Abstract
Nucleoside diphosphate kinase reversibly transfers the γ-phosphate of ATP onto its active site histidine. We have investigated the transition state of histidine phosphorylation with the high-resolution crystal structures of the enzyme from Dictyostelium discoideum with MgADP and either aluminium or beryllium fluoride. The bound aluminium fluoride species is the neutral species AlF3 and not the more common AlF4−. AlF3 forms a trigonal bipyramid that makes it an accurate analog of the transition state of the γ-phosphate of ATP undergoing transfer to the catalytic histidine. Its axial ligands are a histidine nitrogen and a β-phosphate oxygen. Beryllium fluoride also binds at the same position and with the same ligands but in a tetrahedral geometry resembling the Michaelis complex rather than the transition state. The two x-ray structures show explicit enzyme–substrate interactions that discriminate between the ground and the transition states of the reaction. They also illustrate the partially dissociative geometry of the transition state of phosphoryl transfer and demonstrate the potential applications of metallofluorides for the study of kinase mechanisms.
Nucleoside diphosphate kinases (NDPK) phosphorylate nucleoside diphosphates at the expense of ATP with formation of a phospho-histidine covalent intermediate (1). These enzymes are the main cellular supplier of nucleoside triphosphates and are involved in a number of cellular regulations (for review, see ref. 2). The catalytic pathway is outlined by x-ray structures of the enzyme from various sources, including the slime mold Dictyostelium discoideum in native (3) and phosphorylated forms (4) and by dead-end complexes with ADP (5) and dTDP (6). These structures show that the active-site functions as a clip that closes upon the substrate, a moderate motion that positions the γ-phosphate of ATP next to the catalytic residues. Because of the high turnover of the enzyme (greater than 1,000 s−1), analogs of nucleoside triphosphates such as adenosine 5′-[β,γ-imido]triphosphate or adenosine 5′-[γ-thio]triphosphate are still slowly processed and are, therefore, not suitable for a crystallographic study of the phosphoryl transfer. Instead, aluminium and beryllium fluorides, in the presence of nucleoside diphosphates, form, respectively, transition state and ground state analogs for a number of nucleotidases, such as actin and tubulin (7, 8), F1-ATPase (9), Gα proteins (10), and the p21 ras–GAP complex (11). AlF4− and BeF3−, the predominant species in solution at high fluoride concentration, were shown to be the effective metallocomplexes in x-ray structures of Gα proteins (12, 13) and of myosin (14). In the present study, we have crystallized NDPK with MgADP and either aluminium or beryllium fluorides. Unlike the previous structures, we find a trigonal AlF3 species located halfway between the acceptor histidine nitrogen and the donor β-phosphate of ADP in the exact geometry expected for the transition state of the γ-phosphate of the real substrate. This structure, and the complex with BeF3− in which Be2+ is presumably tetrahedrally coordinated, provide a precise picture of the geometry of phosphoryl transfer during NDPK autophosphorylation. They also reveal the general characteristics of metallofluorides as accurate analogs of the transition state and the ground state in the context of kinase mechanisms.
MATERIAL AND METHODS
NDPK from D. discoideum was purified as described (3). NDPK–AlF3–MgADP crystals were grown at 20°C in hanging drops containing NDPK at 5–6 mg/ml, 50 mM Tris·HCl (pH 7.4), 20 mM MgCl2, 25 mM NaF, 0.5 mM Al(NO3)3, 10 mM ADP, and 32% PEG monomethyl ether (Mr = 550). Crystals appeared within 2 weeks. X-ray diffraction data were collected on the W32 station of the LURE-DCI synchrotron radiation center (Orsay, France) with a wavelength of 0.906 Å, using a MAR-Research image plate system. The crystals belong to space group P3121 with cell parameters a = b = 71.3 Å and c = 153.5 Å. The orientation of the NDPK–AlF3–MgADP complex was identified by molecular replacement, using the NDPK–MgADP complex as a search model. The asymmetric unit contains one-half of the 100-kDa hexamer. The structure was refined with MgADP in the active sites and showed extra density for a trigonal bipyramidal compound, which was built as AlF3 with a bond length derived from small molecules. The final model has a R factor of 17.7% at 2-Å resolution (Table 1).
Table 1.
Statistics on crystallographic analysis
| AlF3 | BeF3− | |
|---|---|---|
| Diffraction data | ||
| Space group | P3121 | P3121 |
| Cell parameters, Å | a = b = 71.35; c = 153.49 | a = b = 71.56; c = 153.75 |
| Resolution, Å | 2.0 | 2.3 |
| Measured intensities, no. | 118,259 | 51,271 |
| Unique reflexions, no. | 30,088 | 20,554 |
| Completeness, % | 96.3 | 98 |
| Multiplicity | 5.2 | 2.5 |
| Rmerge, % | 5.9 | 5.3 |
| Refinement | ||
| Resolution, Å | 2.0 | 2.3 |
| Rcryst, % | 17.7 | 19.3 |
| Reflections, no. | 29,606 | 20,058 |
| Protein atoms, no. | 3,537 | 3,537 |
| Solvent atoms, no. | 332 | 220 |
| Average B, Å2 | 30.9 | 28.8 (for solvent) |
| 16.8 | 17.8 (for protein) | |
| Geometry | ||
| Bond length, Å | 0.015 | 0.015 |
| Bond angle, degrees | 2.17 | 2.05 |
| Torsion angle, degrees | 1.86 | 1.88 |
Rmerge = Σh,i |I(h)i − 〈I(h)〉|/Σh,i I(h)i. Rcryst = Σh ∥F0| − |Fc∥/Σh|F0| was calculated with xplor on all reflections with F > 2σ. MgADP and aluminium or beryllium fluorides. For geometry, rms deviation from ideal values are given.
The NDPK–BeF3−–MgADP crystals were obtained by microseeding with NDPK–AlF3–MgADP crystals under the above conditions, except that Al(NO3)3 was replaced by BeCl2. Diffraction data were collected on a Rigaku x-ray generator with a R-axis image plate system. The beryllium-containing crystals are isomorphous to those with aluminium, and the structure was solved by Fourier difference electron density with the NDPK–AlF3–MgADP complex from which AlF3 had been omitted. The structure, refined without BeF3−, showed extra density at the γ-phosphate location that was modeled as BeF3− by using the geometry of small molecules. The final model has a R factor of 19.3% at 2.3-Å resolution (Table 1).
RESULTS AND DISCUSSION
Structure of the Metallofluoride Complexes.
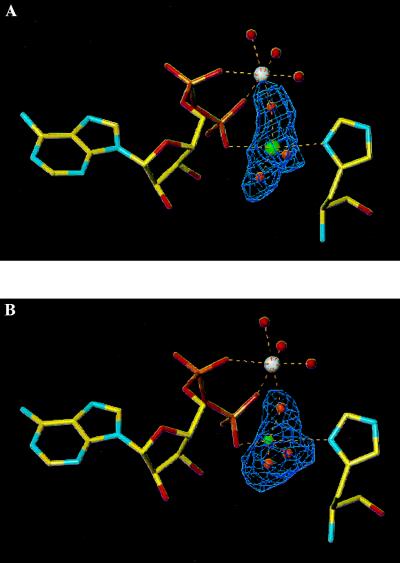
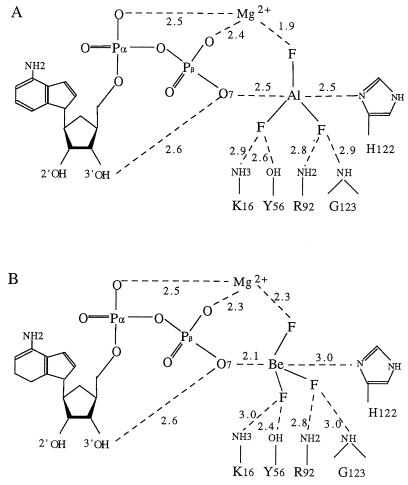
The electronic density of the crystals grown in the presence of aluminium nitrate and sodium fluoride unambiguously defines ADP, Mg2+, and a trigonal planar compound that is wedged between the β-phosphate of ADP and the catalytic histidine (Fig. 1A). The moiety binds with full occupancy at all three independent active sites and makes no crystal contacts. The extra density was attributed to AlF3 on the basis of biochemical and crystallographic data that exclude nitrate as the bound species and make it unlikely that it is MgF3−. The metal cation has the three fluorines on the equator, and two almost colinear axial bonds to an oxygen of the β-phosphate and to the Nδ nitrogen of His-122. The distance from Al3+ to its axial ligands is 2.5 Å, longer than the Al–F distance (1.85 Å). In addition, the fluorines make extensive interactions to the protein and the Mg2+ ion (Fig. 2A). The BeF3− complex is essentially identical, except that Be2+ has only two electrons and cannot be located with confidence from the electronic density (Figs. 1B and 2B). Since beryllium fluorides prefer tetrahedral coordination (15), Be2+ can nevertheless be modeled at two alternative sites about 1.3 Å apart, with O7 of ADP or Nδ from His-122 as the fourth ligand. In either case, the bonded and nonbonded distances are 2.1 Å and 3.0 Å, respectively, and the two situations may coexist in the crystal.
Figure 1.
Simulated annealing Fo–Fc omit maps contoured at 3.5 σ showing ADP (to the left) and His-122 (to the right). (A) AlF3 omit map at 2.0-Å resolution. Al3+ (green) in trigonal bipyramidal configuration is shown coordinating to three equatorial fluorines (orange) and with partial bonds to ADP O7 and to His-122 Nδ. Mg2+ (white) has an octahedral geometry, with a tridendate coordination to the α- and β-phosphates and a fluorine atom and three water molecules (red) as additional ligands. (B) BeF3− omit map at 2.3-Å resolution. Be2+ (green) is modeled in a tetrahedral geometry, with three fluorines and O7 as the fourth ligand. An alternative configuration for Be2+ has His-122 Nδ as the fourth ligand.
Figure 2.
Schematic representation of NDPK–nucleotide interactions. (A) ADP–AlF3 from the x-ray structure. (B) ADP–BeF3− with Be2+ modeled at the β-phosphate O7 as in Fig. 1B. In all cases, there is a short hydrogen bond between the 3′ hydroxyl of the ribose and the β-phosphate O7.
Both the AlF3 and BeF3− complexes compare very well with the MgADP and MgTDP complexes (5, 6). However, although the conformation of ADP is the same, the presence of the metallofluorides has pulled the nucleotide closer to His-122 in a rigid body movement of 0.5–1 Å. As a consequence, the rotation of His-122 side chain, which was speculated to take place to align the attacking group with the substrate (4), is not observed in the present structures. Instead, the histidine retains the orientation that was observed in native and ADP-bound NDPK, suggesting that the catalytic configuration preexists to the binding of the substrate.
AlF3 Is an Analog of the Transition State for Phosphoryl Transfer.
Depending on the Be2+ position, the tetrahedral BeF3− complex is an analog of the phosphate group in either of the two Michaelis complexes that form during the catalytic cycle: enzyme–ATP or phosphoenzyme–ADP. On the other hand, the trigonal planar geometry of Al3+ is the one predicted for the transition state for in-line phosphoryl transfer (16–18). Under the assumption that the AlF3–enzyme interactions are essentially influenced only by the enzyme and the nucleotide, then the AlF3 complex provides an accurate picture of the transition state of the reaction. Alternatively, these interactions may be influenced by the particular properties of the metallic Al3+ ion and the F−, which could result in active-site rearrangements. Because the NDPK active-site residues and the ADP nucleotide constitute a constrained environment, reflected in the absence of induced conformational change at the catalytic residues, it is likely that the AlF3 complex closely mimics the protein and substrate geometries in the transition state. The fluorines of AlF3 provide equivalents of the phosphate equatorial oxygens, except that P–O bonds should be somewhat shorter than the Al–F distance (although the bond length in the transition state is unknown). The formal charge is different, AlF3 being neutral and the transferred PO3 carrying negative charges. However, the actual charge distribution at the ground and the transition state is unknown. In the present structure, AlF3 is most likely polarized. Al3+ forms five bonds, including the bonds to the donor ADP O7 and to the acceptor His-122 Nδ. The 2.5-Å distance to these atoms is longer than expected for a covalent bond but shorter than a van der Waals contact. The nucleophilic attack of the γ-phosphate thus involves partial bond cleavage at the leaving group and partial bond formation at the attacking group. Qualitatively, the mechanism is intermediate between associative (with the phosphorus forming five bonds) and dissociative (with the formation of a PO3− intermediate). The NDPK–AlF3 complex thus shows the existence and the stereochemistry of partial bonds in the active site of the enzyme during phosphoryl transfer.
Substrate-Assisted Stabilization of the Transition State.
To understand enzymic catalysis, it is necessary to estimate the contribution of individual protein groups along the reaction coordinate and to identify those which are responsible for discriminating between the ground and the transition states. They can do this either by numerous small contributions or by a few highly stabilizing determinants, in strong hydrogen bonds for instance (19, 20). The interactions of NDPK with its substrate can readily be modeled in the Michaelis complex from the structure with BeF3− and in the transition state from that with AlF3. At pH 7–8 where NDPK is active, the phosphates of ATP are deprotonated, all the more so because active-site positive charges are likely to lower their pK values. The four negative charges are shielded by Mg2+, which ligates all three phosphates, and by two guanidinium groups. The γ-phosphate also hydrogen bonds to the hydroxyl of Tyr-56 and a main-chain NH. The contribution of Tyr-56 has been measured by site-directed mutagenesis (21). Replacement by Phe or Ala slows catalysis by a factor of 50, indicating a contribution by ≈2.3 kcal/mol (1 cal = 4.184 J) to the activation free energy of the reaction.
This is a significant but minor contribution to catalysis. The AlF3 complex points to another source of stabilization, in the form of a short hydrogen bond between the 3′ hydroxyl of the ribose and the O7 oxygen of the β-phosphate (Fig. 2). In ATP, O7 bridges the β- and γ-phosphates and is expected to be the atom of the substrate that develops the largest change of charge upon the transition state. The hydrogen bond probably exists in the Michaelis complex, since it is present in the MgADP and MgTDP complexes (5, 6), but it should become significantly stronger as the bond to the γ-phosphate breaks and O7 acquires a charge. Its contribution to catalysis has been quantified in a study of nucleoside analogs that lack a hydroxyl in the 3′ position, such as the antiviral agents azidothymidine or dideoxyadenosine. Dideoxy-ATP, for instance, is a factor 104 less active than ATP, suggesting that the 3′ OH contributes some 5.5 kcal/mol to lowering the activation barrier (22). The hydroxyl is in that sense more important than any of the protein groups involved in catalysis (except His-122), since their deletion by site-directed mutagenesis always leaves more than 10−3 of the NDPK activity (21). The catalytic role of the 3′ OH hydrogen bond also points out the importance of interactions made by the β–γ bridge oxygen in stabilizing the transition state of phosphoryl transfer reactions. In NDPK, they are provided by the substrate itself, but they can be provided by protein groups in other enzymes, as recently proposed for p21-ras (23). Ribose hydroxyls have not been proposed a catalytic role in other kinases, but it is well-established in ribonucleases and it is essential to the chemical step in RNA cleavage by the Tetrahymena ribozyme (24).
NDPK provides an example in enzymology where the dynamics of catalysis have been captured in high-resolution x-ray crystal structures. The enzyme exercises its influence by restraining the substrate to the conformer in which substrate-assisted catalysis is made possible. A hydrophobic clamp orients the base; Lys-16 and Asn-119 immobilize the sugar; and Thr-98, Arg-92, Arg-109, and Tyr-56 configure the phosphates next to the catalytic histidine (refs. 5 and 6 and this work). Several of these residues are held by a pair of helices that move upon substrate binding and product release. This breathing movement is of small amplitude (less than 2 Å) and essentially no other structural change occurs during the whole catalytic cycle. However, as this movement simultaneously opens the active site and drags the phosphohistidine out of in-line geometry, it is essential for expelling the intermediary ADP product, which may be the rate-limiting step.
Metallofluorides Are Reaction Coordinate Analogs for Probing Kinases Mechanisms.
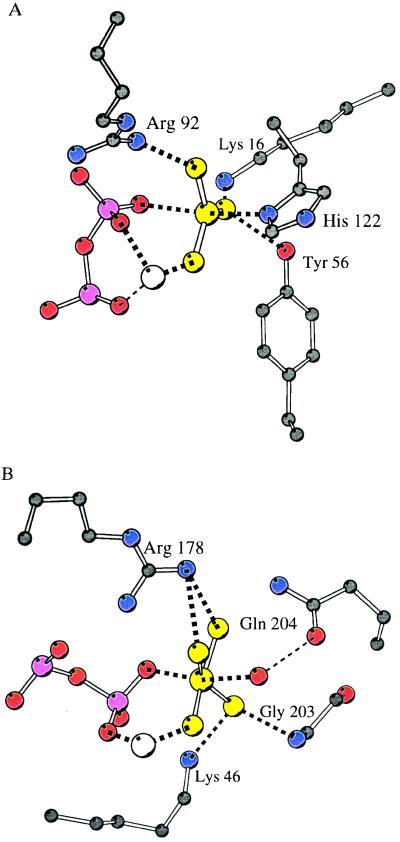
Aluminium and beryllium fluorides are well-established analogs of phosphate in ATPases and GTPases (25), and in nitrogenase (26). Although the NDPK fold is unrelated to Gα proteins, myosin, or nitrogenase, there are counterparts in G proteins for several of the residues that bind the aluminofluoride (Fig. 3). On the other hand, there is no equivalent in NDPK for the glutamine residue postulated to activate the attacking water molecule in G proteins. The reaction does not require one, since the attacking nucleophile is the Nδ of His-122, which is not protonated at pH 7. In return, the GTPase reaction does not involve the ribose hydroxyls.
Figure 3.
Comparison of AlF3 and AlF4− aluminium fluorides species and their binding sites in NDPK and Giα. (A) NDPK–AlF3. (B) Giα–AlF4− (12). The figure shows aluminium fluoride (yellow), the pyrophosphate moiety of ADP (with α- and β-phosphates in pink), the bound Mg2+ (white), and surrounding protein atoms in a similar orientation. Lys-16, Tyr-56, and Arg-92 of NDPK have Lys-46, Gly-203, and Arg-178 as counterparts in Giα. Hydrogen or metal–ligand bonds that are equivalent in both structures are highlighted in thicker dashes.
In x-ray structures of Giα (12) and Gtα GTPases (13) and of myosin (15), Al3+ is present in the AlF4− complex rather than the trigonal AlF3. The octahedral AlF4− is six-coordinated, with a bridging β-phosphate oxygen and a water molecule as axial ligands and four equatorial fluorines. It is therefore not a true analog for the transition state of phosphate. The orientation of the fluorines of AlF4− on the equatorial plane differs between the Giα and Gtα complexes, which may indicate that AlF4− is rearranged relative to the real transition state. The trigonal planar configuration that we see in NDPK is clearly a better mimic of the transition state for phosphoryl transfer. Although this configuration is uncommon in solution in the presence of fluorides, it is found in aluminium derivatives such as AlCl3(NMe3)2 (15). Why AlF3 rather than AlF4− binds to the active site of NDPK may be explained by the nature of the nucleophilic attack, which involves a protein group instead of a water molecule. The fluoroaluminate that binds to hydrolytic nucleotidases is geometrically identical to the AlF4(H2O)2− complexes that are well-described from structures of small molecules. The four fluorines could probably fit in the active site of NDPK, at the price of local protein plasticity as observed between the complexes of MgADP myosin with octahedral AlF4− (14) and trigonal vanadate (VO43−) (27). On the other hand, an interaction of AlF4− with the β-phosphate of ADP and His-122 would require substantial lengthening of the axial bonds, a situation that may be energetically intolerable. In the same way, an additional water could not be fit between Al3+ and His-122. For a similar reason, vanadate does not bind in the active site of NDPK (I. Lascu, personal communication). It is likely that most kinases that do not use water as the phosphate acceptor would encounter the same steric hindrance for AlF4− and for vanadate, whereas AlF3 and BeF3− would be expected to bind with the geometry of the transition state and the Michaelis complexes, respectively. Our results suggest that these metallofluorides can be of general use for the study of phosphokinases, especially protein kinases, and that their potential applications are much wider than previously assumed.
Acknowledgments
We thank M. F. Carlier (Laboratoire d’Enzymologie et de Biochimie Structurales, Centre National de la Recherche Scientifique, Gif-sur-Yvette) and I. Lascu (Institut de Biochimie et Génétique Cellulaire, Centre National de la Recherche Scientifique, Bordeaux) for their expert advice, and M. Véron (Institut Pasteur, Paris), M. Chabre (Centre National de la Recherche Scientifique, Valbonne), and D. Herschlag (Stanford University) for discussions. The survey of the Cambridge Data Base was efficiently performed by L. Tchertanov (Institut de Chimie des Substances Naturelles, Centre National de la Recherche Scientifique, Gif-sur-Yvette). We thank the staff at Laboratoire pour l’Utilisation du Rayonnenent Electromagnetique for making station W32 on the wiggler line of Laboratoire pour l’Utilisation du Rayonnenent Electromagnetique, available to us. Y.-W.X. acknowledges support of the Programme de Recherches Avancées B95-03 between Université Paris-Sud and University of Science and Technology of China (Hefei, Anhui). This work was supported by the Agence Nationale pour la Recherche contre le SIDA and the Association pour la Recherche contre le Cancer.
ABBREVIATION
- NDPK
nucleoside diphosphate kinase
Footnotes
Data deposition: The atomic coordinated and structure factors have been deposited in the Protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1kdn).
References
- 1.Parks R E, Jr, Agarwal R P. Enzymes. 1973;8:307–334. [Google Scholar]
- 2.de la Rosa A, Williams R L, Steeg P S. Bioassays. 1995;17:53–62. doi: 10.1002/bies.950170111. [DOI] [PubMed] [Google Scholar]
- 3.Dumas C, Lascu I, Moréra S, Glaser P, Fourme R, Wallet V, Lacombe M-L, Véron M, Janin J. EMBO J. 1992;11:3203–3208. doi: 10.1002/j.1460-2075.1992.tb05397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moréra S, Chiadmi M, LeBras G, Lascu I, Janin J. Biochemistry. 1995;34:11062–11070. [PubMed] [Google Scholar]
- 5.Moréra S, Lascu I, Dumas C, LeBras G, Briozzo P, Véron M, Janin J. Biochemistry. 1994;33:459–467. doi: 10.1021/bi00168a010. [DOI] [PubMed] [Google Scholar]
- 6.Cherfils J, Moréra S, Lascu I, Véron M, Janin J. Biochemistry. 1994;33:9062–9069. doi: 10.1021/bi00197a006. [DOI] [PubMed] [Google Scholar]
- 7.Combeau C, Carlier M-F. J Biol Chem. 1988;263:17429–17436. [PubMed] [Google Scholar]
- 8.Carlier M-F, Pantaloni D. Handbook of Experimental Pharmacology. 108/I. Berlin: Springer; 1993. pp. 53–62. [Google Scholar]
- 9.Issartel J-P, Dupuis A, Lunardi J, Vignais P. Biochemistry. 1991;30:4726–4733. doi: 10.1021/bi00233a013. [DOI] [PubMed] [Google Scholar]
- 10.Bigay J, Deterre P, Pfister C, Chabre M. EMBO J. 1987;6:2907–2913. doi: 10.1002/j.1460-2075.1987.tb02594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mittal R, Ahmadian M R, Goody R, Wittinghofer A. Science. 1996;273:115–117. doi: 10.1126/science.273.5271.115. [DOI] [PubMed] [Google Scholar]
- 12.Coleman D E, Berghuis A M, Lee E, Linder M E, Gilman A G, Sprang S R. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 13.Sondek J, Lambright D G, Noel J P, Hamm H E, Sigler P B. Nature (London) 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- 14.Fisher A J, Smith C A, Thoden J B, Smith R, Sutoh K, Holden H M, Rayment I. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 15.Allen F A, Kennard O, Taylor R. Acc Chem Res. 1983;16:146–153. [Google Scholar]
- 16.Westmeister F. Acc Chem Res. 1969;1:70–78. [Google Scholar]
- 17.Knowles J R. Annu Rev Biochem. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- 18.Frey P A. Enzymes. 1992;20:141–186. [Google Scholar]
- 19.Fersht A R. Trends Biochem Sci. 1987;12:301–304. [Google Scholar]
- 20.Cleland W W, Kreevoy M M. Science. 1994;264:1887–1890. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]
- 21.Tepper A D, Dammann H, Bominaar A A, Véron M. J Biol Chem. 1994;269:32175–32180. [PubMed] [Google Scholar]
- 22.Bourdais J, Biondi R, Sarfati S, Guerreiro C, Lascu I, Janin J, Véron M. J Biol Chem. 1996;271:7887–7890. doi: 10.1074/jbc.271.14.7887. [DOI] [PubMed] [Google Scholar]
- 23.Maegley K A, Admiraal S J, Herschlag D. Proc Natl Acad Sci USA. 1996;93:8160–8166. doi: 10.1073/pnas.93.16.8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herschlag D, Eckstein F, Cech T R. Biochemistry. 1993;32:8299–8311. doi: 10.1021/bi00083a034. [DOI] [PubMed] [Google Scholar]
- 25.Chabre M. Trends Biochem Sci. 1990;15:6–10. doi: 10.1016/0968-0004(90)90117-t. [DOI] [PubMed] [Google Scholar]
- 26.Renner K A, Howard J B. Biochemistry. 1996;35:5353–5358. doi: 10.1021/bi960441o. [DOI] [PubMed] [Google Scholar]
- 27.Smith C A, Rayment I. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]