Abstract
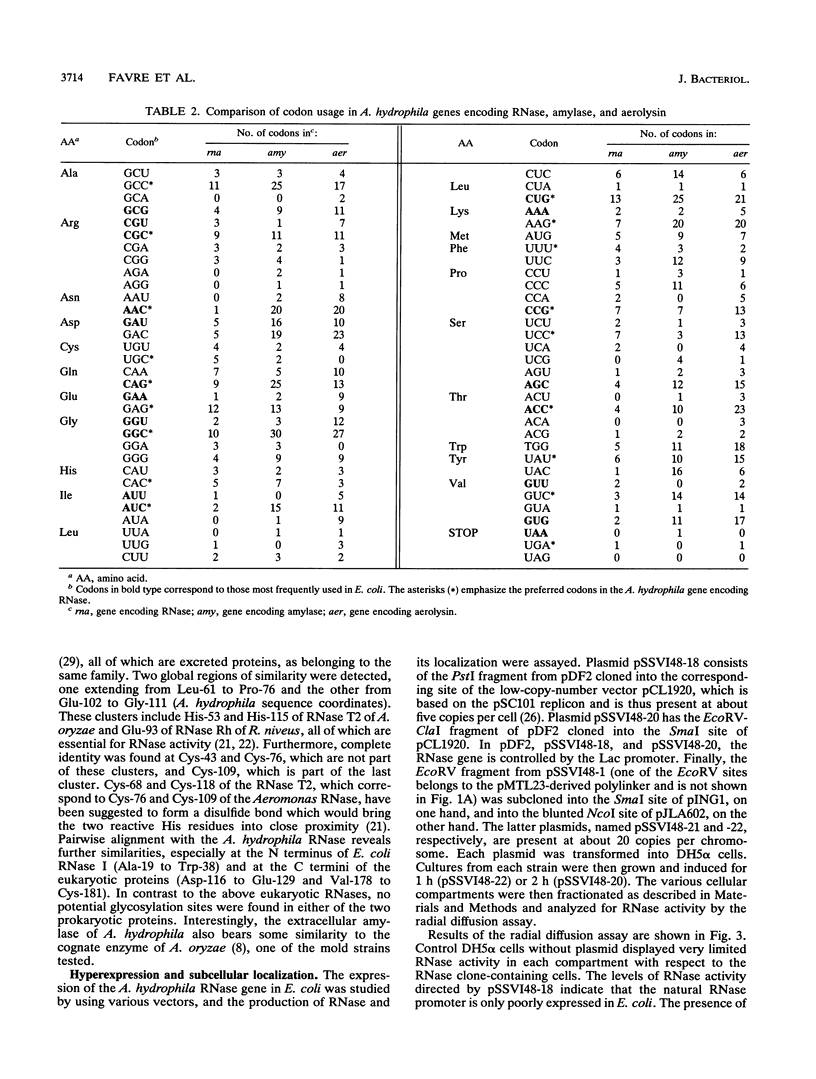
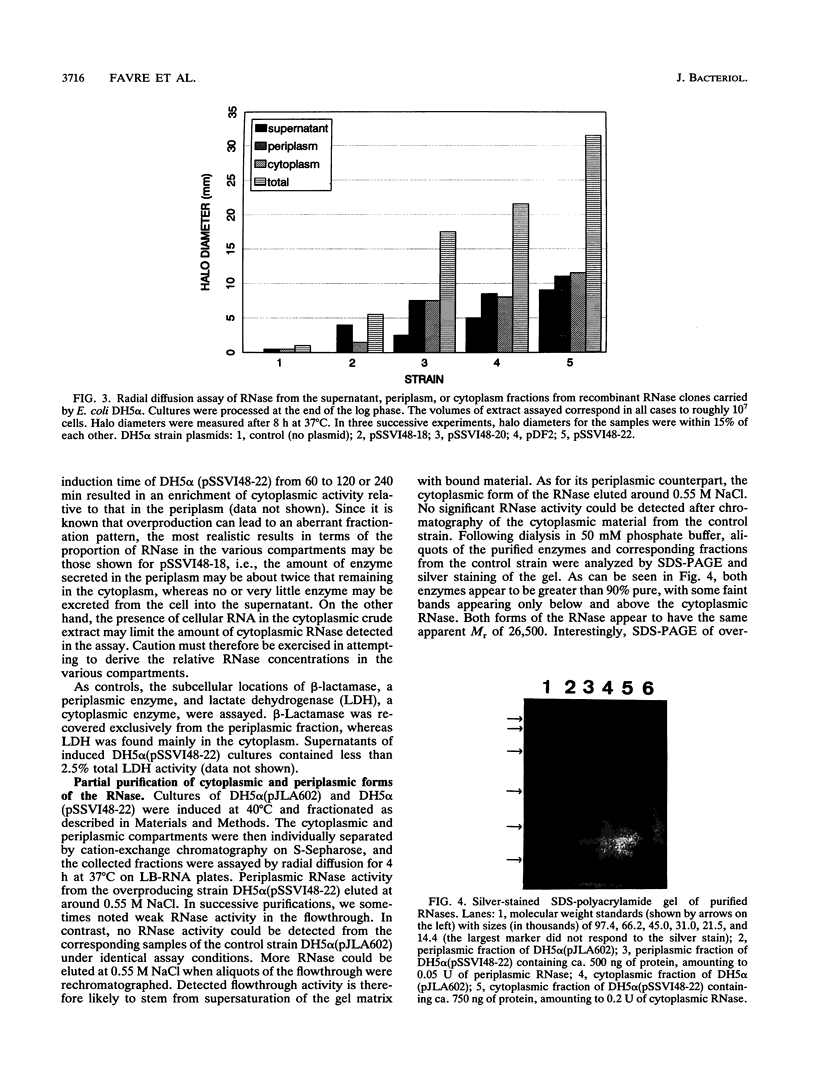
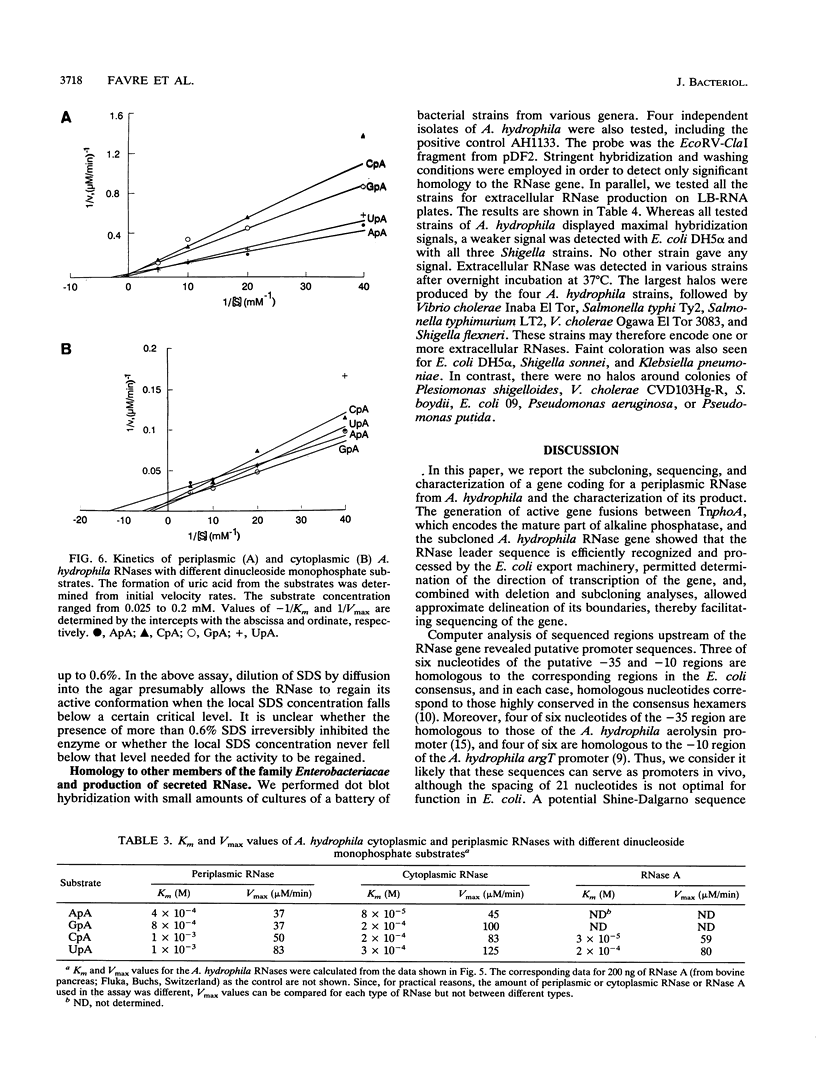
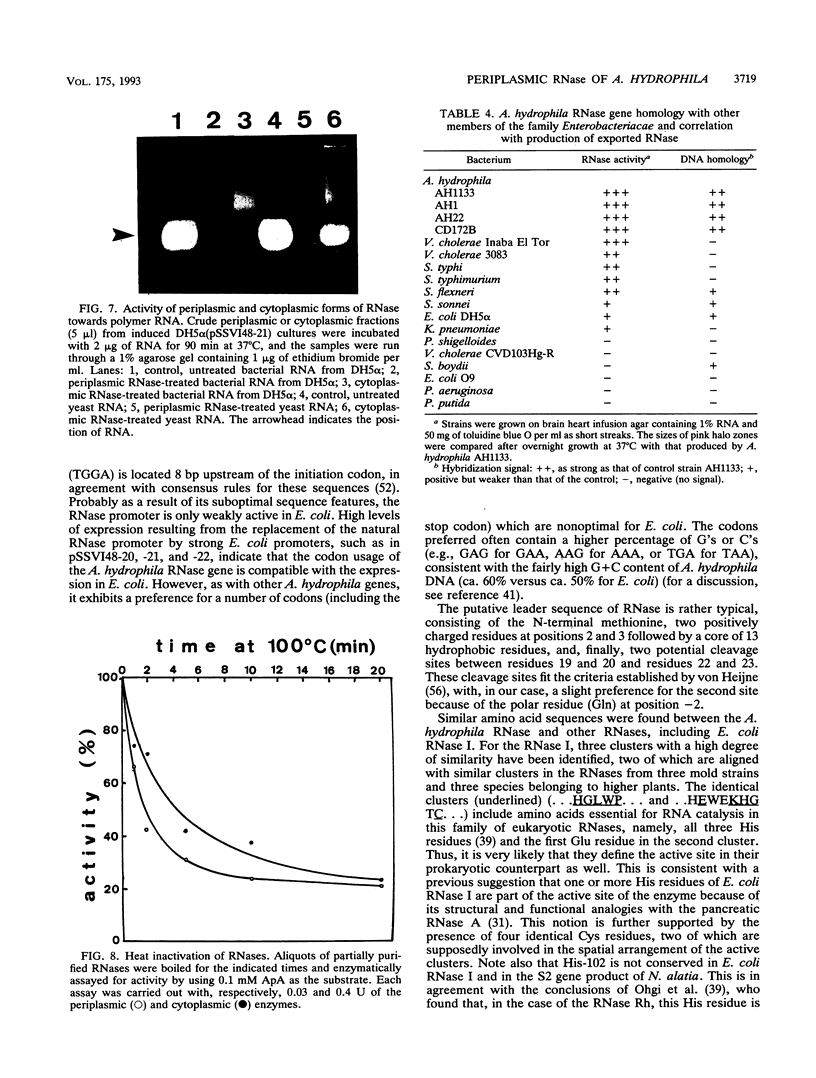
The isolation, sequencing, and characterization of a periplasmic RNase gene from Aeromonas hydrophila AH1133 is described. Following subcloning of the gene on a 2.7-kb PstI fragment, its direction of transcription and approximate location were determined. Analysis of the nucleotide sequence reveals that the gene is 645 bp long, coding for 215 amino acid residues with a total molecular weight of 24,215. A typical leader sequence is present at the beginning of the corresponding protein. Computer analysis revealed strong local similarities to Escherichia coli RNase I and to the active site of a family of eukaryotic RNases. Expression studies indicate that the RNase natural promoter functions poorly in E. coli. In this organism, the enzyme is mainly localized in the cytoplasm and periplasm, although high levels of expression lead to significant release into the extracellular medium. Functional and physical characterizations further indicate that the periplasmic and cytoplasmic enzymes of A. hydrophila are likely to be the counterparts of E. coli RNase I and its cytoplasmic form RNase I*: as for the E. coli enzymes, the A. hydrophila RNase forms have similar sizes and show broad specificity, and the periplasmic form is more active towards natural polymer RNA than its cytoplasmic counterpart. Both forms are relatively thermosensitive and are reversibly inactivated by up to 0.6% sodium dodecyl sulfate. Southern hybridization revealed homology to E. coli K-12 and Shigella sp. genomic DNA, a finding which correlates with the presence of secreted RNases in these organisms. In contrast, species of phylogenetically closer genera, such as Vibrio and Plesiomonas, did not hybridize to the A. hydrophila RNase gene.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baird S. D., Johnson D. A., Seligy V. L. Molecular cloning, expression, and characterization of endo-beta-1,4-glucanase genes from Bacillus polymyxa and Bacillus circulans. J Bacteriol. 1990 Mar;172(3):1576–1586. doi: 10.1128/jb.172.3.1576-1586.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. RNase I*, a form of RNase I, and mRNA degradation in Escherichia coli. J Bacteriol. 1991 Aug;173(15):4653–4659. doi: 10.1128/jb.173.15.4653-4659.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T., Montenegro M. A., Sanyal S. C., Helmuth R., Bulling E., Timmis K. N. Cloning of enterotoxin gene from Aeromonas hydrophila provides conclusive evidence of production of a cytotonic enterotoxin. Infect Immun. 1984 Nov;46(2):435–441. doi: 10.1128/iai.46.2.435-441.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S. P., Prior S. E., Barstow D. A., Minton N. P. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988 Aug 15;68(1):139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Relationships of human protein sequences to those of other organisms. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):447–455. doi: 10.1101/sqb.1986.051.01.054. [DOI] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobius K. S., Pemberton J. M. Molecular cloning, characterization, and nucleotide sequence of an extracellular amylase gene from Aeromonas hydrophila. J Bacteriol. 1988 Mar;170(3):1325–1332. doi: 10.1128/jb.170.3.1325-1332.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X. R., Giroux S., Cedergren R. The nucleotide sequence of the argT locus of Aeromonas hydrophila. Nucleic Acids Res. 1988 Nov 25;16(22):10936–10936. doi: 10.1093/nar/16.22.10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst T. R., Welch R. A. Mechanisms for secretion of extracellular proteins by gram-negative bacteria. Trends Biochem Sci. 1988 Jul;13(7):265–269. doi: 10.1016/0968-0004(88)90160-0. [DOI] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Horiuchi H., Yanai K., Takagi M., Yano K., Wakabayashi E., Sanda A., Mine S., Ohgi K., Irie M. Primary structure of a base non-specific ribonuclease from Rhizopus niveus. J Biochem. 1988 Mar;103(3):408–418. doi: 10.1093/oxfordjournals.jbchem.a122284. [DOI] [PubMed] [Google Scholar]
- Howard S. P., Buckley J. T. Molecular cloning and expression in Escherichia coli of the structural gene for the hemolytic toxin aerolysin from Aeromonas hydrophila. Mol Gen Genet. 1986 Aug;204(2):289–295. doi: 10.1007/BF00425512. [DOI] [PubMed] [Google Scholar]
- Howard S. P., Garland W. J., Green M. J., Buckley J. T. Nucleotide sequence of the gene for the hole-forming toxin aerolysin of Aeromonas hydrophila. J Bacteriol. 1987 Jun;169(6):2869–2871. doi: 10.1128/jb.169.6.2869-2871.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide H., Kimura M., Arai M., Funatsu G. The complete amino acid sequence of ribonuclease from the seeds of bitter gourd (Momordica charantia). FEBS Lett. 1991 Sep 2;289(1):126–126. doi: 10.1016/0014-5793(91)80924-r. [DOI] [PubMed] [Google Scholar]
- Ikemura T., Ozeki H. Codon usage and transfer RNA contents: organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1087–1097. doi: 10.1101/sqb.1983.047.01.123. [DOI] [PubMed] [Google Scholar]
- Janda J. M. Biochemical and exoenzymatic properties of Aeromonas species. Diagn Microbiol Infect Dis. 1985 May;3(3):223–232. doi: 10.1016/0732-8893(85)90034-3. [DOI] [PubMed] [Google Scholar]
- Johnston S., Lee J. H., Ray D. S. High-level expression of M13 gene II protein from an inducible polycistronic messenger RNA. Gene. 1985;34(2-3):137–145. doi: 10.1016/0378-1119(85)90121-0. [DOI] [PubMed] [Google Scholar]
- Jost W., Bak H., Glund K., Terpstra P., Beintema J. J. Amino acid sequence of an extracellular, phosphate-starvation-induced ribonuclease from cultured tomato (Lycopersicon esculentum) cells. Eur J Biochem. 1991 May 23;198(1):1–6. doi: 10.1111/j.1432-1033.1991.tb15978.x. [DOI] [PubMed] [Google Scholar]
- Kawata Y., Sakiyama F., Hayashi F., Kyogoku Y. Identification of two essential histidine residues of ribonuclease T2 from Aspergillus oryzae. Eur J Biochem. 1990 Jan 12;187(1):255–262. doi: 10.1111/j.1432-1033.1990.tb15303.x. [DOI] [PubMed] [Google Scholar]
- Kawata Y., Sakiyama F., Tamaoki H. Amino-acid sequence of ribonuclease T2 from Aspergillus oryzae. Eur J Biochem. 1988 Oct 1;176(3):683–697. doi: 10.1111/j.1432-1033.1988.tb14331.x. [DOI] [PubMed] [Google Scholar]
- Kohli J., Grosjean H. Usage of the three termination codons: compilation and analysis of the known eukaryotic and prokaryotic translation termination sequences. Mol Gen Genet. 1981;182(3):430–439. doi: 10.1007/BF00293932. [DOI] [PubMed] [Google Scholar]
- Kuo S. S., Feng T. Y. Iodometric method for detection of beta-lactamase activity in yeast cells carrying ampicillin resistance gene in chimeric plasmids. Anal Biochem. 1989 Feb 15;177(1):165–167. doi: 10.1016/0003-2697(89)90033-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lerner C. G., Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990 Aug 11;18(15):4631–4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. M., Kaper J. B., Herrington D., Losonsky G., Morris J. G., Clements M. L., Black R. E., Tall B., Hall R. Volunteer studies of deletion mutants of Vibrio cholerae O1 prepared by recombinant techniques. Infect Immun. 1988 Jan;56(1):161–167. doi: 10.1128/iai.56.1.161-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure B. A., Haring V., Ebert P. R., Anderson M. A., Simpson R. J., Sakiyama F., Clarke A. E. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature. 1989 Dec 21;342(6252):955–957. doi: 10.1038/342955a0. [DOI] [PubMed] [Google Scholar]
- Meador J., 3rd, Cannon B., Cannistraro V. J., Kennell D. Purification and characterization of Escherichia coli RNase I. Comparisons with RNase M. Eur J Biochem. 1990 Feb 14;187(3):549–553. doi: 10.1111/j.1432-1033.1990.tb15336.x. [DOI] [PubMed] [Google Scholar]
- Meador J., 3rd, Kennell D. Cloning and sequencing the gene encoding Escherichia coli ribonuclease I: exact physical mapping using the genome library. Gene. 1990 Oct 30;95(1):1–7. doi: 10.1016/0378-1119(90)90406-h. [DOI] [PubMed] [Google Scholar]
- Michaelis S., Hunt J. F., Beckwith J. Effects of signal sequence mutations on the kinetics of alkaline phosphatase export to the periplasm in Escherichia coli. J Bacteriol. 1986 Jul;167(1):160–167. doi: 10.1128/jb.167.1.160-167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miczak A., Srivastava R. A., Apirion D. Location of the RNA-processing enzymes RNase III, RNase E and RNase P in the Escherichia coli cell. Mol Microbiol. 1991 Jul;5(7):1801–1810. doi: 10.1111/j.1365-2958.1991.tb01929.x. [DOI] [PubMed] [Google Scholar]
- Model P., Russel M. Prokaryotic secretion. Cell. 1990 Jun 1;61(5):739–741. doi: 10.1016/0092-8674(90)90180-m. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Ohgi K., Horiuchi H., Watanabe H., Iwama M., Takagi M., Irie M. Evidence that three histidine residues of a base non-specific and adenylic acid preferential ribonuclease from Rhizopus niveus are involved in the catalytic function. J Biochem. 1992 Jul;112(1):132–138. doi: 10.1093/oxfordjournals.jbchem.a123852. [DOI] [PubMed] [Google Scholar]
- Oliver D. B., Beckwith J. Regulation of a membrane component required for protein secretion in Escherichia coli. Cell. 1982 Aug;30(1):311–319. doi: 10.1016/0092-8674(82)90037-x. [DOI] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H., Watanabe K., Muto A. Recent evidence for evolution of the genetic code. Microbiol Rev. 1992 Mar;56(1):229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn M. J., Munson R. Separation of the inner (cytoplasmic) and outer membranes of Gram-negative bacteria. Methods Enzymol. 1974;31:642–653. doi: 10.1016/0076-6879(74)31070-1. [DOI] [PubMed] [Google Scholar]
- Postek K. M., LaDue T., Nelson C., Sandwick R. K. Spectrophotometric ribonuclease assays using dinucleoside monophosphate substrates. Anal Biochem. 1992 May 15;203(1):47–52. doi: 10.1016/0003-2697(92)90041-5. [DOI] [PubMed] [Google Scholar]
- Quaas R., Landt O., Grunert H. P., Beineke M., Hahn U. Indicator plates for rapid detection of ribonuclease T1 secreting Escherichia coli clones. Nucleic Acids Res. 1989 Apr 25;17(8):3318–3318. doi: 10.1093/nar/17.8.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder B., Blöcker H., Frank R., McCarthy J. E. Inducible expression vectors incorporating the Escherichia coli atpE translational initiation region. Gene. 1987;52(2-3):279–283. doi: 10.1016/0378-1119(87)90054-0. [DOI] [PubMed] [Google Scholar]
- Schill W. B., Schumacher G. F. Radial diffusion in gel for micro determination of enzymes. I. Muramidase, alpha-amylase, DNase 1, RNase A, acid phosphatase, and alkaline phosphatase. Anal Biochem. 1972 Apr;46(2):502–533. doi: 10.1016/0003-2697(72)90324-7. [DOI] [PubMed] [Google Scholar]
- Silhavy T. J., Benson S. A., Emr S. D. Mechanisms of protein localization. Microbiol Rev. 1983 Sep;47(3):313–344. doi: 10.1128/mr.47.3.313-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöström M., Wold S., Wieslander A., Rilfors L. Signal peptide amino acid sequences in Escherichia coli contain information related to final protein localization. A multivariate data analysis. EMBO J. 1987 Mar;6(3):823–831. doi: 10.1002/j.1460-2075.1987.tb04825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton J., Howard S. P., Buckley J. T. Molecular cloning of a phospholipid-cholesterol acyltransferase from Aeromonas hydrophila. Sequence homologies with lecithin-cholesterol acyltransferase and other lipases. Biochim Biophys Acta. 1988 Mar 25;959(2):153–159. doi: 10.1016/0005-2760(88)90026-4. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Viret J. F., Bruderer U., Lang A. B. Characterization of the Shigella serotype D (S. sonnei) O polysaccharide and the enterobacterial R1 lipopolysaccharide core by use of mouse monoclonal antibodies. Infect Immun. 1992 Jul;60(7):2741–2747. doi: 10.1128/iai.60.7.2741-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WITZEL H., BARNARD E. A. Mechanism and binding sites in the ribonuclease reaction. II. Kinetic studies on the first step of the reaction. Biochem Biophys Res Commun. 1962 May 4;7:295–299. doi: 10.1016/0006-291x(62)90194-8. [DOI] [PubMed] [Google Scholar]
- Watanabe H., Ohgi K., Irie M. Primary structure of a minor ribonuclease from Aspergillus saitoi. J Biochem. 1982 May;91(5):1495–1509. doi: 10.1093/oxfordjournals.jbchem.a133841. [DOI] [PubMed] [Google Scholar]
- von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984 Feb 25;173(2):243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]