expressed in plants can substitute for DET2 in BR biosynthesis. These data provide further evidence that DET2 is involved in BR biosynthesis and that BRs are important plant hormones that are crucial for growth and development.
Many mammalian steroid hormones contain 21 or fewer carbon atoms, while the known plant steroids contain at least 27 carbon atoms. Therefore, it was surprising to find that the Arabidopsis DET2, which is involved in BR biosynthesis, has similar affinities for animal steroids (C19 testosterone and C21 progesterone) as do the mammalian steroid 5α-reductases, with apparent Km values in the micromolar range. Because hS5R expressed in plants can function in BR biosynthesis and complement the det2-1 mutation, we expect that the human isozymes will have similar affinities for plant steroids as does DET2. Other biochemical properties of the expressed DET2 protein were also comparable to those of the well-characterized mammalian steroid 5α-reductases. Thus, the reaction catalyzed by DET2 was irreversible, showed an absolute requirement for NADPH as cofactor, and was competitively inhibited by the 4-azasteroid, 4-MA. Both the structural and functional conservation between DET2 and mammalian steroid 5α-reductases suggest that they evolved from a common ancestor.
Some differences exist between DET2 and mammalian steroid 5α-reductases. Neither finasteride (a 4-azasteroid) nor LY91704 (a nonsteroidal benzoquinolinone) inhibited the 5α-reductase activity of 293 cell-expressed DET2. DET2 shares approximately the same amount of sequence identity with both type 1 and type 2 mammalian 5α-reductase isozymes (data not shown), and DET2 has a pH optimum that is between the basic pH optimum of the type 1 isozymes and the acidic pH optimum of the type 2 isozymes.
Like its mammalian homologs, the Arabidopsis DET2 5α-reduced several steroids that have a 3-oxo,Δ4,5 structure but was not active against steroids that have a 3β-hydroxyl,Δ5,6 structure. In mammals, a membrane-bound enzyme, 3β-hydroxysteroid dehydrogenase/Δ5,6–Δ4,5 isomerase, catalyzes the oxidation and isomerization of 3β-hydroxyl,Δ5,6 precursors into 3-oxo,Δ4,5 steroids, which is a key step in the biosynthesis of many classes of hormonal steroids. Since the hS5R, which are known to be inactive toward the 3β-hydroxy,Δ5,6 steroids (20), can complement the det2-1 mutation, plants must have a similar enzyme system to convert campesterol and its analogs (sitosterol and stigmasterol) into their 3-oxo,Δ4,5 products that can subsequently be reduced by either hS5R or the DET2 enzyme. In fact, biosynthesis of 5α-sitostanol and its analogs (campestanol and stigmastanol) from their corresponding phytosterols is known to occur in mammalian tissues (22). It has been shown that the major pathway for such a conversion involves a slow direct oxidation of phytosterols to yield 3-oxo,Δ4,5 steroids, followed by a rapid saturation of the Δ4,5 double bond and reduction of the 3-oxo group (23). We suspect that this whole biosynthetic pathway is evolutionarily conserved between plants and mammals.
A conserved steroid metabolic pathway between plants and animals suggests that the use of steroids for signal transduction evolved before plants and animals diverged from protists. Signal substances with steroid structure are also known from algae and fungi, and BRs are found in the green alga, Hydrodyction reticulatum (24). A fungal steroid hormone, antheridiol, regulates sexual reproduction in the water mold, Achlya ambisexualis (25, 26). The observation that steroid hormones from insects, plants, algae and fungi are all polyhydroxylated steroids has been interpreted to mean that modern steroid hormones share a common progenitor steroid that was hydroxylated to render it sufficiently water soluble (27).
Such an evolutionary link might imply a conserved mechanism of steroid signal transduction. In mammals, the major biological actions of steroid hormones are mediated by ligand-dependent transcription factors that comprise the steroid/thyroid hormone receptor superfamily (1). Members of the receptor family share a common modular structure that includes a highly conserved DNA-binding domain and a less conserved carboxyl-terminal region containing ligand binding and dimerization functions (28). These receptors achieve their physiological functions by binding to specific DNA sequences termed hormone response elements, thereby activating or suppressing target gene expression in a ligand-dependent manner (29). Steroid-binding proteins that exhibit high affinity and specificity for animal steroid hormones have been demonstrated in fungi (30), algae (31), and plants (32). Moreover, the cellular machinery that is required to mediate steroid hormone action is evolutionary conserved across many eukaryotes (33–36). However, despite the existence in databases of 37,283 (http://lenti.med.umn.edu/cgi-bin/blast/blastn.cgi) expressed sequence tags from plants, no one gene encoding a predicted steroid/thyroid hormone-type receptor has yet been found. Recently, two Arabidopsis BR-insensitive dwarf mutants, cbb2 (37) and bri1 (38), have been characterized; each is an allele of single locus that maps on the bottom of chromosome 4. The molecular cloning and sequence analysis of this and other Arabidopsis BR-insensitive genes may reveal that plants are very much a part of the phylogenetic tree in which components of the steroid signaling pathway will be shown to have evolved from a common ancestral hormone receptor system.
Acknowledgments
This work was supported by grants from the U.S. Department of Agriculture (J.C.) and the National Institutes of Health (GM43753 to D.W.R.). M.G.B. was supported in part by a Medical Scientist Training Grant (GM08014).
ABBREVIATIONS
- BR
brassinosteroid
- hS5R
human steroid 5α-reductase
- 4-MA
17β-(N,N-diethyl)carbamoyl-4-methyl-4-aza-5α-androstan-3-one
- CaMV
cauliflower mosaic virus
References
- 1.Evans R M. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geuns J M C. Phytochemistry. 1978;17:1–13. [Google Scholar]
- 3.Mandava N B. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:23–52. [Google Scholar]
- 4.Li J, Nagpal P, Vitart V, McMorris C T, Chory J. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 5.Russell D W, Wilson J D. Annu Rev Biochem. 1994;63:25–61. doi: 10.1146/annurev.bi.63.070194.000325. [DOI] [PubMed] [Google Scholar]
- 6.Chory J, Nagpal P, Peto C A. Plant Cell. 1991;3:445–459. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Altmann T, Redei G P, Nagy F, Schell J, Koncz C. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 8.Stryer L. Biochemistry. New York: Freeman; 1988. pp. 565–570. [Google Scholar]
- 9.Fujioka S, Inoue T, Takatsuto S, Yanigasawa T, Yokota T, Sakurai A. Biosci Biotechnol Biochem. 1995;59:1543–1547. [Google Scholar]
- 10.Levy M A, Brandt M, Sheedy K M, Holt D A, Heaslip J I, Trill J J, Ryan P J, Morris R A, Garrison L M, Bergsma D J. J Steroid Biochem Mol Biol. 1995;52:307–319. doi: 10.1016/0960-0760(94)00183-m. [DOI] [PubMed] [Google Scholar]
- 11.Andersson S, Berman D M, Jenkins E P, Russell D W. Nature (London) 1991;354:159–161. doi: 10.1038/354159a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 13.Andersson S, Davis D L, Dahlback H, Jornvall H, Russell D W. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 14.Normington K, Russell D W. J Biol Chem. 1992;267:19548–19554. [PubMed] [Google Scholar]
- 15.Andersson S, Bishop R W, Russell D W. J Biol Chem. 1989;264:16249–16255. [PMC free article] [PubMed] [Google Scholar]
- 16.Thigpen A E, Russell D W. J Biol Chem. 1992;267:8577–8583. [PubMed] [Google Scholar]
- 17.Bechtold N, Ellis J, Pelletier G. CR Acad Sci Paris. 1993;316:1188–1193. [Google Scholar]
- 18.Li J, Chory J. In: Methods in Molecular Biology: Arabidopsis Protocols. Martinez-Zapater J M, Salinas J, editors. Totowa, NJ: Humana; 1997. in press. [Google Scholar]
- 19.Napoli C, Lemieux C, Jorgensen R. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsia S L, Voigt W. J Invest Dermatol. 1974;62:224–227. doi: 10.1111/1523-1747.ep12676791. [DOI] [PubMed] [Google Scholar]
- 21.Dixon M, Webb E C. Enzymes. New York: Academic; 1979. [Google Scholar]
- 22.Skrede B, Bjorkhem I, Bergesen O, Kayden H J, Skrede S. Biochim Biophys Acta. 1985;836:368–375. doi: 10.1016/0005-2760(85)90141-9. [DOI] [PubMed] [Google Scholar]
- 23.Bjorkhem I, Karlmar K-E. Biochim Biophys Acta. 1974;337:129–131. doi: 10.1016/0005-2760(74)90046-0. [DOI] [PubMed] [Google Scholar]
- 24.Yokota T, Kim S-K, Fukui Y, Takahashi N, Takeuchi Y, Takematsu T. Phytochemistry. 1987;26:503–510. [Google Scholar]
- 25.McMorris T C, Barksdale A W. Nature (London) 1967;215:320–321. doi: 10.1038/215320a0. [DOI] [PubMed] [Google Scholar]
- 26.Arsenault G P, Diemann K, Barksdale A W, McMorris T C. J Am Chem Soc. 1968;90:5635–5636. [Google Scholar]
- 27.Karlson P. Hoppe Seyler’s Z Physiol Chem. 1983;364:1067–1071. [PubMed] [Google Scholar]
- 28.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. Cell. 1996;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai M-J, O’Malley B W. Annu Rev Biochem. 1994;64:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 30.Lenard J. Trends Biochem Sci. 1992;17:147–150. doi: 10.1016/0968-0004(92)90323-2. [DOI] [PubMed] [Google Scholar]
- 31.Agarwal M K. FEBS Lett. 1993;322:207–210. doi: 10.1016/0014-5793(93)81570-p. [DOI] [PubMed] [Google Scholar]
- 32.Geuns J M C. Trends Biochem Sci. 1982;7:7–9. [Google Scholar]
- 33.Schena M, Yamamoto K R. Science. 1985;241:965–967. doi: 10.1126/science.3043665. [DOI] [PubMed] [Google Scholar]
- 34.Metzer D, White J H, Chambon P. Nature (London) 1988;334:31–36. doi: 10.1038/334031a0. [DOI] [PubMed] [Google Scholar]
- 35.Yoshinaga S K, Yamamoto K R. Mol Endocrinol. 1991;5:844–853. doi: 10.1210/mend-5-6-844. [DOI] [PubMed] [Google Scholar]
- 36.Schena M, Lloyd A M, Davis R W. Proc Natl Acad Sci USA. 1991;88:10421–10425. doi: 10.1073/pnas.88.23.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Plant J. 1996;9:701–713. [Google Scholar]
- 38.Clouse S D, Langford M, McMorris T C. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]