Abstract
The gene encoding the hormone secretin is expressed only in enteroendocrine S cells and insulin-producing pancreatic β cells during development. A 120-bp enhancer directs cell-specific expression of the rat secretin gene in secretin-expressing cells. The enhancer includes an E-box sequence, CAGCTG, which is important for transcriptional activity. To further characterize the role of the E box, a consensus binding site for basic helix–loop–helix (bHLH) proteins, we have examined factors that interact with this element in the secretin gene. The results suggest that transcription is activated by a recently isolated tissue-specific bHLH protein, BETA2, heterodimerized to the ubiquitously expressed bHLH proteins, Pan 1 and Pan 2, the rodent homologues of E47 and E12. The importance of BETA2 for transcriptional activation of secretin is further illustrated by antisense experiments inhibiting BETA2 expression in secretin-producing cell lines, which resulted in the inhibition of most E box-dependent transcription. Expression of BETA2 in a nonendocrine cell line conferred the ability to express secretin-reporter genes that are transcribed at minimal levels in the absence of BETA2. Secretin-producing enteroendocrine cells in the murine small intestine showed specific immunostaining with BETA2 antibodies, corroborating observations in cell lines. Thus BETA2 is to our knowledge the first transcription factor identified that specifically activates cell type-specific expression of an intestinal hormone gene.
Expression of the peptide hormone secretin is restricted to S-type enteroendocrine cells in the small intestine of most mammalian species. In addition, small numbers of colonic enteroendocrine cells and insulin-producing β cells of the developing pancreas of rodents produce secretin (1). Enteroendocrine cells and pancreatic islets both arise from the primitive embryonic gut endoderm and therefore may be developmentally related (2). Transcription of the secretin gene in secretin-producing islet and intestinal endocrine cell lines is mediated by a positive cis-regulatory domain (1). Mutational analysis of the secretin gene revealed that as little as 174 bp of 5′ flanking sequence was sufficient to direct full transcriptional activity in secretin-producing cell lines. However, the same sequences did not enhance transcription in related endocrine cell lines that did not produce secretin. The secretin gene enhancer contains the E-box sequence, CAGCTG (1). Proteins that interact with the secretin E box also appear to bind to the E-box elements of the rat insulin gene. Replacement of the secretin E box with the mutant sequences TAGTTG reduced transcriptional activity to less than 20% of the native sequence and interfered with the ability to form DNA-protein complexes seen with the wild-type sequence. This suggested that the CAGCTG motif plays an important role in transcriptional regulation.
E boxes bind factors belonging to the basic helix–loop–helix (bHLH) family of transcription factors (3). bHLH transcription factors play an important role in the cell-specific expression of many different genes and, in some cases, the establishment of differentiated cell lineages (4, 5). The bHLH proteins are classified into two groups according to their DNA-binding properties and tissue distribution (6–8). The class A members are ubiquitously expressed. Examples, including E12, E47 (Pan 2 and Pan 1 in rodent species) and related proteins, bind to DNA as homodimers or heterodimers. The class B members are tissue-specific and appear to bind poorly as homodimers (6); however, they heterodimerize with the class A members and bind E boxes with high affinity (7). The myogenic bHLH proteins, which control muscle differentiation, represent one of the best characterized groups of tissue-specific bHLH proteins (9, 10).
E box-binding proteins also appear to be important for transcriptional regulation of the insulin gene (11–14). BETA2, a recently described class B-type bHLH transcription factor cloned from a hamster insulinoma cell line, binds to the insulin E-box sequence as a heterodimer with E47 (Pan 1) to activate insulin gene transcription (15). However the presence of insulin gene E box-binding proteins in islet cell lines that do not express insulin has been interpreted to suggest that other proteins may have greater importance in restricting insulin gene expression to β cells (15–17).
A murine homologue of BETA2, named NeuroD, was cloned independently from a mouse embryonic stem cell tumor library (18). NeuroD is transiently expressed in developing neurons in the central and peripheral nervous system at the time of terminal differentiation, but is not expressed in nearby neuronal progenitors that are still undergoing cell division. This protein induces premature differentiation of increased numbers of neuronal precursors when microinjected into Xenopus embryos and induces neuronal differentiation of some epidermal cells. NeuroD appears to overcome factors that normally inhibit differentiation of embryonic ectoderm into nerve cells and thus may function as a neuronal differentiation factor.
Here we report that BETA2 binds the secretin E-box sequence as a heterodimer with the rodent equivalent of E12/47 to activate secretin gene expression in both secretin-producing cells and a nonendocrine cell line. We also show that BETA2 is expressed in secretin-producing S-type enteroendocrine cells of the murine small intestine, suggesting a potentially important role for BETA2 in directing cell-specific expression of the secretin gene.
MATERIALS AND METHODS
DNA Transfections.
The secretin-luciferase reporter gene constructions for transient expression assays consisted of sequences from −209 to +32 of the rat secretin gene with or without mutations cloned 5′ to the structural gene encoding firefly luciferase. The expression plasmids for Syrian hamster Pan 1 and Pan 2 (shPan1 and shPan2) and BETA2 have been described previously (1, 15, 19). We generated a series of 4-bp block transversion mutants (A↔C and G↔T) every 2 bp in the secretin promoter upstream of the E box and an E box mutant, TAGTTG, by site-directed mutagenesis (20). A hamster BETA2 antisense expression plasmid was generated by cloning a BamHI-XhoI cDNA fragment containing the entire coding sequence into pcDNA1 in reverse orientation. Plasmids were transiently transfected into hamster HIT (HIT T-15 M2.2) or HeLa cells (human) by calcium phosphate precipitation (1). One microgram of secretin reporter gene, expression plasmids, the human growth hormone gene under control of the metallothionein promoter, was added to each plate. Sheared salmon testis carrier DNA was added to adjust the total added DNA to 10 μg. Mouse STC1 enteroendocrine cells were transfected with DEAE dextran using a total of 1 μg DNA. Cell lysates were prepared 20 h after transfection for luciferase assays (21). To account for variations in transfection efficiency, luciferase activity in cell lysates was normalized to the amount of growth hormone secreted into the culture medium. Results from three separate experiments carried out in triplicate were analyzed by ANOVA.
Oligonucleotides.
The following oligonucleotides were used in gel shift experiments as double-stranded sequences (uppercase) with 4-bp single-stranded overhangs (lowercase): wild-type probe, wild-type competitor, methylation interference probe, tcgaGAGGCCGGACGACAGCTGGG (−142 to −123); competitor m5, tcgaGAGGCCGGGTAGCAGCTGGG; competitor mE2, tcgaGAGGCCGGACGATAGTTGGG; and competitor E, tcgaGACAGCTGGG (−132 to −123)
Electrophoretic Mobility Shift Analysis.
Mobility shift assays were performed with 4 μg of crude nuclear protein (22, 23). For competition experiments, competitor DNA was added 15 min before addition of the probe. For antibody immunoneutralization experiments, nuclear extracts were incubated for 10 min at 23°C with 0.02–2 μl of rabbit antisera raised against the C terminus of hamster BETA2 (15), shPan1 (11), shPan2 (11), and human upstream stimulatory factor (USF) (24) before adding the probe. For methylation interference assays, partially methylated probe was used for gel shift experiments with 80 μg of protein. After electrophoresis, the bound and free probes were isolated, cleaved with piperidine, and fragments were resolved on a 20% sequencing gel (25).
Immunohistochemistry.
Tissues obtained from adult CD1 mice were processed as described before (26). Rabbit anti-BETA2 was diluted 1:300 (15). Rabbit antisecretin was used as described previously (27, 28). Secondary antisera were immunoabsorbed for multiple labeling. Controls for specificity of immunostaining included nonimmune primary sera, mismatched secondary antisera, and preadsorption with BETA2 protein.
Coexpression of BETA2 with secretin was determined by multiple label immunostaining. Immunoperoxidase staining for secretin was first carried out using True Blue (Kirkegaard & Perry Laboratories) as a chromogen. BETA2 was subsequently detected by immunofluorescent labeling of the same section. The cy3-conjugated secondary antisera failed to label cells when the BETA2 primary antibody was omitted, indicating that access to the secretin antiserum was blocked after peroxidase detection.
RESULTS
Mutational Analysis of Sequences Around the Secretin Gene E Box.
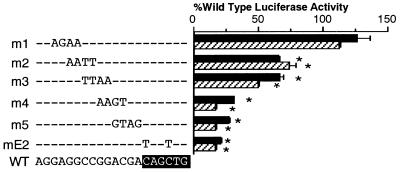
Previous work from this laboratory revealed that the E box in the secretin gene, located between −130 and −125, was important for full transcriptional activity in secretin-producing islet cell lines (1). To further characterize and fine-map important sequences upstream from the E box for transcription of the secretin gene, we carried out a detailed mutational analysis by generating a series of 4-bp block mutations in sequence around the E box of the secretin enhancer from −146 and −131. Each mutant overlapped the adjacent mutant by 2 bp (Fig. 1). Mutation-bearing reporter gene constructions were transiently expressed in the secretin-producing hamster insulinoma cell line, HIT, or secretin expressing STC1 cells. A double-point mutation in the E box (mE2) as well as two mutants, m4 and m5, just 5′ to the E-box, markedly reduced transcriptional activity of the secretin gene in both cell lines (Fig. 1). Mutants m2 and m3, just 5′ to m4 had less effect, modestly reducing transcriptional activity of the secretin gene. However, mutant m1, as well as several mutants 5′ to m1 (not shown), had no effect on transcription (Fig. 1). The relative transcriptional activity of each mutant was essentially the same in STC1 cells.
Figure 1.
Mutational analysis of the secretin gene E box. A series of secretin-luciferase reporter gene constructions with different 4-bp transversion mutations in the secretin E box and upstream sequences was transiently transfected into HIT (solid bars) or STC1 (striped bars) cells. Luciferase activity was measured in cell extracts 20 h later. Results are expressed (in percent) relative to the activity of the wild-type reporter and are the mean ± SEM after correction for relative transfection efficiency for at least three individual transfection experiments done in triplicate. The sequence of the unmodified (WT) is shown at the bottom, with the changed nucleotides in mutants m1 through m5 indicated. The E-box sequence is highlighted. ∗, Values different from the WT, P < 0.001.
Characterization of Nuclear Proteins Binding to Sequences Near the Secretin Gene E Box.
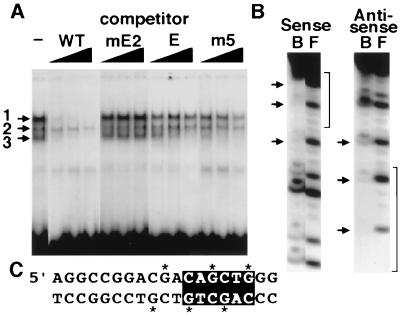
DNA-binding proteins that interact with the sequences around the E box were characterized by electrophoretic mobility shift assays using a radiolabeled probe spanning from −142 to −123, including the CAGCTG element and flanking sequences. Nuclear extracts from secretin-producing HIT cells generated three major DNA-protein complexes (Fig. 2A). The specificity of DNA protein interactions was examined by preincubating the extracts with an excess of unlabeled double-stranded competitor oligonucleotides. Unlabeled wild-type oligonucleotide displaced complexes 1 and 3, indicating sequence-specific protein binding (Fig. 2A). Both required the intact E box for binding, because a double-point mutation in the E box failed to compete with the probe. The two complexes differed in their sensitivity to competitor oligonucleotides containing the mutations tested in HIT cell transfections. The core E-box sequence competed efficiently for complex 3, but only partially displaced complex 1 at the highest concentrations, indicating that additional sequences were necessary to form complex 1. Sequences 5′ to the E box that were necessary for full transcriptional activity also appeared necessary to generate complex 1, because mutant m5 competed poorly for complex 1.
Figure 2.
Analysis of DNA-protein complexes binding to the secretin gene E box. (A) Three complexes denoted 1, 2, and 3 were generated from HIT cell nuclear extracts in gel shift assays. Competitor fragments consisting of the wild-type sequence (WT), a similar fragment with a double-point mutation in the E box (mE2), the core E-box sequence without upstream sequence (E), or 4-bp transversion mutant (m5) were used to determine the binding specificity for each complex. The three lanes shown for each competitor contained a 25-, 50-, or 100-fold mol excess, respectively. (B) Methylation interference analysis of complex 1 binding. Piperidine cleavage patterns of bound (B) and free (F) methylated sense and antisense strands are shown for DNA-protein-binding reactions. Arrowheads denote fragments not present in complex 1. Brackets denote the position of the E box. (C) The corresponding sequence for B with putative contact points necessary for binding indicated by ∗. The E-box sequence is highlighted.
DNA sequences that participate in the generation of complex 1 with the secretin gene E-box regions were further characterized by methylation interference assays (Fig. 2 B and C). Methylation of several G residues 5′ to the E box and G residues in the E box itself interfered with binding of proteins in complex 1. These results indicate that complex 1 contacted not only the sequence CAGCTG, but also the sequence 5′ to this motif important for transcriptional activity. The binding site for complex 1 encompasses the E box and adjacent 5′ sequences, correlating closely with mutationally sensitive sequences identified in transfection experiments.
Identification of Factors Binding to Secretin Gene E Box.
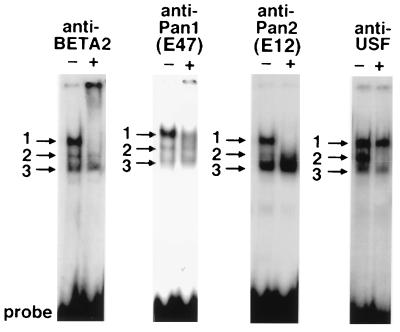
To characterize and identify nuclear factors that interact with the CAGCTG element in the secretin gene, we analyzed the DNA-protein complexes generated in gel shift assays using specific antibodies directed against hamster BETA2 (NeuroD), shPan1 (hamster E47), shPan2 (hamster E12), and human USF. Antibodies against BETA2, Pan1, and Pan2 recognized proteins in complex 1 as shown its disappearance (Fig. 3). These results indicate that complex 1 consists of BETA2 heterodimerized to either Pan1 (E47) or Pan2 (E12). However, antibodies against USF, an unrelated bHLH protein, failed to neutralize proteins in complex 1, confirming the specificity of the shPan1/2 and BETA2 antibodies previously shown with preimmune sera (11, 15). The USF antibody completely immunoneutralized complex 2, which does not appear to correlate with transcriptional activity.
Figure 3.
Identification of bHLH proteins in the activator complex binding to the secretin E box. HIT nuclear extract was assayed by gel mobility shift. Binding reactions, containing 4 μg of nuclear extract, were initiated before the addition of 2 μl of IgG-purified anti-BETA2 antibodies, 0.2 μl of anti-Pan1, 0.02 μl of anti-Pan2, or 0.02 μl of anti-USF. Complex 1 disappears with anti-BETA2, anti-Pan1, or anti-Pan2. Complex 2 disappears with anti-USF.
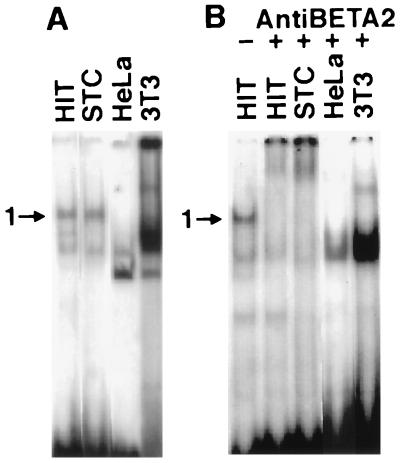
Nuclear extracts from a murine intestinal endocrine cell line (STC1), and two nonendocrine cell lines (HeLa-human, 3T3-mouse) were examined for their ability to generate BETA2 containing complexes with the secretin E-box probe in gel shift assays. Nuclear extracts from STC1 cells generated comparable DNA-protein complexes to HIT cells including complex 1 (Fig. 4A). We confirmed the presence of BETA2 in STC1 cells by the ability of BETA2 antibodies to immunoneutralize this complex (Fig. 4B). Nuclear extracts from the two nonendocrine cell lines failed to generate BETA2-containing complexes.
Figure 4.
Presence of secretin gene E-box binding factors in different cell types. (A) Gel shift analysis of nuclear extracts from the indicated cell lines using the wild-type probe. Position of complex 1 is indicated by an arrowhead. Complex 1 was present in the two endocrine cell lines, HIT and STC1, but not in HeLa or 3T3 cells. (B) Autoradiogram from a separate gel similar to A. The first lanes show the position of complex 1 generated by HIT nuclear extracts without antibody. Complex 1 from each of the endocrine cell lines is entirely immunoneutralized with antibodies against BETA2. Cell lines include STC1, 3T3 (murine), HIT, (hamster), and HeLa (human).
Transactivation of the Secretin Gene by BETA2.
We examined the ability of BETA2, Pan 1 (E47), and Pan 2 (E12) to activate transcription of the secretin gene by transiently cotransfecting mammalian expression vectors for BETA2, shPan1, and shPan2 with the secretin/luciferase reporter plasmid into HIT or STC-1 cells (Fig. 5A). Cotransfection of BETA2, shPan1, or shPan2 alone stimulated expression of the reporter gene only 1.2- to 1.4-fold. However, cotransfection of BETA2 with either shPan1 or shPan2 increased reporter gene expression between 2.5- and 4-fold, respectively, suggesting that BETA2 activates transcription in secretin-producing cells heterodimerized to either Pan1 or Pan2. A BETA2 antisense plasmid inhibited expression of the reporter gene close to level of the E-box mutant plasmid, shown earlier in Fig. 1, confirming the importance of BETA2 in transcriptional activity of the secretin gene E box. The antisense plasmid had no effect on a control reporter, RSV-luciferase, which does interact with BETA2 (not shown), confirming its specificity for the secretin enhancer.
Figure 5.
BETA2 activates the secretin enhancer in both endocrine and nonendocrine cells. Cells were transfected with the secretin-luciferase reporter gene. Expression plasmids for shPan1, shPan2, BETA2, and antisense BETA2 were cotransfected as indicated. (A) Secretin-producing HIT cells (solid bars) and STC-1 (striped bars). (B) HeLa cells. Results are expressed as relative luciferase activity generated compared with the reporter alone and are the mean ± SEM of six independent transfections for each construct. Values significantly different from cells transfected with the reporter gene alone are denoted by ∗, P < 0.05; ∗∗, P < 0.01; and ∗∗∗, P < 0.001.
The secretin reporter gene was expressed at very low levels when transiently expressed in HeLa cells, a nonendocrine epithelial cell line. Cotransfection of BETA2 alone stimulated reporter gene expression nearly 8-fold, whereas the inclusion of BETA2 plus Pan1 increased expression 16-fold (Fig. 5B). These results suggest that BETA2 may confer the ability to express the secretin reporter gene on nonendocrine cells. The ability of cotransfected BETA2 by itself to effectively transactivate the reporter gene suggests that the availability of E protein dimerization partner may not be limiting.
BETA2 Is Expressed in Secretin-Producing Enteroendocrine Cells.
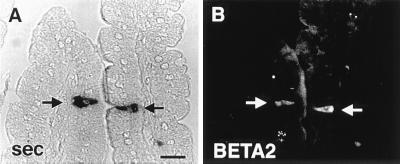
Expression BETA2 is limited to islet cell lines, an enteroendocrine cell line, and neurons whereas Pan1 (E47) and Pan2 (E12), which are expressed in many tissues. Identification of BETA2 in secretin-expressing enteroendocrine cells would be consistent with a role for this factor in tissue-specific expression of the secretin gene. We have examined the mouse small intestine to determine whether BETA2 is expressed in terminally differentiated secretin cells by multiple-label immunohistochemistry. Occasional individual intestinal epithelial cells stained for BETA2. Preadsorption of the antibody with 1 μM glutathione S-transferase fusion protein containing the C-terminal half of BETA2 abolished all staining, confirming the specificity of the antibody. Most of the stained cells appeared to be secretin cells. Virtually all secretin cells in the small intestine showed immunostaining for BETA2 (Fig. 6 A and B).
Figure 6.
BETA2 expression in secretin-producing enteroendocrine cells of the mouse small intestine. Double immunostaining of two cells (arrows) in the same section showing coexpression of secretin detected with immunoperoxidase using True Blue substrate (A) and BETA2 immunofluorescence detected with Cy3-conjugated secondary antibody (B). (Bar = 20 μm.)
DISCUSSION
This work describes the role of a recently described bHLH transcription factor, BETA2, in tissue-specific regulation of the secretin gene (15). In addition, we show for the first time, to our knowledge, that BETA2 is specifically expressed in terminally differentiated secretin-producing enteroendocrine cells of the mouse. BETA2 heterodimerizes with the ubiquitous bHLH proteins E47 (Pan1) and E12 (Pan2), and transactivates insulin gene expression in β cell lines. The restriction of BETA2 mRNA to relatively few tissues, including pancreatic islet cell lines, an enteroendocrine cell line, brain, and the intestine, suggested a potential role for this protein in cell-specific transcriptional regulation (15).
In the present work, mutational analysis has defined sequences just 5′ to the E box in the secretin gene that are important for transcriptional activity besides the E-box motif shown previously (1). Analysis of the proteins binding to the E-box region of the secretin gene, revealed that one DNA-protein complex is dependent on the presence of transcriptionally active sequences 5′ to the core E-box motif. Specific antibodies to BETA2 and the rodent homologues of human E47/E12 (Pan1/Pan2) recognized proteins in this complex, suggesting that BETA2 binds to the secretin enhancer heterodimerized to ubiquitously expressed bHLH proteins. Antisense experiments inhibiting BETA2 expression reduce the activity of the secretin enhancer close to the residual activity of an E-box mutant, suggesting that BETA2 is the major factor activating transcription from the secretin E box.
Cotransfection of BETA2 with a reporter containing one copy of the secretin enhancer minimally increased transcription in the HIT and STC1 cell lines. In these secretin-expressing cells, it appears that BETA2 may not be limiting. Similarly, Pan1 and Pan2 (E47/E12) minimally increased reporter gene expression. However cotransfection of BETA2 with either Pan 1 or Pan2 significantly increased transcription, suggesting that BETA2 activates transcription of the secretin gene as a heterodimer. The ability of BETA2 to activate secretin gene expression in HeLa cells, a nonendocrine cell line that does not express BETA2, provides additional evidence for the importance of BETA2 in secretin expression.
We have localized BETA2-expressing cells in the intestine by immunohistochemistry in secretin-producing S-type enteroendocrine cells. BETA2 was not seen in other enteroendocrine cell types nor in the three other epithelial cell types in the intestine, enterocytes, goblet cells, and Paneth cells. The ability of BETA2 to bind to the secretin E box and activate transcription may explain expression of secretin in the developing pancreas that we observed previously (1, 27). In addition, BETA2 may be the first example of a tissue-specific transcription factor that activates expression of an intestinal hormone gene in its appropriate enteroendocrine cell type.
BETA2/NeuroD appears to be capable of inducing neurogenesis when injected into Xenopus embryos. It is not clear whether BETA2 functions identically to induce terminal differentiation of secretin cells. Gene targeting experiments reveal that mice lacking a functional BETA2 gene fail to develop secretin-expressing enteroendocrine cells (M-J.T., F.J.N., A.B.L., and H.M., unpublished work). However, BETA2 expression in β cells is not sufficient to maintain secretin gene expression after birth. Likewise, the expression of BETA2 in S-type enteroendocrine cells is not sufficient to drive insulin expression in the intestine.
Acknowledgments
We thank M. Sawadogo for anti-USF, and M. German for anti-shPan1, anti-shPan2, and the expression plasmids for shPan1 and shPan2. This work was supported in part by National Institutes of Health Grants DK43673, the GRASP Digestive Disease Center P30-DK34928 to A.B.L. and HD17379 to M.J.T., a Houston Endowment Grant to M.J.T., and by the Juvenile Diabetes Foundation International to A.B.L.
ABBREVIATIONS
- bHLH
basic helix–loop–helix
- USF
upstream stimulatory factor
References
- 1.Wheeler M B, Nishitani J, Buchan A M J, Kopin A S, Chey W Y, Chang T, Leiter A B. Mol Cell Biol. 1992;12:3531–3539. doi: 10.1128/mcb.12.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LeDouarin N. Cell. 1988;53:169–171. [Google Scholar]
- 3.Murre C, McCaw P S, Baltimore D. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 4.Jan Y N, Jan L Y. Cell. 1993;75:827–830. doi: 10.1016/0092-8674(93)90525-u. [DOI] [PubMed] [Google Scholar]
- 5.Weintraub H. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 6.Lassar A B, Davis R L, Wright W E, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 7.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 8.Murre C, Voronova A, Baltimore D. Mol Cell Biol. 1991;11:1156–1160. doi: 10.1128/mcb.11.2.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson E N. Genes Dev. 1990;4:1454–1461. doi: 10.1101/gad.4.9.1454. [DOI] [PubMed] [Google Scholar]
- 10.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell T K, Turner D, Rupp R, Hollenberg S, Zhuang Y, Lassar A. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 11.German M S, Blanar M A, Nelson C, Moss L G, Rutter W J. Mol Endocrinol. 1991;5:292–299. doi: 10.1210/mend-5-2-292. [DOI] [PubMed] [Google Scholar]
- 12.Cordle S R, Henderson E, Masuoka H, Weil P A, Stein R. Mol Cell Biol. 1991;11:1734–1738. doi: 10.1128/mcb.11.3.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shieh S Y, Tsai M J. J Biol Chem. 1991;266:16708–16714. [PubMed] [Google Scholar]
- 14.Park C W, Walker M D. J Biol Chem. 1992;267:15642–15649. [PubMed] [Google Scholar]
- 15.Naya F J, Stellrecht C M M, Tsai M-J. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 16.Ohlsson H, Thor S, Edlund T. Mol Endocrinol. 1991;5:897–904. doi: 10.1210/mend-5-7-897. [DOI] [PubMed] [Google Scholar]
- 17.Robinson G L, Peshavaria M, Henderson E, Shieh S Y, Tsai M J, Teitelman G, Stein R. J Biol Chem. 1994;269:2452–2460. [PubMed] [Google Scholar]
- 18.Lee J E, Hollenberg S M, Snider L, Turner D L, Lipnick N, Weintraub H. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 19.German M S, Moss L G, Wang J, Rutter W J. Mol Cell Biol. 1992;12:1777–1788. doi: 10.1128/mcb.12.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Wet J, Wood K, DeLuca M, Helinski D, Subramani S. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fried M, Crothers D. Nucleic Acids Res. 1983;11:141–158. doi: 10.1093/nar/11.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Dyke M W, Sirito M, Sawadogo M. Gene. 1992;111:99–104. doi: 10.1016/0378-1119(92)90608-r. [DOI] [PubMed] [Google Scholar]
- 25.Rose S D, Kruse F, Swift G H, MacDonald R J, Hammer R E. Mol Cell Biol. 1994;14:2048–2057. doi: 10.1128/mcb.14.3.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Upchurch B H, Aponte G W, Leiter A B. Development. 1994;120:245–252. doi: 10.1242/dev.120.2.245. [DOI] [PubMed] [Google Scholar]
- 27.Lopez M J, Upchurch B H, Rindi G, Leiter A B. J Biol Chem. 1995;270:885–891. doi: 10.1074/jbc.270.2.885. [DOI] [PubMed] [Google Scholar]
- 28.Upchurch B H, Fung B P, Rindi G, Ronco A, Leiter A B. Development. 1996;122:1157–1163. doi: 10.1242/dev.122.4.1157. [DOI] [PubMed] [Google Scholar]