Abstract
A monomeric peptide fragment of GroEL, consisting of residues 191–376, is a mini-chaperone with a functional chaperoning activity. We have solved the crystal structure at 1.7 Å resolution of GroEL(191–376) with a 17-residue N-terminal tag. The N-terminal tag of one molecule binds in the active site of a neighboring molecule in the crystal. This appears to mimic the binding of a peptide substrate molecule. Seven substrate residues are bound in a relatively extended conformation. Interactions between the substrate and the active site are predominantly hydrophobic, but there are also four hydrogen bonds between the main chain of the substrate and side chains of the active site. Although the preferred conformation of a bound substrate is essentially extended, the flexibility of the active site may allow it to accommodate the binding of exposed hydrophobic surfaces in general, such as molten globule-type structures. GroEL can therefore help unfold proteins by binding to a hydrophobic region and exert a binding pressure toward the fully unfolded state, thus acting as an “unfoldase.” The structure of the mini-chaperone is very similar to that of residues 191–376 in intact GroEL, so we can build it into GroEL and reconstruct how a peptide can bind to the tetradecamer. A ring of connected binding sites is noted that can explain many aspects of substrate binding and activity.
Keywords: protein folding, hsp60, cpn60, mini-chaperone, crystallography
The molecular chaperone GroEL assists the folding of many newly synthesized proteins in Escherichia coli and facilitates the refolding in vitro of several proteins that would otherwise misfold or aggregate. GroEL is a cylinder made from two heptameric rings stacked back to back, creating a central cavity ≈45 Å wide (1, 2). Each 57-kDa subunit consists of three domains: the ATP-binding equatorial domain (residues 6–133 and 409–523) connects the two rings; the apical domain (residues 191–376) forms the flexible opening of cylinder and contains the putative polypeptide-binding and GroES-binding sites, which line the inner wall of the cavity; and the intermediate domain (residues 134–190 and 377–408) makes intersubunit contacts within a ring and transmits ATP- and GroES-mediated allosteric effects. The nature of binding of nonnative proteins by GroEL is, so far, unresolved (3–8). But, residues in the region of 199–264 were postulated from a site-directed mutagenesis study to be involved in the binding of polypeptides (9).
The action of GroEL in vivo is multifaceted (10–16). Studies on smaller, isolated domains of the molecule can elucidate the functions of individual components in a detail that may not be achievable when using the intact molecule. For example, isolated monomeric polypeptide-binding fragments of GroEL (residues 191–345 and 191–376) exhibit high chaperone activity in vitro, effecting the refolding of rhodanese and cyclophilin, and the unfolding of barnase (17). These “mini-chaperones” are active without the allosteric properties or the central cavity, which has been proposed to act as a “folding cage” in vivo (18, 19). The crystal structure of GroEL(191–345) at 2.5 Å resolution (17) is very similar to its region in the intact chaperone.
We have investigated the structural details of the polypeptide-binding site of GroEL by solving the crystal structure at 1.7 Å resolution of the mini-chaperone corresponding to residues 191–376 fused to a 17-residue N-terminal tag (which we number as residues −17 to −1). The tail of one molecule is seen to bind in the active site of a neighbor so that we construct a model for the chaperone–substrate complex.
MATERIALS AND METHODS
Protein Expression and Purification.
The mini-chaperone (GroEL191–376) was produced by subcloning the apical domain of GroEL (residues 191–376) by polymerase chain reaction into the polylinker site of a pRSET A vector (Invitrogen), coding for an N-terminal histidine tag, which contained an engineered thrombin cleavage site (17). The histidine tag was composed of 17 amino acids (−17 MRGSHHHHHHGLVPRGS −1). Expression in E. coli TG2 cells and purification of the mini-chaperone was as described (17).
Crystallization.
Crystals of the mini-chaperone were obtained from hanging drops initially containing protein at 23 mg·ml−1/0.5 M NaCl/50 mM Tris·HCl, pH 8.5/25% glycerol, equilibrated against reservoirs consisting of 1.0 M NaCl/100 mM Tris·HCl, pH 8.5/25% glycerol. Crystals grew in space group P212121, with cell dimensions a = 47.72 Å, b = 63.81 Å, and c = 75.10 Å.
Structure Determination and Refinement.
X-ray data were collected from a crystal flash-frozen in liquid N2 at 100 K, using a 15 cm MAR Research image plate detector at Deutsches Elektronen Synchrotron (Hamburg; station X31, λ = 1.07 Å). Data processing, data reduction, electron density syntheses, and structural analyses were carried out using ccp4 software (20). The structure was solved by molecular replacement using the program amore (20) and a search model consisting of residues 191–345 of the refined structure of a recently solved mini-chaperone (17). The asymmetric unit contains one protein monomer. Model rebuilding was performed with the program o (21), and the structure was refined using x-plor (22), followed by refmac (20).
RESULTS
Three-Dimensional Structure of the Mini-Chaperone.
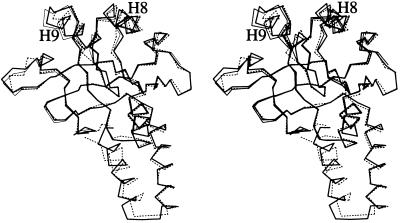
The refined model contained 292 water molecules and was complete. Crystallographic data are summarized in Table 1. The quality of the electron density was excellent throughout (Fig. 1). Overall, the structure was almost identical to the corresponding region of the intact protein (1, 2), and could be described as a β-sandwich scaffold flanked by helical and loop regions (Fig. 2 Top). A least-squares fit of the structures of mini-chaperones GroEL191–376 and GroEL191–345 (17), using backbone atoms of β-sheet residues, gave a root-mean-square deviation (rmsd) of 0.3 Å (for the β-sheet residues). The largest differences were found in helices H8 and H9 (differences in positions of Cα atoms in the range 0.5–1.7 Å) (Fig. 3). A least-squares fit of mini-chaperone GroEL191–376 and residues 191–376 in a subunit (chain A) of intact GroEL (Protein Data Bank code 1OEL), using backbone atoms of β-sheet residues, gave an rmsd of 0.7 Å (for the β-sheet residues). Again, the largest differences were found in helices H8 and H9 (Cα differences in the range 0.8–2.0 Å) (Fig. 3).
Table 1.
Summary of crystallographic data
| Data collection statistics | |
| Resolution, Å | 12.8–1.7 |
| Measured reflections | 260,633 |
| Unique reflections | 25,290 |
| Completeness of data, %* | 97.6 (85.6) |
| Rmerge, %*† | 4.7 (21.7) |
| 〈I/σI〉* | 11.8 (3.2) |
| Multiplicity* | 4.9 (4.4) |
| Refinement statistics | |
| Resolution, Å | 12.8–1.7 |
| R factor/free R factor, %, F > 0‡ | 18.0/22.4 |
| rmsd bond length, Å | 0.016 |
| rmsd bond angle, deg | 2.7 |
rmsd, root-mean-square deviation.
Values given in parentheses are for the highest resolution shell.
Agreement between intensities of repeated measurements of the same reflections and can be defined as: Σ(Ih,i − 〈Ih〉)/ΣIh,i, where Ih,i are individual values and 〈Ih〉 is the mean value of the intensity of reflection h.
The free R factor was calculated with the 10% of data omitted from the refinement.
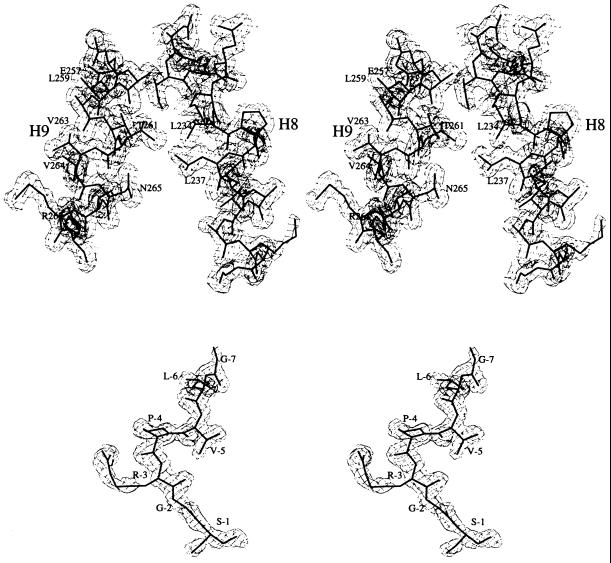
Figure 1.
2Fo-Fc electron density map calculated with SIGMAA coefficients (23), contoured at 1σ (σ is the root-mean-square deviation from the mean electron density in the unit cell). Superimposed is the refined atomic model. (Upper) Helices H8 and H9. (Lower) The N-terminal tag. Drawn with the bobscript (extensions to the program molscript; ref. 24).
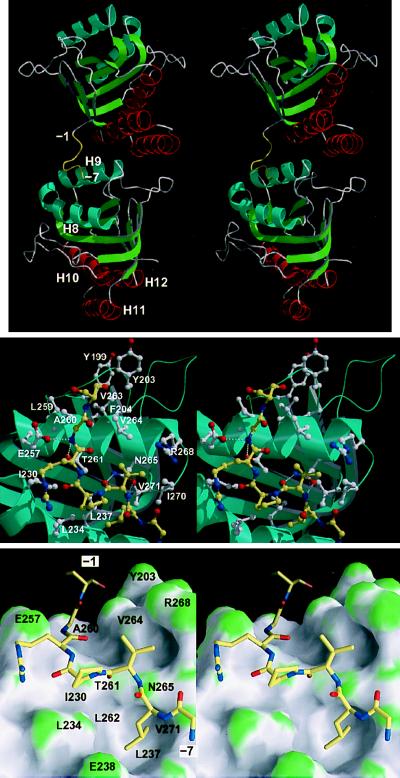
Figure 2.
(Top) Stereo cartoon representation of the structure of the mini-chaperone (GroEL191–376), showing the interaction between the N-terminal tag and a neighboring molecule in the crystal lattice (related by a crystallographic two-fold screw operation along the c axis, positioned approximately vertical and in the plane of the paper). The N-terminal tag (residues −1 to −7) is colored yellow. (Middle) Close-up of peptide-binding site interactions, in stereo. The peptide is represented by yellow bonds; neighboring residues are represented by white bonds. Hydrogen bonds are represented by broken white lines. Drawn with bobscript (extensions to the program molscript; ref. 24) and raster3d (25). (Lower) As in Middle but showing the molecular surface of the mini-chaperone. The surface is colored according to surface curvature to highlight concave surface pockets. Convex, concave, and flat surfaces are colored green, grey, and white, respectively. Residues underlying the surface are labeled. Drawn with grasp (26). All three figures show the model in approximately the same orientation.
Figure 3.
Stereo representation of the overlay of Cα atoms of the apical domain in intact GroEL (residues 191–376; broken bonds), mini-chaperone GroEL191–376 (thick bonds), and mini-chaperone GroEL191–345 (thin bonds) (17). Structures were fitted using backbone atoms from β-sheet residues. Drawn with molscript (24).
Binding of the N-terminal Tag to the Active Site.
Residues −7 to −1 (sequence GLVPRGS) of the N-terminal tag could be built into the electron density map, and adopt a relatively extended conformation (Fig. 1 Lower; Fig. 2). Electron density for the remaining 10 residues of the tag (−17 to −8) was not observed. The tag projected away from the protein surface, and residues −7 to −1 associated with a neighboring molecule in the crystal lattice, binding to residues that have been implicated in the interaction with polypeptide substrates and with GroES (Fig. 2; ref. 9). These residues are nonpolar and are located within a shallow cleft between α-helices H8 and H9, and in an adjacent surface that is formed by the packing between helix H9 and a neighboring loop (residues 199–204) (Fig. 2; ref. 9). We could identify in the crystal structure the binding of residues −7 to −1 of the tag. The side chain of L-6 fit snugly inside a hydrophobic pocket within the interhelical cleft, the remaining residues running diagonally along the cleft and then across the ridge formed by helix H9 (Fig. 2). Most interactions were nonpolar (Table 2), although there were four hydrogen bonds between the main chain of the tag and three side chains (E257, N265, and T261) of helix H9. Model building and an earlier mutagenesis study (9) imply that the substrate-binding site extends further (to include Y199, Y203, F204, and V263). Approximately 10–11 residues may be accommodated in the active site in an extended conformation.
Table 2.
Interactions between mini-chaperone and the N-terminal tag
| Mini-chaperone residue | Closest distance of interaction, Å |
|---|---|
| Residues postulated to interact from sdm (9) | |
| Y199 | 17.3 |
| Y203 | 7.5 |
| F204 | 10.9 |
| L234 | 4.3 |
| L237 | 3.8 |
| L259 | 6.4 |
| V263 | 5.4 |
| V264 | 3.5 |
| Residues not found to interact from sdm (9) | |
| I230 | 3.3 |
| E238 | 3.5 |
| Residues not mutated (9) | |
| A241 | 4.3 |
| E257* | 2.7 |
| A260 | 3.6 |
| T261* | 3.1 |
| N265* | 2.7 |
| R268 | 3.8 |
| I270 | 3.6 |
| V271 | 3.7 |
The table comprises all the residues in the apical domain of GroEL (residues 191–376) that have been found by site-directed mutagenesis (sdm) to appear to be involved in the binding of peptide substrates (9) and all those detected in the crystal structure.
Hydrogen-bonded interaction.
Comparison with Site-Directed Mutagenesis Results.
Fenton et al. (9) postulated that residues Y199, Y203, F204, L234, L237, L259, V263, and V264 were involved in binding polypeptide substrates (Table 2). Residues Y199, Y203, and F204 would be sterically inaccessible to the N-terminal tag, so we did not observe binding close to them. Residues L234, L237, L259, V263, and V264 were in the binding site. Residues I230 and E238 were tested by mutagenesis (9) and found not to affect binding, but we found that they did interact with the tag. Possibly, the binding assay was not sensitive enough to detect the changes on mutation. In addition, we have identified a series of residues that were not mutated but are in the binding site (Table 2) and extend it to residue V271.
DISCUSSION
The Nature of GroEL–Polypeptide Binding.
The most important observation from this study is that the polypeptide tag of one molecule binds in the region of a neighboring molecule that has been identified (9) as being involved in binding polypeptide substrates. The N-terminal tag does not interact with the rest of its own chain. Further, the majority of interactions involved in the packing of crystal molecules are located elsewhere on the protein surface. Accordingly, it is most likely that the nature of the interaction between the tag and the putative polypeptide binding site is not restricted to a crystalline environment. Thus, the mode of binding should provide structural information that is relevant to GroEL–peptide interactions. The binding site is strongly hydrophobic, but polar residues are also available for hydrogen bonding with the main chain of a peptide substrate. The polar residues are arranged such that an extended conformation of the substrate is bound.
Flexibility of the Mini-Chaperone and Unfoldase Activity.
The peptide binding site is the most flexible region in the structure of intact GroEL (refs. 1 and 2; Fig. 3). We propose that the binding site can accommodate a wide range of substrate side chains because of variation of the dimensions of the active site through movements of helices H8 and H9 and surrounding loops. Interestingly, there is a structural similarity between the mini-chaperone and the enzymes pyruvate phosphate dikinase and aconitase, both of which possess a well-ordered β-sandwich surrounded by more flexible helical and loop regions (A. G. Murzin, personal communication). The dynamic behavior of the helical and loop structures of these enzymes is crucial to their function (27, 28) by an induced fit mechanism (29).
Thus, although the preferred polypeptide conformation bound by GroEL is essentially extended, the flexible surface may also accommodate the binding of exposed hydrophobic surfaces in general, such as the face of an amphipathic helix (3, 4, 8) and molten globule-type structures (5–7, 30) as well as the more fully unfolded states, which are implied by hydrogen-exchange studies (11). GroEL can thus help unfold proteins by binding to a hydrophobic region and exert a binding pressure toward the fully unfolded state. By this means, GroEL can act as an unfoldase by a “strain” or “stress” mechanism (31).
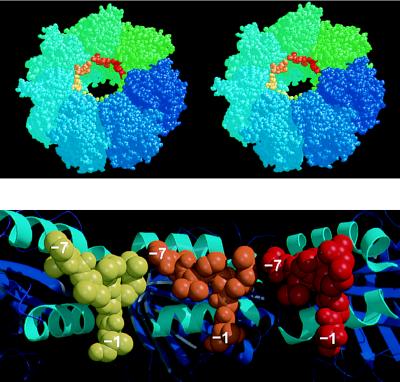
Binding of a Polypeptide Substrate to the GroEL Tetradecamer.
Building this binding site into the structure of intact GroEL reveals a ring of sites around the GroEL apex (Fig. 4). Each site is arranged at approximately 45° to the axis of the ring so that a long denatured protein could bind as a series of parallel segments, connected by loops that extend into the cavity. Rigid-body motions of the apical domains relative to each other are seen by electron microscopy (33, 34) in the ATP-mediated allosteric transitions that regulate the binding, folding, and release of polypeptide substrates (14, 16, 35). Such movements on the binding of ATP would cause the binding sites to move away from each other, weakening binding and resulting in release of polypeptide. It is also possible that direct competitive inhibition between the denatured peptide and the “mobile loop” (residues 16–34) in GroES could displace bound polypeptide from GroEL (36–38), especially as the conserved tripeptide I25-V26-L27 in this loop could bind to GroEL in the mode suggested from our model.
Figure 4.
(Upper) Stereoview of one heptameric ring of the GroEL tetradecamer showing the position of the N-terminal tag bound to each apical domain, near the opening to the central cavity. This model is generated by the superposition of the mini-chaperone GroEL(191–376) with each corresponding apical domain (residues 191–376) in intact GroEL (the second ring of the GroEL cylinder, generated by a two-fold symmetry operation, is not shown, but stacks against the underside of the drawn ring). GroEL subunits are colored around the ring going from blue to green. Superimposed “bound peptides” are colored from green to red. Drawn with rasmol (32). (Lower) Cross-section of the model shown in Upper looking directly at the inner wall of the cavity, and showing the apical domains (cartoon with helices H8 and H9 colored cyan) from three subunits with modeled peptide (shown as space-filling models colored yellow, orange, and red, respectively). Drawn with molscript (24) and raster3d (25). Each of the separate peptides (residues −1 to −7) could be linked together by small fragments of peptides so that a longer peptide could bind from one contiguous site to the next.
Subsites.
The binding of one molecule of denatured barnase to an excess of GroEL fits to a model of very tight 1:1 binding, whereas four molecules of denatured barnase bind when it is present in excess over GroEL (39). This can be interpreted in light of the present model, which has a ring of seven identical sites: in a 1:1 complex, barnase binds to more than one subsite, and therefore has tight binding because of the synergistic effects; whereas in a 4:1 complex there are fewer subsites available for each barnase substrate molecule.
Acknowledgments
A postdoctoral Liebig fellowship to R.Z. is gratefully acknowledged.
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1KID).
References
- 1.Braig K, Otwinowski Z, Hegde R, Boisvert D C, Joachimiak A, Horwich A L, Sigler P B. Nature (London) 1994;371:578–86. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 2.Braig K, Adams P D, Brünger A T. Nat Struct Biol. 1995;2:1083–1094. doi: 10.1038/nsb1295-1083. [DOI] [PubMed] [Google Scholar]
- 3.Landry S J, Gierasch L M. Biochemistry. 1991;30:7359–7362. doi: 10.1021/bi00244a001. [DOI] [PubMed] [Google Scholar]
- 4.Landry S J, Jordan R, McMacken R, Gierasch L M. Nature (London) 1992;355:455–457. doi: 10.1038/355455a0. [DOI] [PubMed] [Google Scholar]
- 5.Hayer-Hartl M K, Ewbank J J, Creighton T E, Hartl F-U. EMBO J. 1994;13:3192–3202. doi: 10.1002/j.1460-2075.1994.tb06618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin J, Langer T, Boteva R, Schramel A, Horwich A L, Hartl F U. Nature (London) 1991;352:36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- 7.Mendoza J A, Butler M C, Horowitz P M. J Biol Chem. 1992;267:24648–24654. [PubMed] [Google Scholar]
- 8.Schmidt M, Buchner J. J Biol Chem. 1992;267:16829–16833. [PubMed] [Google Scholar]
- 9.Fenton W A, Kashi Y, Furtak K, Horwich A L. Nature (London) 1994;371:614–619. doi: 10.1038/371614a0. [DOI] [PubMed] [Google Scholar]
- 10.Ellis R J, Hartl F U. FASEB J. 1996;10:20–26. doi: 10.1096/fasebj.10.1.8566542. [DOI] [PubMed] [Google Scholar]
- 11.Zahn R, Perrett S, Stenberg G, Fersht A R. Science. 1996;271:642–645. doi: 10.1126/science.271.5249.642. [DOI] [PubMed] [Google Scholar]
- 12.Zahn R, Perrett S, Fersht A R. J Mol Biol. 1996;261:43–61. doi: 10.1006/jmbi.1996.0440. [DOI] [PubMed] [Google Scholar]
- 13.Gray T E, Eder J, Bycroft M, Day A G, Fersht A R. EMBO J. 1993;12:4145–4150. doi: 10.1002/j.1460-2075.1993.tb06098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weissman J S, Kashi Y, Fenton W A, Horwich A L. Cell. 1994;78:693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- 15.Todd M J, Viitanen P V, Lorimer G H. Science. 1994;265:659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- 16.Corrales F J, Fersht A R. Proc Natl Acad Sci USA. 1996;93:4509–4512. doi: 10.1073/pnas.93.9.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zahn R, Buckle A M, Perrett S, Johnson C M J, Corrales F J, Golbik R, Fersht A R. Proc Natl Acad Sci USA. 1996;93:15024–15029. doi: 10.1073/pnas.93.26.15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agard D A. Science. 1993;260:1903–1904. doi: 10.1126/science.8100365. [DOI] [PubMed] [Google Scholar]
- 19.Ellis R J. Curr Opin Struct Biol. 1994;4:117–122. [Google Scholar]
- 20.Anonymous Acta Crystallogr. 1994;D50:760–763. [Google Scholar]
- 21.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.Brünger A T. xplor Manual. New Haven, CT: Yale University; 1992. Version 3.0. [Google Scholar]
- 23.Read R. Acta Crystallogr. 1986;A42:140–149. [Google Scholar]
- 24.Kraulis P. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 25.Merrit E A, Murphy M E P. Acta Crystallogr. 1994;D50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 26.Nicholls A. grasp, Graphical Representation and Analysis of Surface Properties. New York; 1992. [Google Scholar]
- 27.Herzberg O, Chen C C, Kapadia G, McGuire M, Carroll L J, Noh S J, Dunaway M D. Proc Natl Acad Sci USA. 1996;93:2652–2657. doi: 10.1073/pnas.93.7.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lauble H, Stout C D. Proteins. 1995;22:1–11. doi: 10.1002/prot.340220102. [DOI] [PubMed] [Google Scholar]
- 29.Koshland D E, Jr, Nemethy G, Filmer D. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 30.Robinson C V, Gross M, Eyles S J, Ewbank J J, Mayhew M, Hartl F U, Dobson C M, Radford S E. Nature (London) 1994;372:646–651. doi: 10.1038/372646a0. [DOI] [PubMed] [Google Scholar]
- 31.Fersht A R. Enzyme Structure and Mechanism. New York: W. H. Freeman and Company; 1985. [Google Scholar]
- 32.Sayle R A, Milnerwhite E J. Trends Biochem Sci. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 33.Langer T, Pfeifer G, Martin J, Baumeister W, Hartl F U. EMBO J. 1992;11:4757–4765. doi: 10.1002/j.1460-2075.1992.tb05581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, Roseman A M, Hunter A S, Wood S P, Burston S G, Ranson N A, Clarke A R, Saibil H R. Nature (London) 1994;371:261–264. doi: 10.1038/371261a0. [DOI] [PubMed] [Google Scholar]
- 35.Todd M J, Viitanen P V, Lorimer G H. Biochemistry. 1993;32:8560–8567. doi: 10.1021/bi00084a024. [DOI] [PubMed] [Google Scholar]
- 36.Zeilstra R J, Fayet O, Georgopoulos C. Annu Rev Microbiol. 1991;45:301–325. doi: 10.1146/annurev.mi.45.100191.001505. [DOI] [PubMed] [Google Scholar]
- 37.Zeilstra R J, Fayet O, Georgopoulos C. J Bacteriol. 1994;176:6558–6565. doi: 10.1128/jb.176.21.6558-6565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landry S J, Zeilstra R J, Fayet O, Georgopoulos C, Gierasch L M. Nature (London) 1993;364:255–258. doi: 10.1038/364255a0. [DOI] [PubMed] [Google Scholar]
- 39.Corrales F J, Fersht A R. Proc Natl Acad Sci USA. 1995;92:5326–5330. doi: 10.1073/pnas.92.12.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]