Abstract
Mini-chaperones (e.g., a peptide consisting of residues 191-345 of GroEL) that are immobilized on agarose have very efficient chaperoning activity with several proteins that are otherwise recalcitrant to renaturation by conventional methods. We have used immobilized mini-chaperones both in column chromatography and batchwise to renature an insoluble protein from an inclusion body, to refold apparently irreversibly denatured proteins, and to recondition enzymes that have lost activity on storage. Refolding chromatography offers an efficient and simple means to renature proteins in high yield and with biological activity.
Keywords: protein, renaturation, folding, GroEL, hsp60
The molecular chaperone GroEL, a tetradecamer of 57 kDa subunits, facilitates the folding in vitro of a number of proteins that would otherwise misfold or aggregate and precipitate. It is cylindrical, consisting of two seven-membered rings that form a large cavity (1, 2) that has been generally considered to be essential for activity (3). In vivo, and for many reactions in vitro, GroEL requires the cochaperonin GroES (7 × 10 kDa subunits) and ATP to be functional. Although there are reports that proteolytic fragments of 34 kDa and 54 kDa have weak chaperoning activity (4, 5), monomers of GroEL that are induced by mutagenesis (6), pressure, or urea are inactive (7–9). A 54-kDa fragment of GroEL is reported as having about 10% activity in chaperoning the refolding of rhodanese when the fragment is covalently attached to a resin (5). Recombinant fragments of GroEL (including the 16.7-kDa fragment GroEL(191–345) and the 21-kDa GroEL(191–376), which were expressed in Escherichia coli) have efficient chaperone activity in facilitating the refolding of cyclophilin A and rhodanese, and the unfolding of barnase (10). Here, we attach the mini-chaperones to agarose gels and find that they can be used for the efficient renaturation of proteins.
MATERIALS AND METHODS
Materials.
The apical domain of GroEL [GroEL(191–376)] and the “core” of the apical domain, GroEL(191–345), were cloned and expressed in E. coli as fusion proteins containing a 17-residue N-terminal histidine tail (10). Glucosamine 6-phosphate deaminase was expressed in and purified from E. coli K12, and assayed as described (11). Indole 3-glycerol phosphate synthase (IGPS) mutants were expressed in E. coli (M.M.A., J. Blackburn, and A.R.F., unpublished data). Cyclophilin A was purchased from Sigma or was a gift from G. Fischer and assayed as described (12). Firefly luciferase was purchased from Sigma and assayed (13).
Immobilization of Mini-Chaperones.
The fragments were immobilized by two methods.
Attachment to Ni-NTA resin.
The Ni-NTA resin (Qiagen, Chatsworth, CA) is a chelating adsorbant composed of a high surface concentration of nitrilotriacetic acid (NTA) ligand attached to Sepharose CL-6B. Proteins containing six-residue His affinity tags, located at either the amino or carboxyl terminus of the protein, bind to the Ni-NTA resin with high affinity (Kd = 10−13 M, pH 7.8). The stability of the 6 × His/Ni-NTA interaction is unaffected by strong denaturants such as 6 M guanidine hydrochloride or 8 M urea, or the presence of low levels of 2-mercaptoethanol (1–10 mM). Ni-NTA resin (3.5 ml) was equilibrated with refolding buffer (0.1 M potassium phosphate at pH 7.8, containing 5 mM 2-mercaptoethanol). The mini-chaperone was added to saturation of the affinity gel (21 mg of protein per 3.5 ml of gel) and incubated at room temperature for 30 min with gentle mixing. The gel was packed in a column suitable for fast protein liquid chromatography (FPLC) (5 × 100 mm; Pharmacia) and thoroughly washed with refolding buffer.
Attachment to CNBr-activated Sepharose 4B.
To minimize the steric effects and preserve the structure of the binding site in the apical domain, the binding capacity of the gel was reduced by controlled hydrolysis of the activated gel before coupling. Freeze-dried powder (300 mg) was suspended in 50 mM NaHCO3 at pH 8.3, washed with the same buffer, and reswollen on a sintered glass filter (G3), then suspended in the buffer and mixed in an end-over-end shaker for 4 hr at room temperature. The mini-chaperone, dissolved in the coupling buffer (0.1 M NaHCO3, pH 8.3/0.5 M NaCl), was added to the gel suspension (10 mg protein per ml gel) and mixed in an end-over-end shaker for 6 hr at room temperature. It was then washed with the coupling buffer. The remaining active groups were blocked by adding 2.5 M ethanolamine at pH 8 and shaking for 4 hr at room temperature. Uncoupled mini-chaperone was removed by washing with five cycles of alternately high and low pH buffer solution (0.1 M Tris·HCl, pH 7.8, containing 0.5 M NaCl) followed by acetate buffer (0.1 M, pH 4, plus 0.5 M NaCl). The gel was finally washed with 5–10 gel volumes of refolding buffer.
Batchwise Renaturation.
A suspension of 200 μl of gel (wet sedimented volume, either bound via the Ni-NTA linkage or covalently linked to mini-chaperone via CNBr activation) was mixed with refolding buffer to give a volume of 990 μl. Cyclophilin A (10 μl; 100 μM stock solution in refolding buffer + 8 M urea = 1 nmol of cyclophilin A) was added to the suspension and mixed in an up-down mixer for 30 min at room temperature. The gel suspension was centrifuged to separate the supernatant (≈800 μl). The gel pellet was washed in empty miniprep columns (Promega) and the eluate added to the supernatant to give a final volume of approximately 900 μl. The concentration of cyclophilin in the supernatant was determined from its absorbance at 280 nm. The gel was regenerated by washing with 3–5 gel volumes of refolding buffer containing 2 M KCl and 2 M urea, followed by 5 gel volumes of refolding buffer.
Column Chromatography.
A column of ht-GroEL 191–345 immobilized on Ni-NTA-agarose (3.0 ml) was loaded with 2 nmol of a mutant [IGPS(49–252), which lacks residues 1–48] in 20 μl of 8 M urea. The column was developed with refolding buffer using a Waters 625 LC HPLC system. After use, the column was regenerated by washing with 3–5 gel volumes of refolding buffer containing 2 M KCl and 2 M urea, followed by 5 gel volumes of refolding buffer.
Refolding Capacity of Mini-Chaperone Agarose.
The mini-chaperone GroEL(191–345) covalently coupled to CNBr-activated Sepharose 4B to 8 mg protein per ml of the swollen gel. The capacity of the resin for refolding was determined from the batchwise renaturation of cyclophilin A. A suspension of 50 μl of preequilibrated gel (wet sedimented volume) was mixed with refolding buffer to give a volume of 192 μl. Denatured cyclophilin A (8 μl; 200 μg of protein) in 8 M urea was added and the suspension was mixed in an up-down mixer for 1 hr at room temperature. The gel suspension was centrifuged to separate the supernatant (≈150 μl). The concentration of cyclophilin in the supernatant was determined from its absorbance at 280 nm, and its activity was assayed as described (12). There was no diminution of the refolding activity toward cyclophilin A at the highest concentrations used of approximately 4 mg substrate per ml drained gel.
RESULTS AND DISCUSSION
Batchwise Renaturation of Proteins.
The fragments GroEL(191–345) or GroEL(191–376) were expressed in E. coli with a 17-residue N-terminal tail containing six histidine residues so that they could be purified using Ni-NTA resin. Initially, the same resin was used to immobilize purified GroEL(191–345) or GroEL(191–376) by their histidine tags for preparative purposes. Subsequently, the purified fragments were also linked covalently to agarose via CNBr activation (14). The immobilized GroEL fragments were found to facilitate the refolding of cyclophilin A with high efficiency. The first experiments used batchwise mixing of materials. For example, a solution of denatured cyclophilin in 8 M urea was diluted 100-fold with Ni-NTA-immobilized GroEL(191–345) in a refolding buffer (Table 1). After gentle mixing for 30 min, the cyclophilin was recovered in 84% yield of protein. The cyclophilin used for the denaturation experiments initially had 88% of the activity of the purest samples previously obtained in this laboratory. The specific activity of the material obtained from the GroEL(191–345)-agarose resin (covalently linked) was 126% of that of the previous purest samples. Further, the total recovery of activity was 25% more than that initially present. Thus, the immobilized GroEL fragment had “reconditioned” the cyclophilin A by converting inactive material into active. The procedure could be scaled up to 4 mg cyclophilin A per ml of drained gel without any loss of recovery.
Table 1.
Batchwise renaturation of cyclophilin A on mini-chaperone-agarose gels
| Gel type | Protein yield, % | Activity yield, % | Specific activity relative to “pure” native, % |
|---|---|---|---|
| GroEL(191–345)-Ni-NTA-agarose | 84 | 98 | 103 |
| GroEL(191–376)-Ni-NTA-agarose | 81 | 105 | 115 |
| GroEL(191–345)-agarose | 87 | 125 | 126 |
| HSA-agarose (control)* | 84 | 33 | 35 |
| Ethanolamine-agarose (control)† | 80 | 25 | 28 |
| Agarose (control)‡ | 72 | 17 | 20 |
| Native cyclophilin (control) | 88§ |
Cyclophilin activity was measured in the supernatant as described (12).
Human serum albumin (HSA) was covalently linked to CNBr-activated agarose.
CNBr-activated agarose was coupled to ethanolamine.
NTA-agarose after removal of the Ni from Ni-NTA agarose.
The sample prior to denaturation had 88% of the specific activity of the highest activity of native cyclophilin previously obtained in this laboratory.
Control experiments (Table 1) in which the protein was denatured in 8 M urea and added to the refolding buffer alone, or mixed with agarose that had not been linked to GroEL fragments, gave cyclophilin that had only 15–20% of the activity of the active material prepared from immobilized GroEL(191–345). Immobilized human serum albumin has weak activity, of about twice the agarose control.
We have applied the procedure to other proteins used in the laboratory. Glucosamine 6-phosphate deaminase (6 × 30 kDa; ref. 15) normally regains only 10% or less activity after renaturation from urea denaturation. A 100% yield was obtained on batchwise treatment with GroEL(191–345)-agarose. Further, a sample that had lost all activity on storage in solution in 50% glycerol/water at −20°C for 5 years also regained 100% activity with this treatment after dissolving in urea.
Refolding Chromatography.
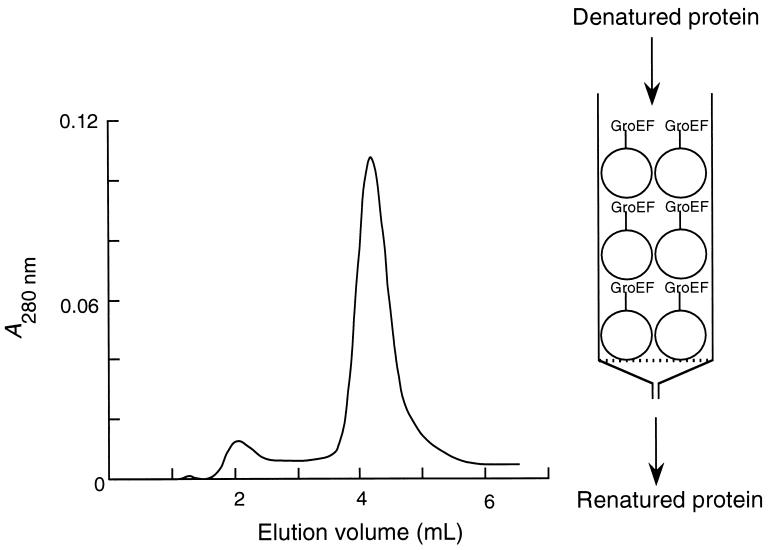
The immobilized fragments could also be used in a chromatography column (Fig. 1). Denatured protein in urea was added to the column and developed with a refolding buffer. Several mutants of IGPS have been expressed in E. coli and isolated as inclusion bodies in this laboratory (M.M.A., J. Blackburn, and A.R.F., unpublished data). They reprecipitate on attempts to renature them after dissolving at high concentrations in urea, and attempts at low protein concentrations yielded soluble material of nonnative conformation. The mutant IGPS(49–252) (1 × 22 kDa), which lacks the first 48 amino acid residues, was obtained by chromatography on the GroEL(191–345) column in a 92% yield of material that had 100% binding activity (Fig. 1). It had a circular dichroism spectrum characteristic of a native α/β protein and bound 3H-rCdRP (tritium-labeled reduced 1[(2-carboxyphenyl)amino]-1-deoxyribulose 5-phosphate), a specific inhibitor of IGPS, with a stoichiometry of 1.0.
Figure 1.
Refolding chromatography of IGPS(49–252) (indole 3-glycerol phosphate synthase lacking residues 1–48).
The protein eluted as from a conventional binding column; its passage was retarded and the peak could be characterized by a retention time. Two peaks are seen in Fig. 1 whose positions may be described by the standard chromatography equation: Kav = (Ve − Vo)/(Vt − Vo), where Vt is the total volume of gel in the column, Vo, the void volume, and Ve, the volume at the maximum of the peak. A value of Kav > 1 indicates that the protein interacts with the support. Peak 1 for IGPS(49–252) has Kav = 1.0 (9.6% of the total area), and peak 2 has Kav = 1.7 (90.4%). The active protein, which was in peak 2, was recovered in a volume of 1.2 ml, and contained 1.85 nmol of truncated IGPS (92.5% yield). Denatured cyclophilin A chromatographed with two peaks, an inactive form at Kav = 0.38 and an active form at Kav = 1.43. We suggest the nomenclature “refolding chromatography” to describe the phenomenon of refolding by passage through the column.
Renaturation May Be Restricted to GroEL Substrates.
Firefly luciferase does not regain activity upon refolding from the denatured form, even in the presence of GroEL/GroES/ATP (13). It did not regain activity on refolding chromatography. This suggests that refolding chromatography may be limited to just some, or all, of the proteins that refold in the presence of GroEL or the complete holoprotein system.
Mechanistic Implications.
The mini-chaperones are monodispersed on the agarose column so that they cannot oligomerize. The efficient activity under these conditions supports the previous evidence that only a 1:1 stoichiometry of mini-chaperone to cyclophilin A is required for activity (10). The cyclophilin folds without needing the central cavity of intact GroEL. But, the monodispersal of the mini-chaperone may have the subsidiary effect of monodispersing the bound substrate molecules, which may allow the substrate to fold in isolation, thereby mimicking some aspects of a “folding cage.” The weak activity of immobilized human serum albumin, which is about twice background (Table 1), may be because of this.
The complex structure of GroEL and the requirement of ATP and GroES in vivo must relate to the control of the activity of GroEL under physiological conditions in the presence of a wide range of competing substrates. Removal of the large fraction of the protein that is responsible for the allosteric interactions and control yields mini-chaperones that are of utility in vitro, especially when they are immobilized. They provide a very convenient and effective system for the renaturation of many proteins with high yield and biological activity.
Acknowledgments
A European Molecular Biology Organization fellowship to M.M.A. and a postdoctoral Liebig fellowship to R.Z. are gratefully acknowledged.
ABBREVIATIONS
- IGPS
indole 3-glycerol phosphate synthase
- NTA
nitrilotriacetic acid
References
- 1.Braig K, Otwinowski Z, Hegde R, Boisvert D C, Joachimiak A, Horwich A L, Sigler P B. Nature (London) 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 2.Braig K, Adams P D, Brünger A T. Nat Struct Biol. 1995;2:1083–1094. doi: 10.1038/nsb1295-1083. [DOI] [PubMed] [Google Scholar]
- 3.Ellis R J, Hartl F U. FASEB J. 1996;10:20–26. doi: 10.1096/fasebj.10.1.8566542. [DOI] [PubMed] [Google Scholar]
- 4.Makino Y, Taguchi H, Yoshida M. FEBS Lett. 1993;336:363–367. doi: 10.1016/0014-5793(93)80838-l. [DOI] [PubMed] [Google Scholar]
- 5.Taguchi H, Makino Y, Yoshida M. J Biol Chem. 1994;269:8529–8534. [PubMed] [Google Scholar]
- 6.White Z W, Fisher K E, Eisenstein E. J Biol Chem. 1995;270:20404–20409. doi: 10.1074/jbc.270.35.20404. [DOI] [PubMed] [Google Scholar]
- 7.Mendoza J A, Demeler B, Horowitz P M. J Biol Chem. 1994;269:2447–2451. [PubMed] [Google Scholar]
- 8.Ybarra J, Horowitz P M. J Biol Chem. 1995;270:22962–22967. doi: 10.1074/jbc.270.39.22962. [DOI] [PubMed] [Google Scholar]
- 9.Gorovits B, Raman C S, Horowitz P M. J Biol Chem. 1995;270:2061–2066. doi: 10.1074/jbc.270.5.2061. [DOI] [PubMed] [Google Scholar]
- 10.Zahn R, Buckle A M, Perrett S, Johnson C M, Corrales F J, Golbik R, Fersht A R. Proc Natl Acad Sci USA. 1996;93:15024–15029. doi: 10.1073/pnas.93.26.15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altamirano M M, Plumbridge J A, Hernandez-Arana A, Calcagno M. Biochim Biophys Acta. 1991;1076:266–272. doi: 10.1016/0167-4838(91)90277-7. [DOI] [PubMed] [Google Scholar]
- 12.Fischer G, Bang H, Mech C. Biomed Biochim Acta. 1984;43:1101–1111. [PubMed] [Google Scholar]
- 13.Buchberger A, Schroder H, Hesterkamp T, Schonfeld H J, Bukau B. J Mol Biol. 1996;261:328–333. doi: 10.1006/jmbi.1996.0465. [DOI] [PubMed] [Google Scholar]
- 14.Axen R, Porath J, Ernback S. Nature (London) 1967;215:1302–1304. doi: 10.1038/2141302a0. [DOI] [PubMed] [Google Scholar]
- 15.Oliva G, Fontes M, Garratt R C, Altamirano M M, Calcagno M L, Horjales E. Structure. 1995;3:1323–1332. doi: 10.1016/s0969-2126(01)00270-2. [DOI] [PubMed] [Google Scholar]